A Novel Sub-Lineage of Chikungunya Virus East/Central/South African Genotype Indian Ocean Lineage Caused Sequential Outbreaks in Bangladesh and Thailand
Abstract
1. Introduction
2. Materials and Methods
2.1. CHIKV Samples
2.2. CHIKV Genome Sequencing
2.3. Nucleotide Sequence Accession Numbers
2.4. Phylogenetic Analyses
2.5. Selection Analyses
3. Results
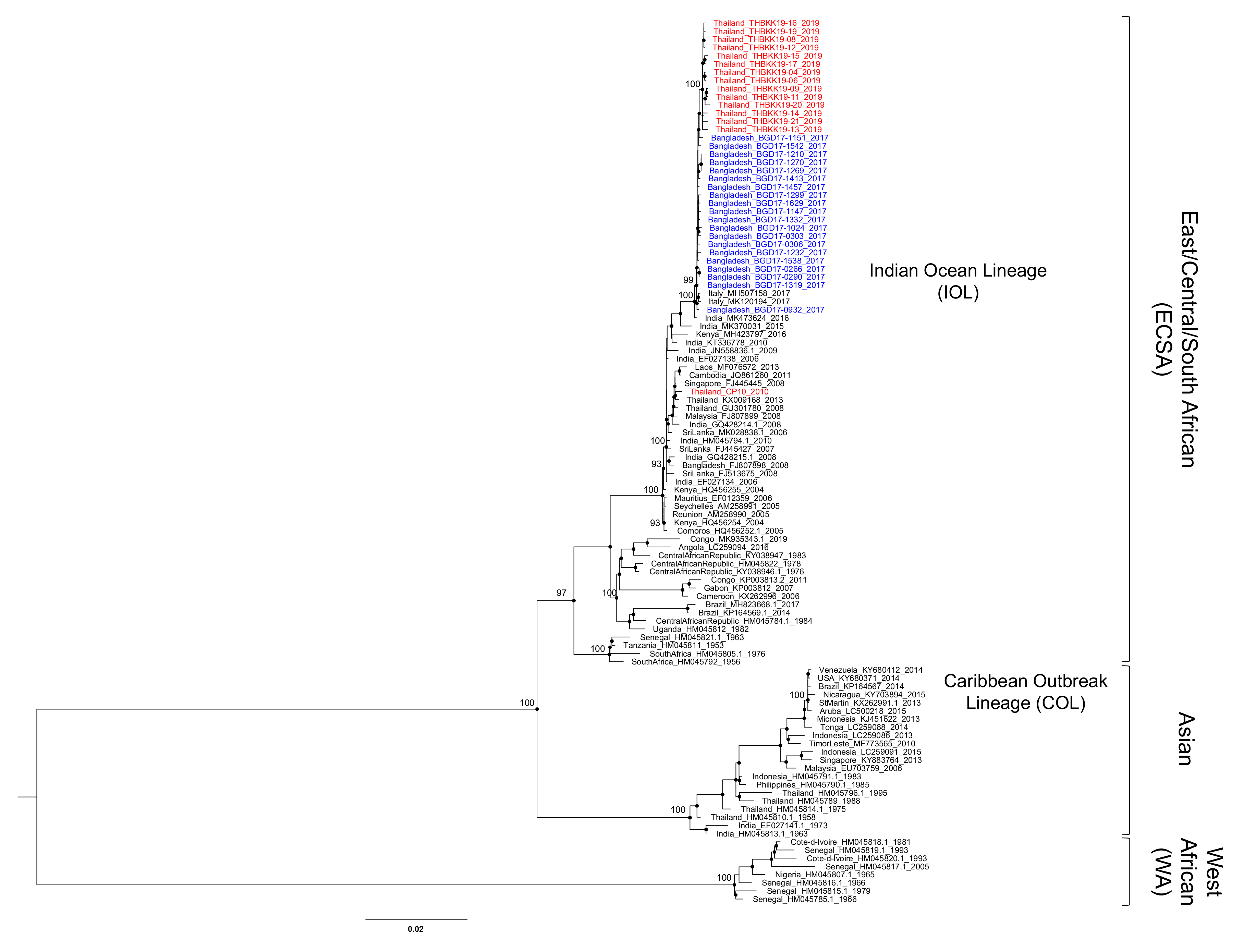
3.1. ECSA-IOL CHIKVs in Bangladesh and Thailand During 2017–2019
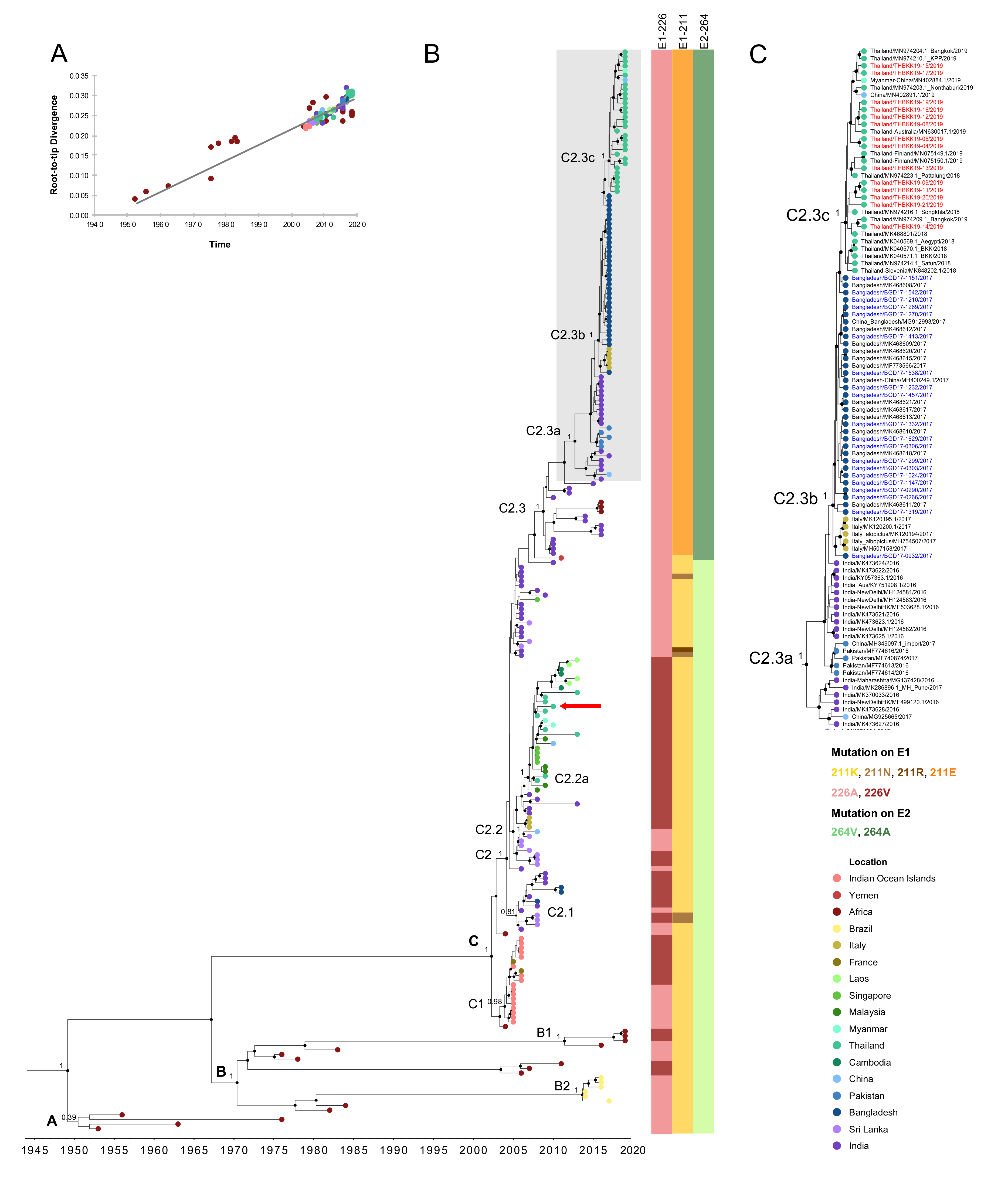
3.2. Evolution of the ECSA-CHIKV IOL Sub-Lineage
3.3. Signature Mutations Related to CHIKV ECSA-IOL Sub-Lineages among CHIKV Outbreak Strains
3.4. Selection Analyses
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Halstead, S.B. Reappearance of chikungunya, formerly called dengue, in the Americas. Emerg. Infect. Dis. 2015, 21, 557–561. [Google Scholar] [CrossRef] [PubMed]
- Volk, S.M.; Chen, R.; Tsetsarkin, K.A.; Adams, A.P.; Garcia, T.I.; Sall, A.A.; Nasar, F.; Schuh, A.J.; Holmes, E.C.; Higgs, S.; et al. Genome-scale phylogenetic analyses of chikungunya virus reveal independent emergences of recent epidemics and various evolutionary rates. J. Virol. 2010, 84, 6497–6504. [Google Scholar] [CrossRef] [PubMed]
- Weaver, S.C.; Chen, R.; Diallo, M. Chikungunya Virus: Role of Vectors in Emergence from Enzootic Cycles. Annu. Rev. Entomol. 2020, 65, 313–332. [Google Scholar] [CrossRef] [PubMed]
- Weaver, S.C. Arrival of chikungunya virus in the new world: Prospects for spread and impact on public health. PLoS Negl. Trop. Dis. 2014, 8, e2921. [Google Scholar] [CrossRef] [PubMed]
- Schuffenecker, I.; Iteman, I.; Michault, A.; Murri, S.; Frangeul, L.; Vaney, M.C.; Lavenir, R.; Pardigon, N.; Reynes, J.M.; Pettinelli, F.; et al. Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med. 2006, 3, e263. [Google Scholar] [CrossRef]
- Tsetsarkin, K.A.; Vanlandingham, D.L.; McGee, C.E.; Higgs, S. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 2007, 3, e201. [Google Scholar] [CrossRef]
- Tan, Y.; Pickett, B.E.; Shrivastava, S.; Gresh, L.; Balmaseda, A.; Amedeo, P.; Hu, L.; Puri, V.; Fedorova, N.B.; Halpin, R.A.; et al. Differing epidemiological dynamics of Chikungunya virus in the Americas during the 2014-2015 epidemic. PLoS Negl. Trop. Dis. 2018, 12, e0006670. [Google Scholar] [CrossRef]
- Nunes, M.R.; Faria, N.R.; de Vasconcelos, J.M.; Golding, N.; Kraemer, M.U.; de Oliveira, L.F.; Azevedo Rdo, S.; da Silva, D.E.; da Silva, E.V.; da Silva, S.P.; et al. Emergence and potential for spread of Chikungunya virus in Brazil. BMC Med. 2015, 13, 102. [Google Scholar] [CrossRef]
- Jain, J.; Kaur, N.; Haller, S.L.; Kumar, A.; Rossi, S.L.; Narayanan, V.; Kumar, D.; Gaind, R.; Weaver, S.C.; Auguste, A.J.; et al. Chikungunya Outbreaks in India: A Prospective Study Comparing Neutralization and Sequelae during Two Outbreaks in 2010 and 2016. Am. J. Trop. Med. Hyg. 2020, 102, 857–868. [Google Scholar] [CrossRef]
- Shrinet, J.; Jain, S.; Sharma, A.; Singh, S.S.; Mathur, K.; Rana, V.; Bhatnagar, R.K.; Gupta, B.; Gaind, R.; Deb, M.; et al. Genetic characterization of Chikungunya virus from New Delhi reveal emergence of a new molecular signature in Indian isolates. Virol. J. 2012, 9, 100. [Google Scholar] [CrossRef]
