The International Virus Bioinformatics Meeting 2020
Abstract
1. Introduction
2. Sessions and Oral Presentations
2.1. Proteome and RNAome of RNA Viruses
2.1.1. Computational Methods for Identifying Functional RNA Structure Features—By Irmtraud M. Meyer
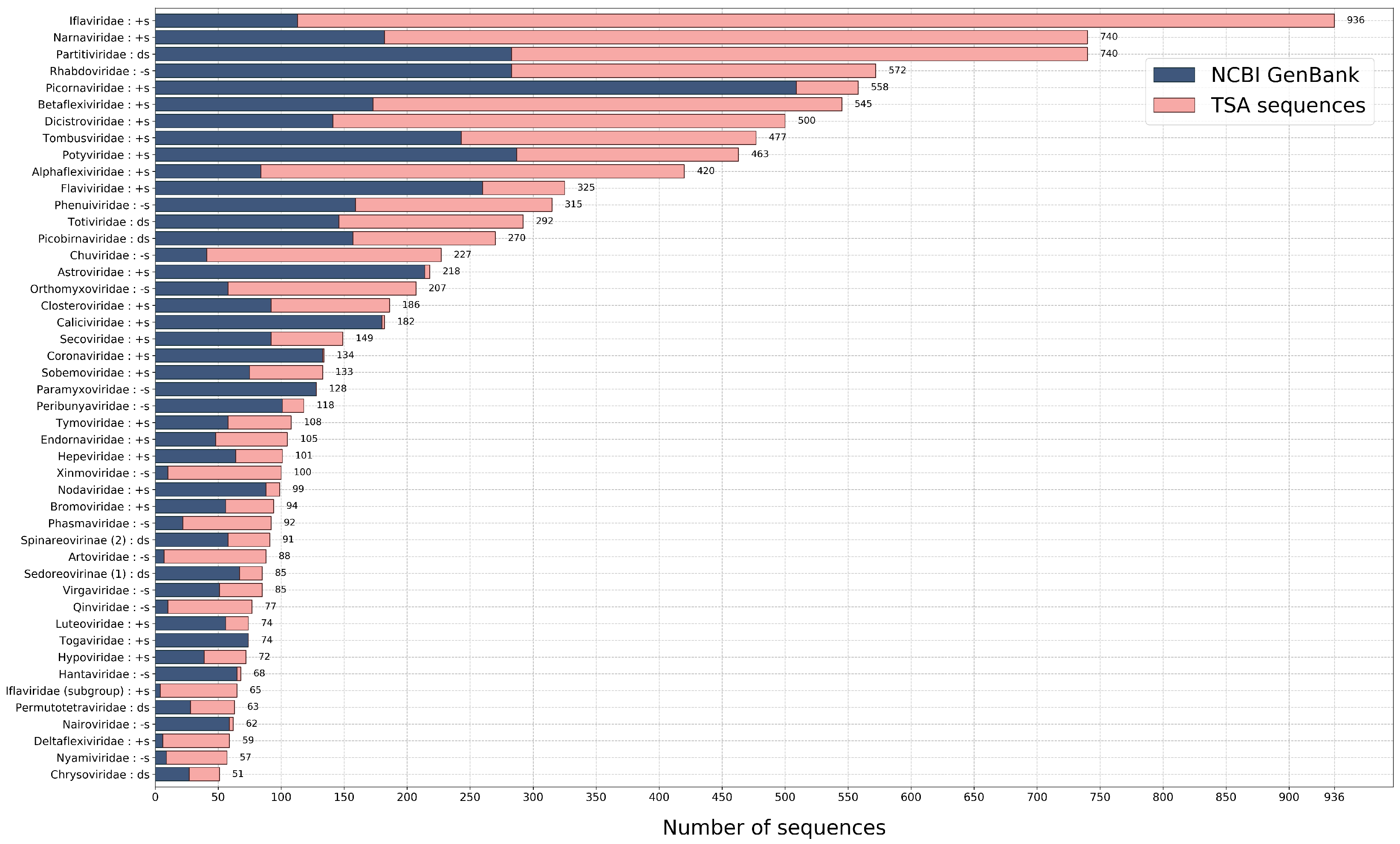
2.1.2. Expanding Diversity and Molecular Biology of RNA Viruses—By Ingrida Olendraite
2.1.3. Using Ribosome Profiling (RiboSeq) as a Tool to Analyse Virus Gene Expression—By Georgia Cook
2.2. Viral Metagenomics and Ecology
2.2.1. Viral Ecogenomics: Exploring Viral Diversity and Virus–Host Interactions from Metagenomes—By Simon Roux
2.2.2. viromeBrowser: A Shiny App for Browsing Virome Sequencing Analysis Results—By David Nieuwenhuijse
2.3. Virus Evolution and Classification
2.3.1. Leveraging High-Throughput Sequencing Data to Investigate Viral Diversity—By Niko Beerenwinkel
2.3.2. Parallel and Scalable Workflow for the Identification and Analysis of Phages in Sequencing Data—By Mike Marquet
2.3.3. Re-Assessing the Diversity of Negative Strand RNA Viruses in Insects—By Sofia Paraskevopoulou
2.3.4. Reducing Haystacks to Needles: Comparative Genomics Based on Viral Clusters—By Kevin Lamkiewicz
2.4. Viral Infections and Immunology
2.4.1. Machine Learning Approach to Predicting Host Taxonomic Information from Viral Genomes: Combining Feature Representations—By Francesca Young
2.4.2. Recombination Networks and Endogenous Viral Anchors for High-Throughput Host Identification—Cormac M. Kinsella
3. Poster Session
4. EVBC Annual Meeting
5. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Ibrahim, B.; McMahon, D.P.; Hufsky, F.; Beer, M.; Deng, L.; Mercier, P.L.; Palmarini, M.; Thiel, V.; Marz, M. A new era of virus bioinformatics. Virus Res. 2018, 251, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Hufsky, F.; Ibrahim, B.; Beer, M.; Deng, L.; Mercier, P.L.; McMahon, D.P.; Palmarini, M.; Thiel, V.; Marz, M. Virologists-Heroes need weapons. PLoS Pathog. 2018, 14, e1006771. [Google Scholar] [CrossRef]
- Ibrahim, B.; Arkhipova, K.; Andeweg, A.; Posada-Céspedes, S.; Enault, F.; Gruber, A.; Koonin, E.; Kupczok, A.; Lemey, P.; McHardy, A.; et al. Bioinformatics Meets Virology: The European Virus Bioinformatics Center’s Second Annual Meeting. Viruses 2018, 10, 256. [Google Scholar] [CrossRef] [PubMed]
- Hufsky, F.; Ibrahim, B.; Modha, S.; Clokie, M.R.J.; Deinhardt-Emmer, S.; Dutilh, B.E.; Lycett, S.; Simmonds, P.; Thiel, V.; Abroi, A.; et al. The Third Annual Meeting of the European Virus Bioinformatics Center. Viruses 2019, 11, 420. [Google Scholar] [CrossRef]
- Watts, J.M.; Dang, K.K.; Gorelick, R.J.; Leonard, C.W.; Bess, J.W.; Swanstrom, R.; Burch, C.L.; Weeks, K.M. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 2009, 460, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, J.S.; Forsberg, R.; Meyer, I.M.; Hein, J. An evolutionary model for protein-coding regions with conserved RNA structure. Mol. Biol. Evol. 2004, 21, 1913–1922. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, J.S.; Meyer, I.M.; Forsberg, R.; Simmonds, P.; Hein, J. A comparative method for finding and folding RNA secondary structures within protein-coding regions. Nucleic Acids Res. 2004, 32, 4925–4936. [Google Scholar] [CrossRef]
