Abstract
Infectious laryngotracheitis virus (ILTV) is a herpes virus that causes an acute respiratory disease of poultry known as infectious laryngotracheitis (ILT). Chicken embryo origin (CEO) and tissue culture origin (TCO) live attenuated vaccines are routinely used for the control of ILT. However, vaccine virus is known to revert to virulence, and it has been recently shown that ILT field viral strains can undergo recombination with vaccinal ILTV and such recombinant ILT viruses possess greater transmission and pathogenicity potential. Based on complete or partial genes of the ILTV genome, few studies genotyped ILTV strains circulating in Canada, and so far, information is scarce on whole-genome sequencing or the presence of recombination in Canadian ILTV isolates. The objective of this study was to genetically characterize the 14 ILTV isolates that originated from three provinces in Canada (Alberta, British Columbia and Quebec). To this end, a phylogenetic analysis of 50 ILTV complete genome sequences, including 14 sequences of Canadian origin, was carried out. Additional phylogenetic analysis of the unique long, unique short and inverted repeat regions of the ILTV genome was also performed. We observed that 71%, 21% and 7% of the ILTV isolates were categorized as CEO revertant, wild-type and TCO vaccine-related, respectively. The sequences were also analyzed for potential recombination events, which included evidence in the British Columbia ILTV isolate. This event involved two ILTV vaccine (CEO) strains as parental strains. Recombination analysis also identified that one ILTV isolate from Alberta as a potential parental strain for a United States origin ILTV isolate. The positions of the possible recombination breakpoints were identified. These results indicate that the ILTV wild-type strains can recombine with vaccinal strains complicating vaccine-mediated control of ILT. Further studies on the pathogenicity of these ILTV strains, including the recombinant ILTV isolate are currently ongoing.
1. Introduction
The infectious laryngotracheitis (ILT) was first reported in Canada in 1925 [1]. The causative virus, infectious laryngotracheitis virus (ILTV), is a member of the family Herpesviridae, subfamily alphaherpesvirinae and genus Iltovirus, known as Gallid herpesvirus-1 (GaHV-1). Peafowls and pheasants are able to contract this virus, but chickens are considered the main host [2,3]. ILTV is an enveloped virus with glycoprotein projections on its surface. It contains an icosahedral nucleocapsid with linear double-stranded deoxyribonucleic acid (DNA) genome of an approximate length of 150 kilobase pairs (kbps). The genome is comprised of a Unique Long (UL) region and a Unique Sort (US) region, flanked by two inverted repeat regions, an internal repeat (IR) region and a terminal repeat region [4]. The ILTV genome contains 76 open reading frames (ORFs), from which 63 are homologous to those of the herpes simplex virus (HSV) 1, 6 are Iltovirus genus-specific, and one ORF (UL 0) is only found in the ILTV genome [4].
ILTV is known to have tropism for the tracheal mucosa and conjunctiva, and it causes an acute upper respiratory tract disease. Presentation of clinical signs can go from mild to severe, ranging from nasal and eye secretions to conjunctivitis, reduced weight gain and decrease in egg production to the most characteristic bloody mucus expectoration, gasping and death [1]. ILTV can establish lifelong infections by establishing latency in the trachea [5] and trigeminal ganglia coinciding with the development of the adaptive immune response [6]. Latent ILTV can be periodically reactivated when the affected birds are exposed to stressful episodes, such as extreme weather conditions, nutritional imbalances, transportation, the onset of laying or relocation to a new flock [7].
Control of ILT relies largely on biosecurity measures and vaccination [8]. The commonly used vaccines for ILTV control are recombinant ILT vaccines with either herpesvirus of turkeys (HVT) or fowlpox virus (FPV) as vectors, as well as live attenuated vaccines. Based on the method of attenuation, attenuated vaccines can be categorized into either chicken embryo origin (CEO) or tissue culture origin (TCO) [9]. Attenuated vaccines are usually preferred over recombinant vaccines, given the fact that they provide fast and long-lasting protection and induce stronger cellular immunity [8]. However, vaccine viruses present in attenuated vaccines can still go into a state of latency only to be shed later by the infected birds. Attenuated vaccine virus is also prone to reversion to virulence by multiple passages through bird-to-bird transmission within the flock. This event is reported more often among CEO vaccines given the lower degree of attenuation in comparison to TCO vaccines [10,11,12].