- Rahman, M.; Yamagishi, J.; Rahim, R.; Hasan, A.; Sobhan, A. East/Central/South African Genotype in a Chikungunya Outbreak, Dhaka, Bangladesh, 2017. Emerg. Infect. Dis. 2019, 25, 370–372. [Google Scholar] [CrossRef] [PubMed]
- Aamir, U.B.; Badar, N.; Salman, M.; Ahmed, M.; Alam, M.M. Outbreaks of chikungunya in Pakistan. Lancet Infect. Dis. 2017, 17, 483. [Google Scholar] [CrossRef]
- Venturi, G.; Di Luca, M.; Fortuna, C.; Remoli, M.E.; Riccardo, F.; Severini, F.; Toma, L.; Del Manso, M.; Benedetti, E.; Caporali, M.G.; et al. Detection of a chikungunya outbreak in Central Italy, August to September 2017. Eurosurveillance 2017, 22, 1700646. [Google Scholar] [CrossRef] [PubMed]
- Maljkovic Berry, I.; Eyase, F.; Pollett, S.; Konongoi, S.L.; Joyce, M.G.; Figueroa, K.; Ofula, V.; Koka, H.; Koskei, E.; Nyunja, A.; et al. Global Outbreaks and Origins of a Chikungunya Virus Variant Carrying Mutations Which May Increase Fitness for Aedes aegypti: Revelations from the 2016 Mandera, Kenya Outbreak. Am. J. Trop. Med. Hyg. 2019, 100, 1249–1257. [Google Scholar] [CrossRef]
- BOE, National Disease Surveillance (Report 506) Chikungunya: Bureau of Epidemiology, Ministry of Public Health, Thailand. Available online: http://www.boe.moph.go.th/boedb/surdata/disease.php?ds=84 (accessed on 10 July 2020).
- Suzuki, K.; Huits, R.; Phadungsombat, J.; Tuekprakhon, A.; Nakayama, E.E.; van den Berg, R.; Barbe, B.; Cnops, L.; Rahim, R.; Hasan, A.; et al. Promising application of monoclonal antibody against chikungunya virus E1-antigen across genotypes in immunochromatographic rapid diagnostic tests. Virol. J. 2020, 17, 90. [Google Scholar] [CrossRef]
- Sasayama, M.; Benjathummarak, S.; Kawashita, N.; Rukmanee, P.; Sangmukdanun, S.; Masrinoul, P.; Pitaksajjakul, P.; Puiprom, O.; Wuthisen, P.; Kurosu, T.; et al. Chikungunya virus was isolated in Thailand, 2010. Virus Genes 2014, 49, 485–489. [Google Scholar] [CrossRef]
- Phadungsombat, J.; Tuekprakhon, A.; Cnops, L.; Michiels, J.; van den Berg, R.; Nakayama, E.E.; Shioda, T.; Arien, K.K.; Huits, R. Two distinct lineages of chikungunya virus cocirculated in Aruba during the 2014-2015 epidemic. Infect. Genet. Evol. 2020, 78, 104129. [Google Scholar] [CrossRef]
- Pyke, A.T.; McMahon, J.; Burtonclay, P.; Nair, N.; De Jong, A. Genome Sequences of Chikungunya Virus Strains from Bangladesh and Thailand. Microbiol. Resour. Announc. 2020, 9, e01452-19. [Google Scholar] [CrossRef]
- Pickett, B.E.; Sadat, E.L.; Zhang, Y.; Noronha, J.M.; Squires, R.B.; Hunt, V.; Liu, M.; Kumar, S.; Zaremba, S.; Gu, Z.; et al. ViPR: An open bioinformatics database and analysis resource for virology research. Nucleic. Acids Res. 2012, 40, D593–D598. [Google Scholar] [CrossRef]
- Arankalle, V.A.; Shrivastava, S.; Cherian, S.; Gunjikar, R.S.; Walimbe, A.M.; Jadhav, S.M.; Sudeep, A.B.; Mishra, A.C. Genetic divergence of Chikungunya viruses in India (1963-2006) with special reference to the 2005–2006 explosive epidemic. J. Gen. Virol. 2007, 88, 1967–1976. [Google Scholar] [CrossRef]