- Bogdanow, B.; Wang, X.; Eichelbaum, K.; Sadewasser, A.; Husic, I.; Paki, K.; Budt, M.; Hergeselle, M.; Vetter, B.; Hou, J.; et al. The dynamic proteome of influenza A virus infection identifies M segment splicing as a host range determinant. Nat. Commun. 2019, 10, 5518. [Google Scholar] [CrossRef]
- Lai, D.; Proctor, J.R.; Meyer, I.M. On the importance of cotranscriptional RNA structure formation. RNA 2013, 19, 1461–1473. [Google Scholar] [CrossRef] [PubMed]
- Proctor, J.R.; Meyer, I.M. COFOLD: An RNA secondary structure prediction method that takes co-transcriptional folding into account. Nucleic Acids Res. 2013, 41, e102. [Google Scholar] [CrossRef]
- Meyer, I.M. In silico methods for co-transcriptional RNA secondary structure prediction and for investigating alternative RNA structure expression. Methods 2017, 120, 3–16. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wiebe, N.J.P.; Meyer, I.M. TRANSAT—Method for detecting the conserved helices of functional RNA structures, including transient, pseudo-knotted and alternative structures. PLoS Comput. Biol. 2010, 6, e1000823. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.Y.A.; Steif, A.; Proctor, J.R.; Meyer, I.M. Transient RNA structure features are evolutionarily conserved and can be computationally predicted. Nucleic Acids Res. 2013, 41, 6273–6285. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhu, J.Y.A.; Meyer, I.M. Four RNA families with functional transient structures. RNA Biol. 2015, 12, 5–20. [Google Scholar] [CrossRef]
- Lai, D.; Proctor, J.R.; Zhu, J.Y.A.; Meyer, I.M. R-CHIE: A web server and R package for visualizing RNA secondary structures. Nucleic Acids Res. 2012, 40, e95. [Google Scholar] [CrossRef]
- Lu, Z.; Zhang, Q.C.; Lee, B.; Flynn, R.A.; Smith, M.A.; Robinson, J.T.; Davidovich, C.; Gooding, A.R.; Goodrich, K.J.; Mattick, J.S.; et al. RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell 2016, 165, 1267–1279. [Google Scholar] [CrossRef]
- Aw, J.G.A.; Shen, Y.; Wilm, A.; Sun, M.; Lim, X.N.; Boon, K.L.; Tapsin, S.; Chan, Y.S.; Tan, C.P.; Sim, A.Y.L.; et al. In vivo Mapping of Eukaryotic RNA Interactomes Reveals Principles of Higher-Order Organization and Regulation. Mol. Cell 2016, 62, 603–617. [Google Scholar] [CrossRef]
- Sharma, E.; Sterne-Weiler, T.; O’Hanlon, D.; Blencowe, B.J. Global Mapping of Human RNA-RNA Interactions. Mol. Cell 2016, 62, 618–626. [Google Scholar] [CrossRef]
- Stefanov, S.R.; Meyer, I.M. Deciphering the Universe of RNA Structures and trans RNA–RNA Interactions of Transcriptomes in vivo: From Experimental Protocols to Computational Analyses. In RNA Technologies; Springer: Cham, Switzerland, 2018; pp. 173–216. [Google Scholar] [CrossRef]
- Lai, D.; Meyer, I.M. e-RNA: A collection of web servers for comparative RNA structure prediction and visualisation. Nucleic Acids Res. 2014, 42, W373–W376. [Google Scholar] [CrossRef][Green Version]
