Increased HIV Subtype Diversity Reflecting Demographic Changes in the HIV Epidemic in New South Wales, Australia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics
2.2. Sequence Phylogeny and Identification of NSW-Specific Clusters
2.3. Demographic Factors
2.4. Data Availability
3. Results
3.1. Subtype B and CRF01_AE Dominate the HIV-1 Epidemic in New South Wales
3.2. Subtype C is Strongly Associated with Hetereosexual Transmission
3.3. Changes in Associations between Demographics and Subtype between 2009 and 2018
3.4. Demographic Differences in Clusters and Singleton Sequences
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bbosa, N.; Kaleebu, P.; Ssemwanga, D. HIV subtype diversity worldwide. Curr. Opin. HIV AIDS 2019, 14, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Angelis, K.; Albert, J.; Mamais, I.; Magiorkinis, G.; Hatzakis, A.; Hamouda, O.; Struck, D.; Vercauteren, J.; Wensing, A.M.; Alexiev, I.; et al. Global dispersal pattern of HIV type 1 subtype CRF01_AE: A genetic trace of human mobility related to heterosexual sexual activities centralized in Southeast Asia. J. Infect. Dis. 2015, 211, 1735–1744. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Hora, B.; DeMarco, T.; Berba, R.; Register, H.; Hood, S.; Carter, M.; Stone, M.; Pappas, A.; Sanchez, A.M.; et al. Increased predominance of HIV-1 CRF01_AE and its recombinants in the Philippines. J. Gen. Virol. 2019, 100, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Kirby Institute. HIV, Viral Hepatitis and Sexually Transmissible Infections in Australia: Annual surveillance Report 2018; Kirby Institute, UNSW Sydney: Sydney, Australia, 2018. [Google Scholar]
- Dauwe, K.; Mortier, V.; Schauvliege, M.; Van Den Heuvel, A.; Fransen, K.; Servais, J.Y.; Bercoff, D.P.; Seguin-Devaux, C.; Verhofstede, C. Characteristics and spread to the native population of HIV-1 non-B subtypes in two European countries with high migration rate. BMC Infect. Dis. 2015, 15, 524. [Google Scholar] [CrossRef] [Green Version]
- Oster, A.M.; Switzer, W.M.; Hernandez, A.L.; Saduvala, N.; Wertheim, J.O.; Nwangwu-Ike, N.; Ocfemia, M.C.; Campbell, E.; Hall, H.I. Increasing HIV-1 subtype diversity in seven states, United States, 2006-2013. Ann. Epidemiol. 2017, 27, 244–251.e1. [Google Scholar] [CrossRef]
- Hauser, A.; Hofmann, A.; Meixenberger, K.; Altmann, B.; Hanke, K.; Bremer, V.; Bartmeyer, B.; Bannert, N. Increasing proportions of HIV-1 non-B subtypes and of NNRTI resistance between 2013 and 2016 in Germany: Results from the national molecular surveillance of new HIV-diagnoses. PLoS ONE 2018, 13, e0206234. [Google Scholar] [CrossRef]
- Fabeni, L.; Santoro, M.M.; Lorenzini, P.; Rusconi, S.; Gianotti, N.; Costantini, A.; Sarmati, L.; Antinori, A.; Ceccherini-Silberstein, F.; d’Arminio Monforte, A.; et al. Evaluation of HIV transmission clusters among natives and foreigners living in Italy. Viruses 2020, 12, 791. [Google Scholar] [CrossRef]
- Xiao, P.; Zhou, Y.; Lu, J.; Yan, L.; Xu, X.; Hu, H.; Li, J.; Ding, P.; Qiu, T.; Fu, G.; et al. HIV-1 genotype diversity and distribution characteristics among heterosexually transmitted population in Jiangsu province, China. Virol. J. 2019, 16, 51. [Google Scholar] [CrossRef]
- Delgado, E.; Benito, S.; Montero, V.; Cuevas, M.T.; Fernandez-Garcia, A.; Sanchez-Martinez, M.; Garcia-Bodas, E.; Diez-Fuertes, F.; Gil, H.; Canada, J.; et al. Diverse large HIV-1 non-subtype B clusters are spreading among men who have wex with men in Spain. Front. Microbiol 2019, 10, 655. [Google Scholar] [CrossRef]