- Cherian, S.S.; Walimbe, A.M.; Jadhav, S.M.; Gandhe, S.S.; Hundekar, S.L.; Mishra, A.C.; Arankalle, V.A. Evolutionary rates and timescale comparison of Chikungunya viruses inferred from the whole genome/E1 gene with special reference to the 2005-07 outbreak in the Indian subcontinent. Infect. Genet. Evol. 2009, 9, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef] [PubMed]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic. Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587. [Google Scholar] [CrossRef]
- Rambaut, A.; Lam, T.T.; Max Carvalho, L.; Pybus, O.G. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus. Evol. 2016, 2, vew007. [Google Scholar] [CrossRef]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018, 4, vey016. [Google Scholar] [CrossRef]
- Baele, G.; Lemey, P.; Suchard, M.A. Genealogical Working Distributions for Bayesian Model Testing with Phylogenetic Uncertainty. Syst. Biol. 2016, 65, 250–264. [Google Scholar] [CrossRef]
- Shapiro, B.; Rambaut, A.; Drummond, A.J. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol. Biol. Evol. 2006, 23, 7–9. [Google Scholar] [CrossRef]
- Gill, M.S.; Lemey, P.; Faria, N.R.; Rambaut, A.; Shapiro, B.; Suchard, M.A. Improving Bayesian population dynamics inference: A coalescent-based model for multiple loci. Mol. Biol. Evol. 2013, 30, 713–724. [Google Scholar] [CrossRef]
- Weaver, S.; Shank, S.D.; Spielman, S.J.; Li, M.; Muse, S.V.; Kosakovsky Pond, S.L. Datamonkey 2.0: A Modern Web Application for Characterizing Selective and Other Evolutionary Processes. Mol. Biol. Evol. 2018, 35, 773–777. [Google Scholar] [CrossRef] [PubMed]
- Murrell, B.; Wertheim, J.O.; Moola, S.; Weighill, T.; Scheffler, K.; Kosakovsky Pond, S.L. Detecting individual sites subject to episodic diversifying selection. PLoS Genet 2012, 8, e1002764. [Google Scholar] [CrossRef] [PubMed]
- Murrell, B.; Moola, S.; Mabona, A.; Weighill, T.; Sheward, D.; Kosakovsky Pond, S.L.; Scheffler, K. FUBAR: A fast, unconstrained bayesian approximation for inferring selection. Mol. Biol. Evol. 2013, 30, 1196–1205. [Google Scholar] [CrossRef] [PubMed]
- Kosakovsky Pond, S.L.; Frost, S.D. Not so different after all: A comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 2005, 22, 1208–1222. [Google Scholar] [CrossRef]
- Kariuki Njenga, M.; Nderitu, L.; Ledermann, J.P.; Ndirangu, A.; Logue, C.H.; Kelly, C.H.L.; Sang, R.; Sergon, K.; Breiman, R.; Powers, A.M. Tracking epidemic Chikungunya virus into the Indian Ocean from East Africa. J. Gen. Virol. 2008, 89, 2754–2760. [Google Scholar] [CrossRef]
- Kabir, I.; Dhimal, M.; Muller, R.; Banik, S.; Haque, U. The 2017 Dhaka chikungunya outbreak. Lancet Infect. Dis. 2017, 17, 1118. [Google Scholar] [CrossRef]
- Diaz-Menendez, M.; Esteban, E.T.; Ujiie, M.; Calleri, G.; Rothe, C.; Malvy, D.; Nicastri, E.; Bissinger, A.L.; Grandadam, M.; Alpern, J.D.; et al. Travel-associated chikungunya acquired in Myanmar in 2019. Eurosurveillance 2020, 25, 1900721. [Google Scholar] [CrossRef]
- Venturi, G.; Aberle, S.W.; Avsic-Zupanc, T.; Barzon, L.; Batejat, C.; Burdino, E.; Carletti, F.; Charrel, R.; Christova, I.; Connell, J.; et al. Specialist laboratory networks as preparedness and response tool the Emerging Viral Diseases-Expert Laboratory Network and the Chikungunya outbreak, Thailand, 2019. Eurosurveillance 2020, 25, 1900438. [Google Scholar] [CrossRef]