Mutations are very rare in viral DNA genomes [13]; however, recombination among herpesviruses has been well documented and is considered to be a key evolutionary strategy [14]. Recombination can occur more frequently with the use of attenuated vaccines when coinfection with a vaccine and/or wild-type strain takes place. Furthermore, studies have provided evidence confirming recombination between ILTV strains [15,16,17], with recombination breaking points mostly identified within the UL and IR regions of the genome [18,19]. Recombination has been reported mostly among strains related to the CEO vaccines, resulting in new viruses with higher virulence than that of their parental strains [20]. Studies conducted in Australia [21], United States (US) [9], Italy [20], China [22] and Korea [23] have indicated that circulating ILTV strains were genetically related to attenuated CEO vaccine viruses, and in some cases, vaccine recombinants [24].
ILTV infection is endemic in backyard flocks in Canada, and sporadic ILT outbreaks are commonly observed in commercial flocks [25]. CEO and TCO, as well as recombinant vaccines, are currently available in Canada. For more than 10 years, backyard flock owners were urged to vaccinate their birds with TCO vaccines in some of the provinces, and it is possible that backyard flocks vaccinated with live attenuated vaccines could be infected with wild-type virus. Such situations may facilitate co-circulation of two different ILTV strains, which could potentially enable recombination, as has been seen in other countries [19]. Few studies have addressed the molecular nature of ILTV circulating in Canada using either complete or partial genes of the ILTV genome (UL 47, US 8, ORF a and ORF b). These studies have been successful in differentiating between wild-type and vaccine-related ILTV strains and have reported the presence of both CEO vaccine virus and wild-type ILTV in the Canadian chicken flocks [26,27]. However, to this date, there are neither whole-genome sequences of Canadian-origin ILTV strains nor any available information to suggest that ILTV recombination has ever occurred in Canada. The objective of the study was to genetically characterize the ILTV isolates obtained from ILTV clinical cases in Alberta (AB), Quebec (QC) and British Columbia (BC) through whole-genome sequencing (WGS) as well as to provide evidence for recombination events if present in these isolates.
2. Materials and Methods
2.1. Samples
Quantitative polymerase chain reaction (qPCR) assays targeting the ICP4 (primers, F: 5′-GGGTCGGTTCAGTCAGTAA-3′; R: 5′-GGTCATCGACCAAAGACTGT-3′ and probe 5′-GAGGTCGACGGCCAACGC-FL-3′) and glycoprotein (g) G genes (primers, F: 5′ -ATTGCCACCGTTTCCCTAG-3′; R: 5′-CCATTTCACCTCGACTGACACT-3′ and probe, 5’-CAACCGCACCACGATTGAGG-FL-3′) have been used to quantify ILTV genome in nucleic acids of tracheal swabs and tissues (n = 46) originated from backyard flocks of AB. A qPCR assay targeting ILTV thymidine kinase gene (primers, F: 5′- CGA GAA CGA TGA CTC CGA CTT -3′; R: 5′- GGC CCG TCG ACG TAA AGA -3′ and TaqMan probe, 5′-6-FAM-CGC CGC GTT GTA C-MGBNFQ-3′) has been used to detect ILTV genome in clinical samples (n = 9) originated from commercial poultry operations in BC. The qPCR reactions have been carried out using LC FastStart DNA Master Hybridization Probes Kit (Roche Diagnostics, Laval, QC, Canada) or One-Step RT–PCR Master Mix (Life Technologies Inc, Austin, TX, USA) according to manufacturers recommendation and run using LightCycler thermocycler (Roche Diagnostics GmbH, Mannheim, State of Baden-Wurttemberg, Germany) or an Applied Biosystems Inc (ABI) 7500 Fast Real-Time PCR unit (ABI, Foster City, CA, USA). The amplification conditions consisted of initial denaturation of 50 °C for 2 min followed by denaturation of 95 °C for 10 min, then 40 cycles of denaturation of 95 °C for 15 s and annealing at 60 °C for 1 min. A qPCR assay targeting gC gene (primers, F: 5′-CCTTGCGTTTGAATTTTTCTGT-3’; R: 5′-TTCGTGGGTTAGAGGTCTGT-3′ and TaqMan probe, 5′-6-FAM-CAGCTCGGTGACCCCATTCTA-BHQ1-3′) has been used to screen clinical samples (n = 3) originated in QC commercial operations as has been described previously [28]. These ILTV-positive samples (n = 58) were obtained from the Agri Food Laboratories, Alberta Agriculture and Forestry in Edmonton, AB, the Animal Health Center in Abbotsford, BC and the Laboratoire de Diagnostic Moléculaire (LDM), University of Montreal, in St-Hyacinthe, QC, respectively. In order to get enough viral DNA for WGS, the ILTV samples were propagated in either chorioallantoic membrane (CAM) of 9- to 11-day-old specific-pathogen-free (SPF) chicken embryos or monolayers of hepatocellular carcinoma cell lines (LMH) to increase virus titer. Only 14 of the samples were successfully propagated and each sample required between one to three passages before submission for WGS.