- Tsybulskyi, V.; Mounir, M.; Meyer, I.M. R-chie: A web server and R package for visualizing cis and trans RNA–RNA, RNA–DNA and DNA–DNA interactions. Nucleic Acids Res. 2020, 48, e105. [Google Scholar] [CrossRef]
- Wolf, Y.I.; Kazlauskas, D.; Iranzo, J.; Lucía-Sanz, A.; Kuhn, J.H.; Krupovic, M.; Dolja, V.V.; Koonin, E.V. Origins and Evolution of the Global RNA Virome. mBio 2018, 9. [Google Scholar] [CrossRef]
- Olendraite, I.; Lukhovitskaya, N.I.; Porter, S.D.; Valles, S.M.; Firth, A.E. Polycipiviridae: A proposed new family of polycistronic picorna-like RNA viruses. J. Gen. Virol. 2017, 98, 2368–2378. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T.; Brar, G.A.; Rouskin, S.; McGeachy, A.M.; Weissman, J.S. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 2012, 7, 1534–1550. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T.; Ghaemmaghami, S.; Newman, J.R.S.; Weissman, J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 2009, 324, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Treffers, E.E.; Li, Y.; Tas, A.; Sun, Z.; van der Meer, Y.; de Ru, A.H.; van Veelen, P.A.; Atkins, J.F.; Snijder, E.J.; et al. Efficient -2 frameshifting by mammalian ribosomes to synthesize an additional arterivirus protein. Proc. Natl. Acad. Sci. USA 2012, 109, E2920–E2928. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Treffers, E.E.; Napthine, S.; Tas, A.; Zhu, L.; Sun, Z.; Bell, S.; Mark, B.L.; van Veelen, P.A.; van Hemert, M.J.; et al. Transactivation of programmed ribosomal frameshifting by a viral protein. Proc. Natl. Acad. Sci. USA 2014, 111, E2172–E2181. [Google Scholar] [CrossRef] [PubMed]
- Napthine, S.; Treffers, E.E.; Bell, S.; Goodfellow, I.; Fang, Y.; Firth, A.E.; Snijder, E.J.; Brierley, I. A novel role for poly(C) binding proteins in programmed ribosomal frameshifting. Nucleic Acids Res. 2016, 44, 5491–5503. [Google Scholar] [CrossRef]
- Finch, L.K.; Ling, R.; Napthine, S.; Olspert, A.; Michiels, T.; Lardinois, C.; Bell, S.; Loughran, G.; Brierley, I.; Firth, A.E. Characterization of Ribosomal Frameshifting in Theiler’s Murine Encephalomyelitis Virus. J. Virol. 2015, 89, 8580–8589. [Google Scholar] [CrossRef]
- Hill, C.H.; Cook, G.; Napthine, S.; Kibe, A.; Brown, K.; Caliskan, N.; Firth, A.E.; Graham, S.C.; Brierley, I. Structural and molecular basis for protein-stimulated ribosomal frameshifting in Theiler’s murine encephalomyelitis virus. bioRxiv 2020. [Google Scholar] [CrossRef]
- Breitbart, M.; Bonnain, C.; Malki, K.; Sawaya, N.A. Phage puppet masters of the marine microbial realm. Nat. Microbiol. 2018, 3, 754–766. [Google Scholar] [CrossRef]
- Brum, J.R.; Sullivan, M.B. Rising to the challenge: Accelerated pace of discovery transforms marine virology. Nat. Rev. Microbiol. 2015, 13, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Ogilvie, L.A.; Jones, B.V. The human gut virome: A multifaceted majority. Front. Microbiol. 2015, 6, 918. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Brito, B.; Li, L.; Wegley, L.; Furlan, M.; Angly, F.; Breitbart, M.; Buchanan, J.; Desnues, C.; Dinsdale, E.; Edwards, R.; et al. Viral and microbial community dynamics in four aquatic environments. ISME J. 2010, 4, 739–751. [Google Scholar] [CrossRef] [PubMed]