- Zakotnik, S.; Korva, M.; Knap, N.; Robnik, B.; Gorisek Miksic, N.; Avsic Zupanc, T. Complete Coding Sequence of a Chikungunya Virus Strain Imported into Slovenia from Thailand in Late 2018. Microbiol. Resour. Announc. 2019, 8, e00581-19. [Google Scholar] [CrossRef]
- Chen, R.; Puri, V.; Fedorova, N.; Lin, D.; Hari, K.L.; Jain, R.; Rodas, J.D.; Das, S.R.; Shabman, R.S.; Weaver, S.C. Comprehensive Genome Scale Phylogenetic Study Provides New Insights on the Global Expansion of Chikungunya Virus. J. Virol. 2016, 90, 10600–10611. [Google Scholar] [CrossRef]
- Vazeille, M.; Moutailler, S.; Coudrier, D.; Rousseaux, C.; Khun, H.; Huerre, M.; Thiria, J.; Dehecq, J.S.; Fontenille, D.; Schuffenecker, I.; et al. Two Chikungunya isolates from the outbreak of La Reunion (Indian Ocean) exhibit different patterns of infection in the mosquito, Aedes albopictus. PLoS ONE 2007, 2, e1168. [Google Scholar] [CrossRef] [PubMed]
- Naresh Kumar, C.V.; Sivaprasad, Y.; Sai Gopal, D.V. Genetic diversity of 2006-2009 Chikungunya virus outbreaks in Andhra Pradesh, India, reveals complete absence of E1:A226V mutation. Acta Virol. 2016, 60, 114–117. [Google Scholar] [CrossRef] [PubMed]
- Sumathy, K.; Ella, K.M. Genetic diversity of Chikungunya virus, India 2006-2010: Evolutionary dynamics and serotype analyses. J. Med. Virol. 2012, 84, 462–470. [Google Scholar] [CrossRef] [PubMed]
- Abraham, R.; Manakkadan, A.; Mudaliar, P.; Joseph, I.; Sivakumar, K.C.; Nair, R.R.; Sreekumar, E. Correlation of phylogenetic clade diversification and in vitro infectivity differences among Cosmopolitan genotype strains of Chikungunya virus. Infect. Genet Evol. 2016, 37, 174–184. [Google Scholar] [CrossRef]
- Santhosh, S.R.; Dash, P.K.; Parida, M.M.; Khan, M.; Tiwari, M.; Lakshmana Rao, P.V. Comparative full genome analysis revealed E1: A226V shift in 2007 Indian Chikungunya virus isolates. Virus Res. 2008, 135, 36–41. [Google Scholar] [CrossRef]
- Taraphdar, D.; Chatterjee, S. Molecular characterization of chikungunya virus circulating in urban and rural areas of West Bengal, India after its re-emergence in 2006. Trans. R Soc. Trop Med. Hyg. 2015, 109, 197–202. [Google Scholar] [CrossRef]
- Rianthavorn, P.; Prianantathavorn, K.; Wuttirattanakowit, N.; Theamboonlers, A.; Poovorawan, Y. An outbreak of chikungunya in southern Thailand from 2008 to 2009 caused by African strains with A226V mutation. Int. J. Infect. Dis. 2010, 14, e161–e165. [Google Scholar] [CrossRef]
- Hapuarachchi, H.C.; Bandara, K.B.; Sumanadasa, S.D.; Hapugoda, M.D.; Lai, Y.L.; Lee, K.S.; Tan, L.K.; Lin, R.T.; Ng, L.F.; Bucht, G.; et al. Re-emergence of Chikungunya virus in South-east Asia: Virological evidence from Sri Lanka and Singapore. J. Gen. Virol. 2010, 91, 1067–1076. [Google Scholar] [CrossRef]
- Newase, P.; More, A.; Patil, J.; Patil, P.; Jadhav, S.; Alagarasu, K.; Shah, P.; Parashar, D.; Cherian, S.S. Chikungunya phylogeography reveals persistent global transmissions of the Indian Ocean Lineage from India in association with mutational fitness. Infect. Genet. Evol. 2020, 82, 104289. [Google Scholar] [CrossRef]
- Fahmy, N.T.; Klena, J.D.; Mohamed, A.S.; Zayed, A.; Villinski, J.T. Complete Genome Sequence of Chikungunya Virus Isolated from an Aedes aegypti Mosquito during an Outbreak in Yemen, 2011. Genome Announc. 2015, 3, e00789-15. [Google Scholar] [CrossRef]