2.2. ILTV Propagation on CAM and LMH Cells
The use of embryos was approved by the Health Science Animal Care Committee (HSACC) of the University of Calgary (Protocol number: AC19-0013). The ILTV isolates were propagated on the CAM of SPF eggs on embryo day 10, obtained from the Canadian Food Inspection Agency (CFIA) in Ottawa, Canada.
On viable embryonated eggs, the air cell was located by candling. A small mark was placed in the eggshell 0.5 cm right below the bottom edge of the air cell. Carefully, while holding the previously disinfected egg in a horizontal position, and with the previously mentioned mark looking upwards, a puncture was made in the eggshell above the mark using a 25-gauge needle without introducing the needle too deep to avoid damaging the embryo. Another puncture was made in the shell at the top of the air cell while still maintaining the egg in a horizontal position. Using a rubber bulb covering the orifice made in the air cell, air was taken out of the air cell to create an artificial air cell on the first puncture via negative pressure [29]. The inoculum was then deposited in the artificial air cell and 5 days post-inoculation the CAM is collected from the eggs. CAMs were placed in 2 milliliter (mL) tubes containing sterile zirconium beads (Benchmark Scientific, NJ, USA), 1 mL of Dulbecco’s phosphate-buffered saline (DPBS) with 1% of penicillin–streptomycin (Gibco Life Technologies, Burlington, ON, Canada). Shortly after, the CAMs were homogenized using a microtube homogenizer Bedbug d1030 by Benchmark Scientific. The supernatant was collected, aliquoted and kept at −80 °C for further use.
For propagation of the ILTV isolates on cell monolayers, T-75 tissue culture flasks (Greiner Bio One, Kremsmünster, Austria) precoated with 2% gelatin and containing a confluent monolayer of LMH cells were used. Each flask contained 15 mL of Dulbecco’s Modified Eagle Medium (DMEM) containing 10% fetal bovine serum and 1% penicillin/streptomycin (Gibco Life Technologies, Burlington, ON, Canada). The monolayer of LMH cells was inoculated with the ILTV Canadian isolates and incubated at 37 °C in 5% CO2 for 72 h. After the incubation period, flasks were submitted to three freeze- (−80 °C) and thaw- (37 °C) cycles of 30 min each, after which cells and supernatant were collected and taken for ultracentrifugation on a high-speed centrifuge Avanti J-26 (Beckman Coulter Life Sciences, IN, USA) at 50,000× g for three hours. After ultracentrifugation, the supernatant was discarded, and the cell pellets were resuspended in 500 microliters (µL) of DMEM and aliquoted in small vials and stored at −80 °C until further use.
2.3. Viral DNA Purification from Homogenized LMH Cells and qPCR
DNA purification was carried out using QIAmp DNA Mini Kit (QIAGEN GmbH, Hilden, Mettmann, Germany), according to the manufacturer’s instructions, assessed for purity and quantified using a Nanodrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA).
A quantitative PCR assay was carried out using a CFX96-c1000 Thermocycler (Bio-Rad laboratories, Mississauga, ON, Canada) targeting the proteinase K (PK) gene, as described previously [30,31]. The total volume per reaction was 20 µL, which included 20 nanograms (ng) of genomic DNA, the Fast SYBR Green Master Mix (Invitrogen, Burlington, ON, Canada), 10 µL of SYBR Green, 0.5 µL of forward and reverse specific primers with a final concentration of 10 picomolar (pmol)/ µL targeting the ILTV PK gene (F: 5′-TAC GAT GAA GCG TTC GAC TG -3′ and R: 5′-AGG CGT GAC AGT TCC AAA GT -3′) and DNAse/RNAse-free water (Thermo Scientific, Wilmington, DE, USA). Thermocycler conditions were 95 °C for 20 s for initial denaturation, then 40 cycles of denaturation to 95 °C for 3 s, annealing at 60 °C for 30 s and elongation at 95 °C for 10 s.