- Hemelaar, J.; Elangovan, R.; Yun, J.; Dickson-Tetteh, L.; Fleminger, I.; Kirtley, S.; Williams, B.; Gouws-Williams, E.; Ghys, P.D.; WHO–UNAIDS Network for HIV Isolation Characterisation. Global and regional molecular epidemiology of HIV-1, 1990-2015: A systematic review, global survey, and trend analysis. Lancet Infect. Dis. 2019, 19, 143–155. [Google Scholar] [CrossRef]
- Castley, A.; Sawleshwarkar, S.; Varma, R.; Herring, B.; Thapa, K.; Dwyer, D.; Chibo, D.; Nguyen, N.; Hawke, K.; Ratcliff, R.; et al. A national study of the molecular epidemiology of HIV-1 in Australia 2005–2012. PLoS ONE 2017, 12, e0170601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- SA Health. Surveillance of Sexually Transmitted Infections and Blood-Borne Viruses in South Australia, 2018; Communicable Disease Control Branch SA Healht: Rundle Mall, SA, Australia, 2018. [Google Scholar]
- Di Giallonardo, F.; Pinto, A.N.; Keen, P.; Shaik, A.; Carrera, A.; Salem, H.; Telfer, B.; Cooper, C.; Price, K.; Selvey, C.; et al. Limited sustained local transmission of HIV-1 CRF01_AE in New South Wales, Australia. Viruses 2019, 11, 482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Giallonardo, F.; Pinto, A.N.; Keen, P.; Shaik, A.; Carrera, A.; Salem, H.; Selvey, C.; Nigro, S.J.; Fraser, N.; Price, K.; et al. Subtype specific differences in transmission cluster dynamics of HIV-1 B and CRF01_AE in New South Wales, Australia. J. Int. AIDS Soc. 2020. accepted for publication. [Google Scholar]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- REGA HIV-1 Subtyping Tool v3.0. Available online: http://dbpartners.stanford.edu:8080/RegaSubtyping/stanford-hiv/typingtool/ (accessed on 14 June 2019).
- Turk, T.; Bachmann, N.; Kadelka, C.; Boni, J.; Yerly, S.; Aubert, V.; Klimkait, T.; Battegay, M.; Bernasconi, E.; Calmy, A.; et al. Assessing the danger of self-sustained HIV epidemics in heterosexuals by population based phylogenetic cluster analysis. Elife 2017, 6. [Google Scholar] [CrossRef] [Green Version]
- Kuraku, S.; Zmasek, C.M.; Nishimura, O.; Katoh, K. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 2013, 41, W22–W28. [Google Scholar] [CrossRef] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Lipscomb, J.T.; Switzer, W.M.; Li, J.F.; Masciotra, S.; Owen, S.M.; Johnson, J.A. HIV reverse-transcriptase drug resistance mutations during early infection reveal greater transmission diversity than in envelope sequences. J. Infect. Dis. 2014, 210, 1827–1837. [Google Scholar] [CrossRef] [Green Version]
- Callander, D.; Mooney-Somers, J.; Keen, P.; Guy, R.; Duck, T.; Bavinton, B.R.; Grulich, A.E.; Holt, M.; Prestage, G. Australian ‘gayborhoods’ and ‘lesborhoods’: A new method for estimating the number and prevalence of adult gay men and lesbian women living in each Australian postcode. Int. J. Geogr. Inf. Sci. 2020, 34, 2160–2176. [Google Scholar] [CrossRef]
- NSW Ministry of Health. NSW Ministry of Health. NSW HIV strategy 2016–2020: Quarter 4 & annual 2019 data report. In Centre for Population Health; NSW Ministry of Health: Sydney, NSW, Australia, 2020. [Google Scholar]
- R Core Team. R: A language and environment for statistical computing. In R Foundation for Statistical Computing; R Core Team: Vienna, Austria, 2019; Available online: https://www.R-project.org/ (accessed on 13 November 2020).