- Nyari, N.; Maan, H.S.; Sharma, S.; Pandey, S.N.; Dhole, T.N. Identification and genetic characterization of chikungunya virus from Aedes mosquito vector collected in the Lucknow district, North India. Acta Trop. 2016, 158, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.; Sharma, A.K.; Sukumaran, D.; Parida, M.; Dash, P.K. Two novel epistatic mutations (E1:K211E and E2:V264A) in structural proteins of Chikungunya virus enhance fitness in Aedes aegypti. Virology 2016, 497, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Haque, F.; Rahman, M.; Banu, N.N.; Sharif, A.R.; Jubayer, S.; Shamsuzzaman, A.; Alamgir, A.; Erasmus, J.H.; Guzman, H.; Forrester, N.; et al. An epidemic of chikungunya in northwestern Bangladesh in 2011. PLoS ONE 2019, 14, e0212218. [Google Scholar] [CrossRef] [PubMed]
- Khatun, S.; Chakraborty, A.; Rahman, M.; Nasreen Banu, N.; Rahman, M.M.; Hasan, S.M.; Luby, S.P.; Gurley, E.S. An Outbreak of Chikungunya in Rural Bangladesh, 2011. PLoS Negl. Trop. Dis. 2015, 9, e0003907. [Google Scholar] [CrossRef]
- Anwar, S.; Taslem Mourosi, J.; Khan, M.F.; Ullah, M.O.; Vanakker, O.M.; Hosen, M.J. Chikungunya outbreak in Bangladesh (2017): Clinical and hematological findings. PLoS Negl. Trop. Dis. 2020, 14, e0007466. [Google Scholar] [CrossRef]
- Hossain, M.S.; Hasan, M.M.; Islam, M.S.; Islam, S.; Mozaffor, M.; Khan, M.A.S.; Ahmed, N.; Akhtar, W.; Chowdhury, S.; Arafat, S.M.Y.; et al. Chikungunya outbreak (2017) in Bangladesh: Clinical profile, economic impact and quality of life during the acute phase of the disease. PLoS Negl. Trop. Dis. 2018, 12, e0006561. [Google Scholar] [CrossRef]
- Paul, K.K.; Dhar-Chowdhury, P.; Haque, C.E.; Al-Amin, H.M.; Goswami, D.R.; Kafi, M.A.H.; Drebot, M.A.; Lindsay, L.R.; Ahsan, G.U.; Brooks, W.A. Risk factors for the presence of dengue vector mosquitoes, and determinants of their prevalence and larval site selection in Dhaka, Bangladesh. PLoS ONE 2018, 13, e0199457. [Google Scholar] [CrossRef]
- Saha, S.; Ramesh, A.; Kalantar, K.; Malaker, R.; Hasanuzzaman, M.; Khan, L.M.; Mayday, M.Y.; Sajib, M.S.I.; Li, L.M.; Langelier, C.; et al. Unbiased Metagenomic Sequencing for Pediatric Meningitis in Bangladesh Reveals Neuroinvasive Chikungunya Virus Outbreak and Other Unrealized Pathogens. mBio 2019, 10, e02877-19. [Google Scholar] [CrossRef]
- Pan, J.; Fang, C.; Yan, J.; Yan, H.; Zhan, B.; Sun, Y.; Liu, Y.; Mao, H.; Cao, G.; Lv, L.; et al. Chikungunya Fever Outbreak, Zhejiang Province, China, 2017. Emerg. Infect. Dis. 2019, 25, 1589–1591. [Google Scholar] [CrossRef]
- Rezza, G.; Nicoletti, L.; Angelini, R.; Romi, R.; Finarelli, A.C.; Panning, M.; Cordioli, P.; Fortuna, C.; Boros, S.; Magurano, F.; et al. Infection with chikungunya virus in Italy: An outbreak in a temperate region. Lancet 2007, 370, 1840–1846. [Google Scholar] [CrossRef]
- Fortuna, C.; Toma, L.; Remoli, M.E.; Amendola, A.; Severini, F.; Boccolini, D.; Romi, R.; Venturi, G.; Rezza, G.; Di Luca, M. Vector competence of Aedes albopictus for the Indian Ocean lineage (IOL) chikungunya viruses of the 2007 and 2017 outbreaks in Italy: A comparison between strains with and without the E1:A226V mutation. Eurosurveillance 2018, 23, 1800246. [Google Scholar] [CrossRef] [PubMed]