- Suttle, C.A. Viruses in the sea. Nature 2005, 437, 356–361. [Google Scholar] [CrossRef]
- Zimmerman, A.E.; Howard-Varona, C.; Needham, D.M.; John, S.G.; Worden, A.Z.; Sullivan, M.B.; Waldbauer, J.R.; Coleman, M.L. Metabolic and biogeochemical consequences of viral infection in aquatic ecosystems. Nat. Rev. Microbiol. 2020, 18, 21–34. [Google Scholar] [CrossRef]
- Feiner, R.; Argov, T.; Rabinovich, L.; Sigal, N.; Borovok, I.; Herskovits, A.A. A new perspective on lysogeny: Prophages as active regulatory switches of bacteria. Nat. Rev. Microbiol. 2015, 13, 641–650. [Google Scholar] [CrossRef]
- Paez-Espino, D.; Roux, S.; Chen, I.M.A.; Palaniappan, K.; Ratner, A.; Chu, K.; Huntemann, M.; Reddy, T.B.K.; Pons, J.C.; Llabrés, M.; et al. IMG/VR v.2.0: An integrated data management and analysis system for cultivated and environmental viral genomes. Nucleic Acids Res. 2019, 47, D678–D686. [Google Scholar] [CrossRef]
- Roux, S.; Brum, J.R.; Dutilh, B.E.; Sunagawa, S.; Duhaime, M.B.; Loy, A.; Poulos, B.T.; Solonenko, N.; Lara, E.; Poulain, J.; et al. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature 2016, 537, 689–693. [Google Scholar] [CrossRef]
- Emerson, J.B.; Roux, S.; Brum, J.R.; Bolduc, B.; Woodcroft, B.J.; Jang, H.B.; Singleton, C.M.; Solden, L.M.; Naas, A.E.; Boyd, J.A.; et al. Host-linked soil viral ecology along a permafrost thaw gradient. Nat. Microbiol. 2018, 3, 870–880. [Google Scholar] [CrossRef]
- Henry, K.A.; Arbabi-Ghahroudi, M.; Scott, J.K. Beyond phage display: Non-traditional applications of the filamentous bacteriophage as a vaccine carrier, therapeutic biologic, and bioconjugation scaffold. Front. Microbiol. 2015, 6, 755. [Google Scholar] [CrossRef]
- Salmond, G.P.C.; Fineran, P.C. A century of the phage: Past, present and future. Nat. Rev. Microbiol. 2015, 13, 777–786. [Google Scholar] [CrossRef] [PubMed]
- Roux, S.; Adriaenssens, E.M.; Dutilh, B.E.; Koonin, E.V.; Kropinski, A.M.; Krupovic, M.; Kuhn, J.H.; Lavigne, R.; Brister, J.R.; Varsani, A.; et al. Minimum Information about an Uncultivated Virus Genome (MIUViG). Nat. Biotechnol. 2019, 37, 29–37. [Google Scholar] [CrossRef]
- Roux, S.; Krupovic, M.; Daly, R.A.; Borges, A.L.; Nayfach, S.; Schulz, F.; Sharrar, A.; Matheus Carnevali, P.B.; Cheng, J.F.; Ivanova, N.N.; et al. Cryptic inoviruses revealed as pervasive in bacteria and archaea across Earth’s biomes. Nat. Microbiol. 2019, 4, 1895–1906. [Google Scholar] [CrossRef] [PubMed]
- Roux, S. A Viral Ecogenomics Framework to Uncover the Secrets of Nature’s “Microbe Whisperers”. mSystems 2019, 4. [Google Scholar] [CrossRef]
- Roux, S.; Brum, J.R. A viral reckoning: Viruses emerge as essential manipulators of global ecosystems. Environ. Microbiol. Rep. 2019, 11, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Emerson, J.B. Soil Viruses: A New Hope. mSystems 2019, 4. [Google Scholar] [CrossRef]