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Liaw, W.H.A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S.; Schwartz, M.; et al. Gplots: Various R Programming Tools for Plotting Data. R Package Version 3.0.1.2. 2020. Available online: https://CRAN.R-project.org/package-gplots (accessed on 14 June 2019).
- Wei, T.; Simko, V. R package “Corrplot”: Visualization of a Correlation Matrix (Version 0.84). 2017. Available online: https://github.com/taiyun/corrplot (accessed on 14 June 2019).
- MATLAB, 2020, version R2020b; The MathWorks Inc.: Natick, MA, USA, 2010.
- Chibo, D.; Birch, C. Increasing diversity of human immunodeficiency virus type 1 subtypes circulating in Australia. AIDS Res. Hum. Retrovir. 2012, 28, 578–583. [Google Scholar] [CrossRef]
- Australian Government, Department of Education, Skills and Employment. 2018 Full Year Higher Education Statistics. Available online: https://docs.education.gov.au/system/files/doc/other/2018_student_data_infographic.pdf (accessed on 15 November 2020).
- Grulich, A.E.; Guy, R.; Amin, J.; Jin, F.; Selvey, C.; Holden, J.; Schmidt, H.A.; Zablotska, I.; Price, K.; Whittaker, B.; et al. Population-level effectiveness of rapid, targeted, high-co, doi:verage roll-out of HIV pre-exposure prophylaxis in men who have sex with men: The EPIC-NSW prospective cohort study. Lancet HIV 2018, 5, e629–e637. [Google Scholar] [CrossRef]
- Holt, M.; Lee, E.; Lea, T.; Bavinton, B.; Broady, T.; Mao, L.; MacGibbon, J.; Keen, P.; Murphy, D.; Bear, B.; et al. HIV Preexposure Prophylaxis Cascades to Assess Implementation in Australia: Results From Repeated, National Behavioral Surveillance of Gay and Bisexual Men, 2014-2018. J. Acquir. Immune Defic. Syndr. 2020, 83, e16–e22. [Google Scholar] [CrossRef] [PubMed]
- Kouyos, R.D.; von Wyl, V.; Yerly, S.; Boni, J.; Taffe, P.; Shah, C.; Burgisser, P.; Klimkait, T.; Weber, R.; Hirschel, B.; et al. Molecular epidemiology reveals long-term changes in HIV type 1 subtype B transmission in Switzerland. J. Infect. Dis. 2010, 201, 1488–1497. [Google Scholar] [CrossRef] [PubMed]
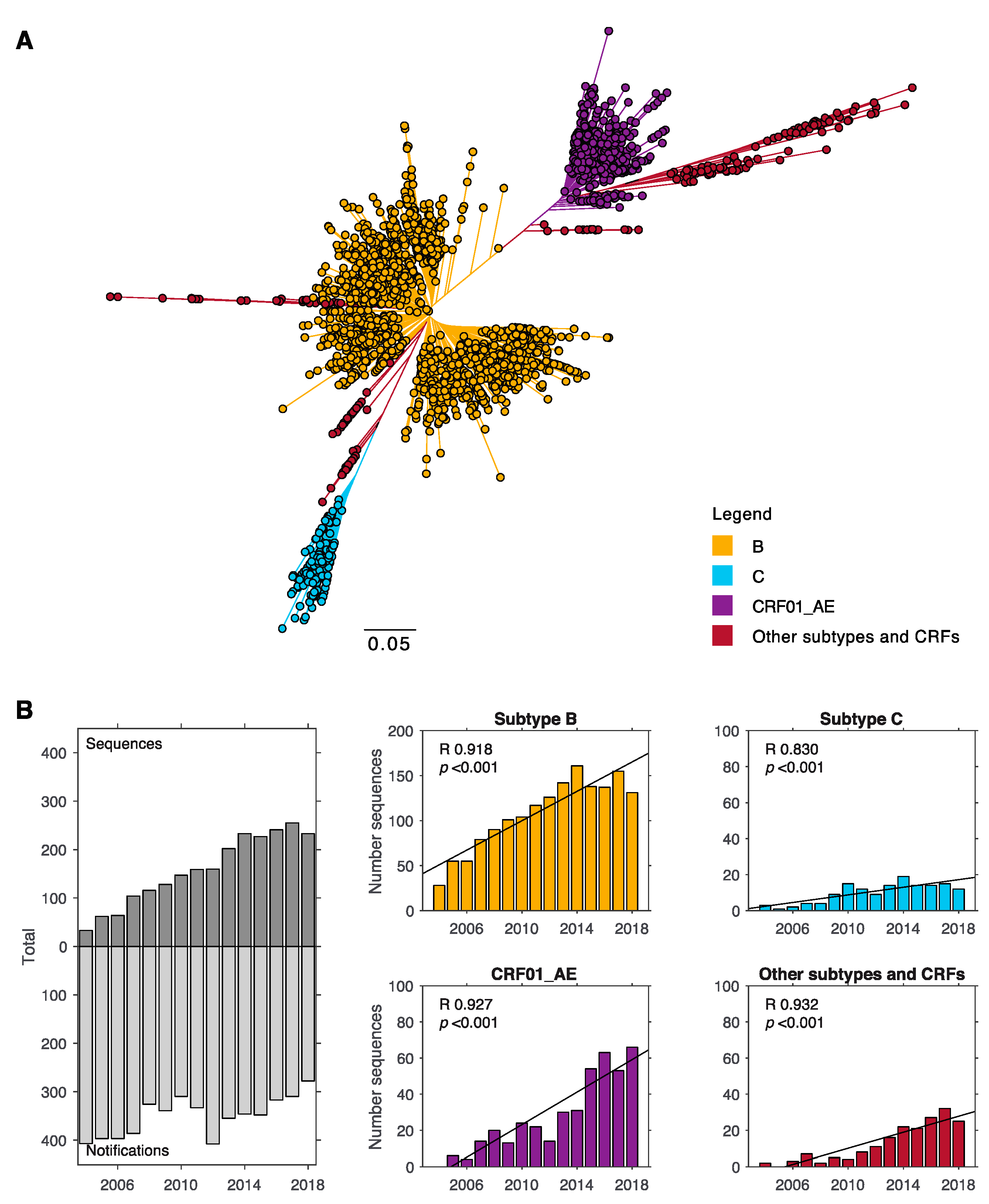
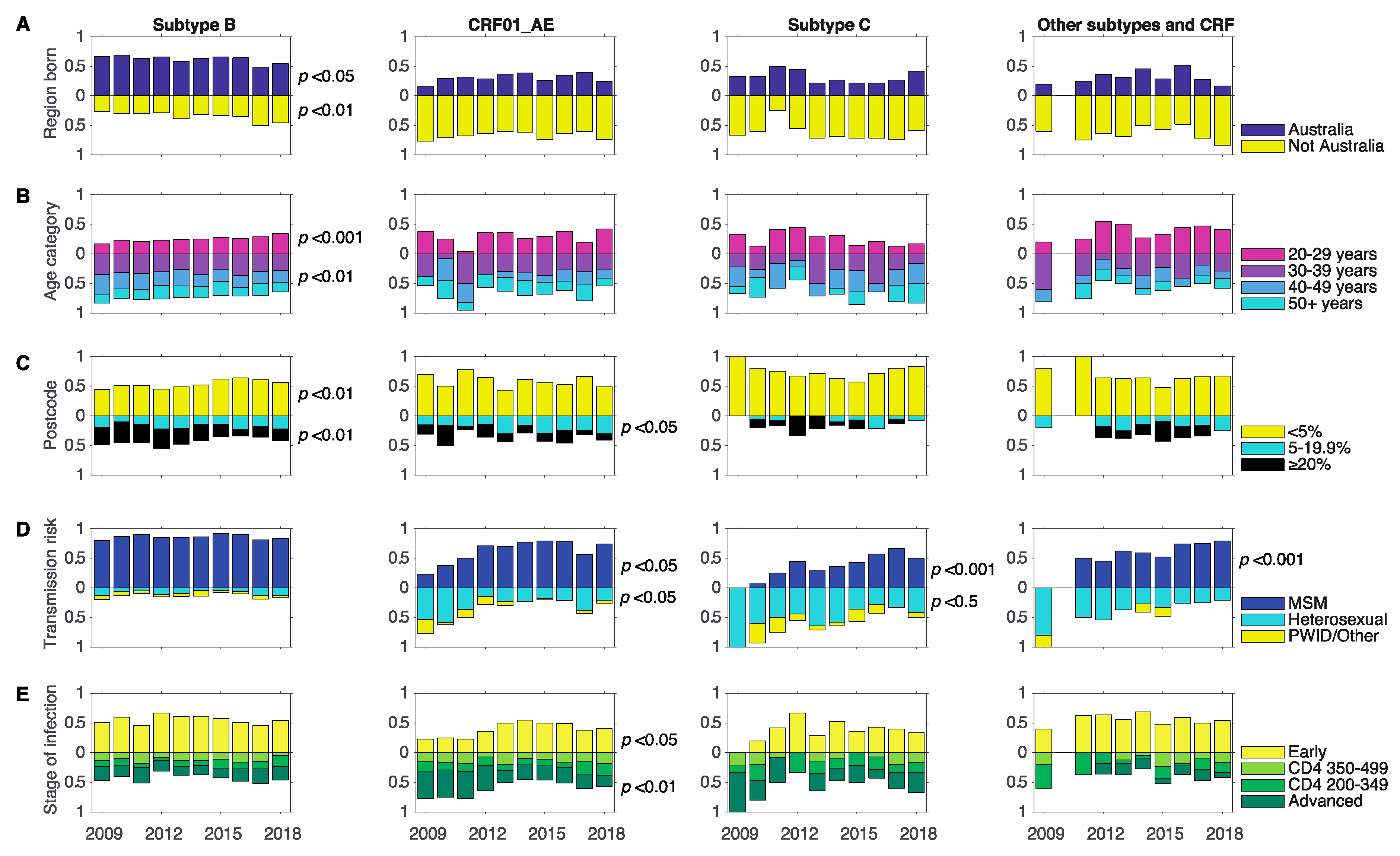
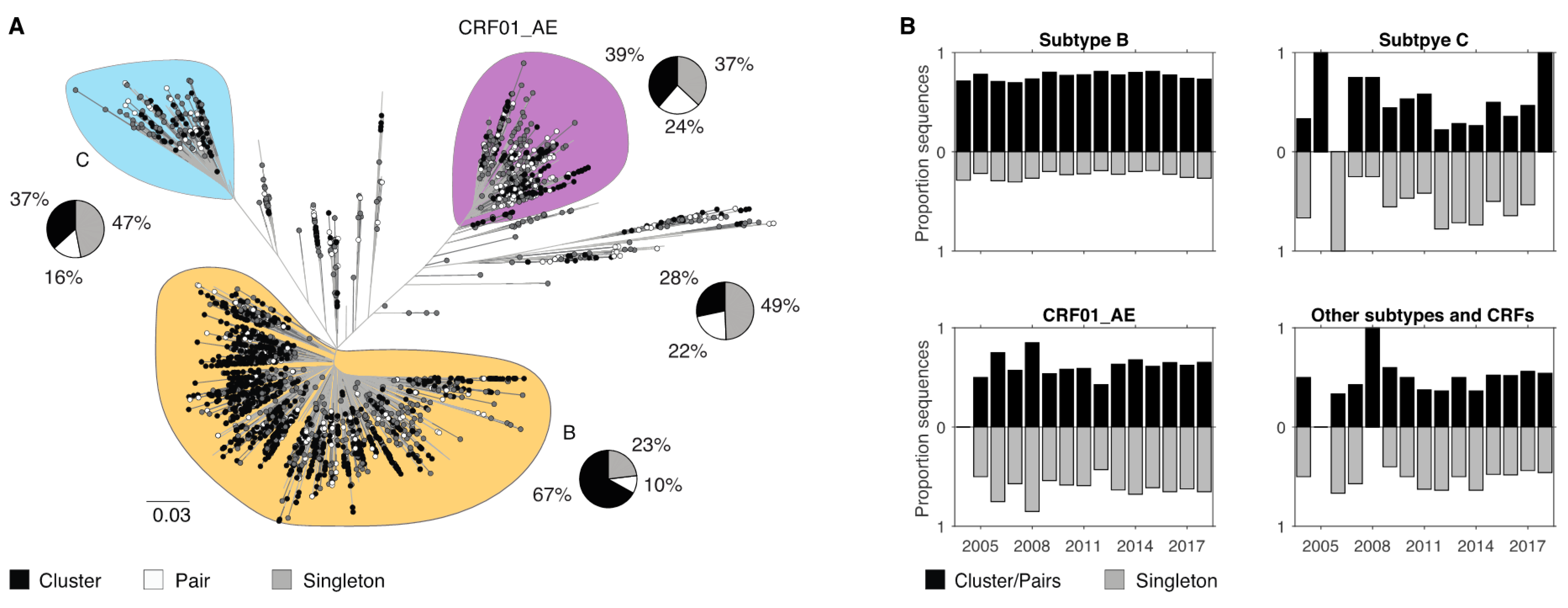
| Demographic (n) | Subtype B | All non-B | CRF01_AE | Subtype C | Other Subtypes and CRFs |
|---|---|---|---|---|---|
| Sex | |||||