- Intayot, P.; Phumee, A.; Boonserm, R.; Sor-Suwan, S.; Buathong, R.; Wacharapluesadee, S.; Brownell, N.; Poovorawan, Y.; Siriyasatien, P. Genetic Characterization of Chikungunya Virus in Field-Caught Aedes aegypti Mosquitoes Collected during the Recent Outbreaks in 2019, Thailand. Pathogens 2019, 8, 121. [Google Scholar] [CrossRef] [PubMed]
- Thavara, U.; Tawatsin, A.; Pengsakul, T.; Bhakdeenuan, P.; Chanama, S.; Anantapreecha, S.; Molito, C.; Chompoosri, J.; Thammapalo, S.; Sawanpanyalert, P.; et al. Outbreak of chikungunya fever in Thailand and virus detection in field population of vector mosquitoes, Aedes aegypti (L.) and Aedes albopictus Skuse (Diptera: Culicidae). Southeast Asian J. Trop. Med. Public Health 2009, 40, 951–962. [Google Scholar] [PubMed]
| Strain | Collection Date | Location | Country | Sample Type | Passage History | Accession No. |
|---|---|---|---|---|---|---|
| BGD17-0266 | July 2017 | Maghbazar, Dhaka | Bangladesh | serum | 0 | LC580236 |
| BGD17-0290 | July 2017 | Uttara, Dhaka | Bangladesh | isolate | C6/36 | LC580237 |
| BGD17-0303 | July 2017 | Bashundhara, Dhaka | Bangladesh | serum | 0 | LC580238 |
| BGD17-0306 | July 2017 | Maghbazar, Dhaka | Bangladesh | serum | 0 | LC580239 |
| BGD17-0932 | August 2017 | Bashundhara, Dhaka | Bangladesh | serum | 0 | LC580240 |
| BGD17-1024 | September 2017 | Bashundhara, Dhaka | Bangladesh | serum | 0 | LC580241 |
| BGD17-1147 | September 2017 | Bashundhara, Dhaka | Bangladesh | serum | 0 | LC580242 |
| BGD17-1151 | September 2017 | Bashundhara, Dhaka | Bangladesh | isolate | C6/36 | LC580243 |
| BGD17-1210 | September 2017 | Bashundhara, Dhaka | Bangladesh | isolate | C6/36 | LC580244 |
| BGD17-1232 | September 2017 | Bashundhara, Dhaka | Bangladesh | isolate | C6/36 | LC580245 |
| BGD17-1269 | September 2017 | Bashundhara, Dhaka | Bangladesh | isolate | C6/36 | LC580246 |
| BGD17-1270 | September 2017 | Bashundhara, Dhaka | Bangladesh | isolate | C6/36 | LC580247 |
| BGD17-1299 | October 2017 | Rampura, Dhaka | Bangladesh | isolate | C6/36 | LC580248 |
| BGD17-1319 | October 2017 | Chittagon | Bangladesh | isolate | C6/36 | LC580249 |
| BGD17-1332 | October 2017 | Mirpur, Dhaka | Bangladesh | serum | 0 | LC580250 |
| BGD17-1413 | October 2017 | Mirpur, Dhaka | Bangladesh | serum | 0 | LC580251 |
| BGD17-1457 | October 2017 | Gulshan, Dhaka | Bangladesh | isolate | C6/36 | LC580252 |
| BGD17-1538 | November 2017 | Uttara, Dhaka | Bangladesh | serum | 0 | LC580253 |
| BGD17-1542 | November 2017 | Gulshan, Dhaka | Bangladesh | serum | 0 | LC580254 |
| BGD17-1629 | December 2017 | Uttara, Dhaka | Bangladesh | serum | 0 | LC580255 |
| THBKK19-04 | October 2019 | Bangkok | Thailand | serum | 0 | LC580256 |
| THBKK19-06 | October 2019 | Bangkok | Thailand | isolate | C6/36 | LC580257 |
| THBKK19-08 | October 2019 | Bangkok | Thailand | serum | 0 | LC580258 |
| THBKK19-09 | October 2019 | Bangkok | Thailand | isolate | C6/36 | LC580259 |
| THBKK19-11 | October 2019 | Bangkok | Thailand | isolate | C6/36 | LC580260 |
| THBKK19-12 | October 2019 | Bangkok | Thailand | isolate | C6/36 | LC580261 |