- Kortright, K.E.; Chan, B.K.; Koff, J.L.; Turner, P.E. Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria. Cell Host Microbe 2019, 25, 219–232. [Google Scholar] [CrossRef]
- Huson, D.H.; Beier, S.; Flade, I.; Górska, A.; El-Hadidi, M.; Mitra, S.; Ruscheweyh, H.J.; Tappu, R. MEGAN Community Edition—Interactive Exploration and Analysis of Large-Scale Microbiome Sequencing Data. PLoS Comput. Biol. 2016, 12, e1004957. [Google Scholar] [CrossRef]
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Interactive metagenomic visualization in a Web browser. BMC Bioinf. 2011, 12, 385. [Google Scholar] [CrossRef]
- Eren, A.M.; Esen, Ö.C.; Quince, C.; Vineis, J.H.; Morrison, H.G.; Sogin, M.L.; Delmont, T.O. Anvi’o: An advanced analysis and visualization platform for ’omics data. PeerJ 2015, 3, e1319. [Google Scholar] [CrossRef]
- Posada-Céspedes, S.; Seifert, D.; Topolsky, I.; Metzner, K.J.; Beerenwinkel, N. V-pipe: A computational pipeline for assessing viral genetic diversity from high-throughput sequencing data. bioRxiv 2020. [Google Scholar] [CrossRef]
- Kuipers, J.; Batavia, A.A.; Jablonski, K.P.; Bayer, F.; Borgsmüller, N.; Dondi, A.; Drăgan, M.A.; Ferreira, P.; Jahn, K.; Lamberti, L.; et al. Within-patient genetic diversity of SARS-CoV-2. bioRxiv 2020. [Google Scholar] [CrossRef]
- Käfer, S.; Paraskevopoulou, S.; Zirkel, F.; Wieseke, N.; Donath, A.; Petersen, M.; Jones, T.C.; Liu, S.; Zhou, X.; Middendorf, M.; et al. Re-assessing the diversity of negative strand RNA viruses in insects. PLoS Pathog. 2019, 15, e1008224. [Google Scholar] [CrossRef]
- Misof, B.; Liu, S.; Meusemann, K.; Peters, R.S.; Donath, A.; Mayer, C.; Frandsen, P.B.; Ware, J.; Flouri, T.; Beutel, R.G.; et al. Phylogenomics resolves the timing and pattern of insect evolution. Science 2014, 346, 763–767. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Tommaso, P.D.; Chatzou, M.; Floden, E.W.; Barja, P.P.; Palumbo, E.; Notredame, C. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 2017, 35, 316–319. [Google Scholar] [CrossRef]
- McInnes, L.; Healy, J.; Melville, J. Umap: Uniform manifold approximation and projection for dimension reduction. arXiv 2018, arXiv:1802.03426. [Google Scholar]
- McInnes, L.; Healy, J.; Astels, S. HDBSCAN: Hierarchical density based clustering. J. Open Source Softw. 2017, 2, 205. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Young, F.; Rogers, S.; Robertson, D.L. Predicting host taxonomic information from viral genomes: A comparison of feature representations. PLoS Comput. Biol. 2020, 16, e1007894. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, P.; Adams, M.J.; Benkő, M.; Breitbart, M.; Brister, J.R.; Carstens, E.B.; Davison, A.J.; Delwart, E.; Gorbalenya, A.E.; Harrach, B.; et al. Consensus statement: Virus taxonomy in the age of metagenomics. Nat. Rev. Microbiol. 2017, 15, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Kinsella, C.M.; Bart, A.; Deijs, M.; Broekhuizen, P.; Kaczorowska, J.; Jebbink, M.F.; van Gool, T.; Cotten, M.; van der Hoek, L. Entamoeba and Giardia parasites implicated as hosts of CRESS viruses. Nat. Commun. 2020, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kazlauskas, D.; Varsani, A.; Krupovic, M. Pervasive Chimerism in the Replication-Associated Proteins of Uncultured Single-Stranded DNA Viruses. Viruses 2018, 10, 187. [Google Scholar] [CrossRef]