| Male (1825) | 0.73 (1330) | 0.27 (495) | 0.60 (298) | 0.16 (80) | 0.24 (117) |
| Female (139) | 0.27 (37) | 0.73 (102) | 0.36 (37) | 0.38 (39) | 0.25 (26) |
| No data (397) | 0.63 (252) | 0.37 (145) | 0.52 (76) | 0.19 (28) | 0.28 (41) |
| Region Born | |||||
| Australia (1249) | 0.81 (1007) | 0.19 (242) | 0.56 (135) | 0.18 (44) | 0.26 (63) |
| Not Australia (1012) | 0.53 (533) | 0.47 (479) | 0.57 (271) | 0.19 (93) | 0.24 (115) |
| No data (103) | 0.77 (79) | 0.23 (24) | 0.33 (8) | 0.42 (10) | 0.25 (6) |
| Age Category | |||||
| <20 years (37) | 0.51 (19) | 0.49 (18) | 0.39 (7) | 0.33 (6) | 0.28 (5) |
| 20–29 years (632) | 0.63 (400) | 0.37 (232) | 0.53 (124) | 0.16 (36) | 0.31 (72) |
| 30–39 years (742) | 0.70 (518) | 0.30 (224) | 0.58 (129) | 0.20 (44) | 0.23 (51) |
| 40–49 years (534) | 0.74 (394) | 0.26 (140) | 0.50 (70) | 0.27 (38) | 0.23 (32) |
| 50+ years (419) | 0.69 (288) | 0.31 (131) | 0.64 (84) | 0.18 (23) | 0.18 (24) |
| Postcode Category according to the Proportion of Adult Gay Men | |||||
| <5% (1281) | 0.65 (827) | 0.35 (454) | 0.50 (228) | 0.24 (109) | 0.26 (117) |
| 5–19.9% (425) | 0.67 (284) | 0.33 (141) | 0.72 (101) | 0.08 (11) | 0.21 (29) |