| THBKK19-13 | October 2019 | Bangkok | Thailand | serum | 0 | LC580262 |
| THBKK19-14 | October 2019 | Bangkok | Thailand | serum | 0 | LC580263 |
| THBKK19-15 | October 2019 | Bangkok | Thailand | isolate | C6/36 | LC580264 |
| THBKK19-16 | October 2019 | Bangkok | Thailand | isolate | C6/36 | LC580265 |
| THBKK19-17 | October 2019 | Bangkok | Thailand | isolate | C6/36 | LC580266 |
| THBKK19-19 | October 2019 | Bangkok | Thailand | serum | 0 | LC580267 |
| THBKK19-20 | October 2019 | Bangkok | Thailand | serum | 0 | LC580268 |
| THBKK19-21 | October 2019 | Bangkok | Thailand | serum | 0 | LC580269 |
| CP10 | 2010 | Ratchaburi | Thailand | isolate | C6/36 and Vero | LC580270 |
| Node | Lineage/Clade | Detected Areas/Countries | PP | tMRCA (95% HPD) | Substitution Rate × 10−4 (95% HPD) |
|---|---|---|---|---|---|
| Root | Root ECSA | 1 | 1949.17 (1944.14–1957.42) | 5.74 (4.96–6.60) | |
| A | Enzootic ECSA | South Africa, Central African Republic, Senegal, and Tanzania | 0.39 | 1950.48 (1948.58–1952.53) | 5.70 (0.80–12.62) |
| B | African ECSA | Africa and Brazil | 1 | 1970.39 (1967.68–1973.27) | 5.60 (1.37–11.68) |
| B1 | West African ECSA | Congo and Angola | 1 | 2011.39 (2008.69–2014.26) | 1.02 (0.69–1.36) |
| B2 | South American ECSA | Brazil | 1 | 2013.66 (2013.29–2014.18) | 3.27 (2.59–3.92) |
| C | Indian Ocean Lineage (IOL) | Coastal Kenya, Indian Ocean Islands, Indian subcontinent, SEA countries, Italy, and France | 1 | 2002.26 (2001.14–2003.59) | 2.98 (2.31–3.67) |
| C1 | Indian Ocean Islands Sub-lineage | Indian Ocean Islands, France | 0.98 | 2003.92 (2003.43–2004.45) | 2.88 (5.93–6.10) |
| C2 | Indian subcontinent and SEA Sub-lineage | Indian subcontinent, SEA countries, and Italy | 1 | 2004.16 (2003.52–2004.88) | 4.72 (1.34–8.93) |
| C2.1 | Indian subcontinent | India, Sri Lanka, and Bangladesh | 0.81 | 2005.33 (2004.74–2006.15) | 4.16 (0.61–9.57) |
| C2.2 | Indian subcontinent/SEA | Indian Continent, SEA countries, and Italy | 1 | 2004.97 (2004.46–2005.52) | 6.91 (1.49–14.50) |
| C2.2a | SEA clade | Malaysia, Singapore, Thailand, Cambodia, and Laos | 1 | 2006.82 (2006.42–2007.25) | 4.91 (1.17–10.07) |
| C2.3 | Northern Indian subcontinent/SEA | Northern Indian subcontinent, SEA countries, and Italy | 1 | 2008.75 (2008.18–2009.37) | 6.29 (1.57–13.03) |
| C2.3a | India clade | India, Pakistan, Bangladesh, Italy, Myanmar, and Thailand | 1 | 2012.71 (2011.72–2013.74) | 8.52 (2.52–16.22) |
| C2.3b | Bangladesh clade | Bangladesh, Italy, Myanmar, and Thailand | 1 | 2015.61 (2015.25–2016.01) | 6.79 (1.75–13.76) |
| C2.3c | Thailand clade | Thailand and Myanmar | 1 | 2016.98 (2016.73–2017.25) | 10.75 (3.07–20.41) |
| Polyproteins | Position* | Protein | Position** | Ancestor IOL | Indian Ocean Islands (C1***) | Indian Sub-Continent (C2.1***) | Indian Sub-Continent/SEA (C2.2a***) | Northern Indian Subcontinent/SEA (C2.3***) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kenya 2004 | Reunion 2005 | India 2006 | Thailand 2010 | India 2010 | India 2016 | Pakistan 2016 | Bangladesh 2017 | Italy 2017 | Thailand 2019 | ||||
| Non-structural protein | 665 | nsP2 | 130 | H | H | H | H | H | H/Y | Y | Y | Y | Y |
| 680 | nsP2 | 145 | E | E | E | E | E | E/D | D | D | D | D | |