- Roux, S.; Enault, F.; Bronner, G.; Vaulot, D.; Forterre, P.; Krupovic, M. Chimeric viruses blur the borders between the major groups of eukaryotic single-stranded DNA viruses. Nat. Commun. 2013, 4, 2700. [Google Scholar] [CrossRef] [PubMed]







| Date | Location | No. of Participants | Key Changes and Outcomes | |
|---|---|---|---|---|
| 6–8 March 2017 | Jena, Germany | ~100 | Founding of the center; Discussion of the role of EVBC; Election of the first Board of Directors; Insights into EU policy and funding opportunities. | |
| 9–10 April 2018 | Utrecht, The Netherlands | ~120 | Extending the EVBC network to include America and Asia;Discussion and design of joint projects;Insights on first applied European fund among EVBC members. | [3] |
| 28–29 March 2019 | Glasgow, United Kingdom | ~110 | Inclusion of contributed talks in themed sections in the scientific programme;Establishment of travel, poster and best contributed talk awards for junior scientists;Need for greater coordination and communication within the European virology community. | [4] |
| 8–9 October 2020 | virtually in Bern, Switzerland | ~120 | Renaming to International Virus Bioinformatics Meeting;Online format;Re-election of Board of Directors;Presentation of the newly EU-funded MSCA ITN VIROINF. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hufsky, F.; Beerenwinkel, N.; Meyer, I.M.; Roux, S.; Cook, G.M.; Kinsella, C.M.; Lamkiewicz, K.; Marquet, M.; Nieuwenhuijse, D.F.; Olendraite, I.; et al. The International Virus Bioinformatics Meeting 2020. Viruses 2020, 12, 1398. https://doi.org/10.3390/v12121398
Hufsky F, Beerenwinkel N, Meyer IM, Roux S, Cook GM, Kinsella CM, Lamkiewicz K, Marquet M, Nieuwenhuijse DF, Olendraite I, et al. The International Virus Bioinformatics Meeting 2020. Viruses. 2020; 12(12):1398. https://doi.org/10.3390/v12121398
Chicago/Turabian StyleHufsky, Franziska, Niko Beerenwinkel, Irmtraud M. Meyer, Simon Roux, Georgia May Cook, Cormac M. Kinsella, Kevin Lamkiewicz, Mike Marquet, David F. Nieuwenhuijse, Ingrida Olendraite, and et al. 2020. "The International Virus Bioinformatics Meeting 2020" Viruses 12, no. 12: 1398. https://doi.org/10.3390/v12121398
APA StyleHufsky, F., Beerenwinkel, N., Meyer, I. M., Roux, S., Cook, G. M., Kinsella, C. M., Lamkiewicz, K., Marquet, M., Nieuwenhuijse, D. F., Olendraite, I., Paraskevopoulou, S., Young, F., Dijkman, R., Ibrahim, B., Kelly, J., Le Mercier, P., Marz, M., Ramette, A., & Thiel, V. (2020). The International Virus Bioinformatics Meeting 2020. Viruses, 12(12), 1398. https://doi.org/10.3390/v12121398