| >20% (538) | 0.80 (430) | 0.20 (108) | 0.58 (63) | 0.13 (14) | 0.29 (31) |
| No data (120) | 0.65 (78) | 0.35 (42) | 0.52 (22) | 0.31 (13) | 0.17 (7) |
| Transmission Risk Factor | |||||
| MSM (1815) | 0.76 (1385) | 0.24 (430) | 0.62 (266) | 0.12 (50) | 0.27 (114) |
| Heterosexual (393) | 0.35 (137) | 0.65 (256) | 0.46 (118) | 0.30 (78) | 0.23 (60) |
| PWID/Other (156) | 0.62 (97) | 0.38 (59) | 0.51 (31) | 0.32 (19) | 0.17 (10) |
| Stage of Infection at Diagnosis | |||||
| Early (1224) | 0.74 (903) | 0.26 (321) | 0.52 (168) | 0.17 (53) | 0.31 (100) |
| CD4 350 to 499 (278) | 0.64 (179) | 0.36 (99) | 0.60 (59) | 0.17 (17) | 0.23 (23) |
| CD4 200 to 349 (278) | 0.62 (172) | 0.38 (106) | 0.52 (55) | 0.24 (25) | 0.25 (26) |
| Advanced (495) | 0.62 (309) | 0.38 (186) | 0.63 (118) | 0.22 (41) | 0.15 (27) |
| No data (89) | 0.63 (56) | 0.37 (33) | 0.42 (14) | 0.33 (11) | 0.24 (8) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Giallonardo, F.; Pinto, A.N.; Keen, P.; Shaik, A.; Carrera, A.; Salem, H.; Selvey, C.; Nigro, S.J.; Fraser, N.; Price, K.; et al. Increased HIV Subtype Diversity Reflecting Demographic Changes in the HIV Epidemic in New South Wales, Australia. Viruses 2020, 12, 1402. https://doi.org/10.3390/v12121402