| 1030 | nsP2 | 495 | N | N | N | N | N | N | N | N | N | S | |
| 1328 | nsP2 | 793 | V | V | V | V | V | V/I | V | V/A | V | A | |
| 1391 | nsP3 | 58 | V | V | V | V | V | V | V | V/I | V | V | |
| 1705 | nsP3 | 372 | D | D | D | D | D | D | D | E | E | E | |
| 1918 | nsP4 | 55 | S | S | S | S | S | S/N | N | N | N | N | |
| 1948 | nsP4 | 85 | R | R | R | R | R | R/G | G | G | G | G | |
| Structural protein | 73 | Capsid | 73 | K | K | K | K | K | K | K | K | K | R |
| 530 | E2 | 205 | G | G | G | G | G | G | G | S | S | S | |
| 546 | E2 | 221 | K | K | K | K | K | K | K | K/R | K | K | |
| 589 | E2 | 264 | V | V | V | V | A | A | A | A | A | A | |
| 1020 | E1 | 211 | K | K | K | K | E | E | E | E | E | E | |
| 1035 | E1 | 226 | A | A/V | A/V | V | A | A | A | A | A | A | |
| 1126 | E1 | 317 | I | I | I | I | I | I/V | V | V | V | V | |
| Dataset | FEL | FUBAR | MEME | |
|---|---|---|---|---|
| ECSA dataset | 171: nsP1-R171Q 488: nsP1-Q488R 1550: nsP3-H217Y 1695: nsP3-A362V 2418: nsP4-V555I 1020: E1-E211K | 171: nsP-R171Q 1550: nsP3-H217Y 1661: nsP3-Q328P 2418: nsP4-V555I 471: E2-Q146R 1020: E1-E211K | 82: nsP1-C82S 171: nsP1-R171Q 178: nsP1-F178V 753: nsP2-T218I 992: nsP2-I457S 1426: nsP3-A93P 1550: nsP3-H217Y 1661: nsP3-Q328P 1768: nsP3-C435R 1849: nsP3-C516L 2325: nsP4-A462G 2330: nsP4-D467Y | 147: C-A147R 156: C-R156A 467: E2-H142Y 471: E2-Q146R 632: E2-Q307R 795: 6K-A47G/V 955: E1-A146N 1020: E1-E211K 1100: E1-V291I 1191: E1-P382F |
| The present study dataset | 1030: nsP2-N495S 1391: nsP3-V58I 1550: nsP3-H217Y 1695: nsP3-A362V 2228: nsP4-T365A 73: C-K73R 546: E2-K221R | 1391: nsP3-V58I 73: C-K73R | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phadungsombat, J.; Imad, H.; Rahman, M.; Nakayama, E.E.; Kludkleeb, S.; Ponam, T.; Rahim, R.; Hasan, A.; Poltep, K.; Yamanaka, A.; et al. A Novel Sub-Lineage of Chikungunya Virus East/Central/South African Genotype Indian Ocean Lineage Caused Sequential Outbreaks in Bangladesh and Thailand. Viruses 2020, 12, 1319. https://doi.org/10.3390/v12111319
Phadungsombat J, Imad H, Rahman M, Nakayama EE, Kludkleeb S, Ponam T, Rahim R, Hasan A, Poltep K, Yamanaka A, et al. A Novel Sub-Lineage of Chikungunya Virus East/Central/South African Genotype Indian Ocean Lineage Caused Sequential Outbreaks in Bangladesh and Thailand. Viruses. 2020; 12(11):1319. https://doi.org/10.3390/v12111319
Chicago/Turabian StylePhadungsombat, Juthamas, Hisham Imad, Mizanur Rahman, Emi E. Nakayama, Sajikapon Kludkleeb, Thitiya Ponam, Rummana Rahim, Abu Hasan, Kanaporn Poltep, Atsushi Yamanaka, and et al. 2020. "A Novel Sub-Lineage of Chikungunya Virus East/Central/South African Genotype Indian Ocean Lineage Caused Sequential Outbreaks in Bangladesh and Thailand" Viruses 12, no. 11: 1319. https://doi.org/10.3390/v12111319
APA StylePhadungsombat, J., Imad, H., Rahman, M., Nakayama, E. E., Kludkleeb, S., Ponam, T., Rahim, R., Hasan, A., Poltep, K., Yamanaka, A., Matsee, W., Piyaphanee, W., Phumratanaprapin, W., & Shioda, T. (2020). A Novel Sub-Lineage of Chikungunya Virus East/Central/South African Genotype Indian Ocean Lineage Caused Sequential Outbreaks in Bangladesh and Thailand. Viruses, 12(11), 1319. https://doi.org/10.3390/v12111319