Di Giallonardo F, Pinto AN, Keen P, Shaik A, Carrera A, Salem H, Selvey C, Nigro SJ, Fraser N, Price K, et al. Increased HIV Subtype Diversity Reflecting Demographic Changes in the HIV Epidemic in New South Wales, Australia. Viruses. 2020; 12(12):1402. https://doi.org/10.3390/v12121402
Chicago/Turabian StyleDi Giallonardo, Francesca, Angie N. Pinto, Phillip Keen, Ansari Shaik, Alex Carrera, Hanan Salem, Christine Selvey, Steven J. Nigro, Neil Fraser, Karen Price, and et al. 2020. "Increased HIV Subtype Diversity Reflecting Demographic Changes in the HIV Epidemic in New South Wales, Australia" Viruses 12, no. 12: 1402. https://doi.org/10.3390/v12121402
APA StyleDi Giallonardo, F., Pinto, A. N., Keen, P., Shaik, A., Carrera, A., Salem, H., Selvey, C., Nigro, S. J., Fraser, N., Price, K., Holden, J., Lee, F. J., Dwyer, D. E., Bavinton, B. R., Grulich, A. E., Kelleher, A. D., & on behalf of the NSW HIV Prevention Partnership Project. (2020). Increased HIV Subtype Diversity Reflecting Demographic Changes in the HIV Epidemic in New South Wales, Australia. Viruses, 12(12), 1402. https://doi.org/10.3390/v12121402