2.4. Sample Submission for WGS and Genome Reconstruction
The samples with high ILTV genomic content were directly submitted for WGS (LDM, Faculty of Veterinary Medicine, University of Montreal, QC, Canada) and sequenced using the MiSeq platform (Illumina Corp, San Diego, CA, USA). To remove Illumina adaptors, automatic adaptor trimming was selected on the MiSeq spreadsheet following CLC Genomic Workbench (QIAGEN, Redwood, CA, USA). Right before de novo assembly, another step of trimming was performed with the following settings: minus strand only (3′ end trimming); allow internal and end matches with a minimum internal score of 10 and a minimum end score at 4 and alignment scores cost (Mismatch cost at 2 and Gap cost at 3).
For quality filter, settings were set at Q30 reads. Further, using CLC Genomic Workbench Quality Trimming, with the Trim using quality scores with the limit set at 0.05, the Trim ambiguous nucleotides with a maximum number of ambiguities set at 2 and the discard reads below 15 selected.
For genome reconstruction, all reads were mapped to a complete genome ILTV GenBank reference (Supplementary Table S1) with the Map Reads to Reference tool in CLC Genomic Workbench, with no reference masking selected, match score at 1, mismatch cost at 2, linear gap cost selected, autodetect paired distance selected, and the map randomly selected in the nonspecific match handling box.
The sequence with the most and longest reads mapped against was used as the reference to perform the mapping of all reads with the same setting as described previously. A second mapping was then used to obtain the consensus sequence with the Extract Consensus Sequence tool with the options as following: low coverage definition threshold set at 5, insert N ambiguity symbols selected, in the conflict resolution vote selected, and the use quality score checked.
Consensus sequences with the Extract Consensus Sequence tool with the options were set as follows: Low coverage definition Threshold set at 5, Insert N ambiguity symbols selected, in the Conflict resolution vote selected, and the Use quality score checked. Thus, the threshold was set at 5 (Supplementary Table S1).
2.5. Genotyping Based on Complete ILTV Genome
For phylogenetic analysis, the obtained sequences were aligned with ILTV sequences available in the National Center for Biotechnology Information (NCBI) database. Sequences known to be a product of experimental studies on recombination were not deemed necessary to fulfill the ends of this research and hence, were not included in this study. The sequences included in this analysis mostly comprised of field strains and live attenuated CEO and TCO vaccine sequences from different geographic backgrounds (Supplementary Table S2). Multiple Sequence Alignment was done with Fast Fourier Transform (MAFFT v7.450 [32]) in Geneious® v10.2.6 [33]. Sequences and alignments were visually inspected. Sites where ambiguous nucleotides were present in the AB ILTV sequences were replaced by the consensus sequence only in sites with no nucleotide variability in the consensus. Next, a phylogenetic tree of the complete sequence alignment was generated using a Bayesian inference method using MrBayes 3.2.6 [34] with default settings. It should also be mentioned that phylogenetic analysis using maximum-likelihood resulted in very similar inferences as the Bayesian inference method. Additional phylogenetic trees were constructed using the UL, US and IR regions of the ILTV genome of the 50 sequences to gather key information on the presence of recombination in any of the sequences. Posterior probability values (given as decimals) indicating the support for any given branch are displayed as branches labels in each of the phylogenetic trees.
2.6. Recombination Analysis
Recombination analysis and detection of crossover points in the 50 aligned genome sequences were conducted in the Recombination Detection Program (RDP4 v.4.80) [35] using default settings. Recombination events suggested by RDP4 software were confirmed by using Similarity Plotting (Simplot, 3.5.1, SCRoftware, Baltimore, MD, USA) [36], with a window size of 6000 and a step size of 200.
3. Results
3.1. Flock Background Information
qPCR-confirmed ILT-positive clinical samples originated from AB belonged to small backyard flocks, while the ILT samples originated from QC, and BC belonged to larger commercial flocks. Only two flocks had a history of ILT vaccination: one of the flocks in QC and the other one from AB. All of the relevant flock history and clinicopathological findings are summarized in Table 1 and Supplementary Table S3.

Table 1.
Relevant background information of the poultry farms from which the 14 Canadian infectious laryngotracheitis virus (ILTV) samples that yielded full genome sequences originated. Dash lines fill slots where information could not be obtained.
3.2. ILTV Whole-Genome Sequences
Ten samples from AB, 3 samples from QC and 1 sample from BC yielded enough viral DNA and were directly submitted for WGS (Table 2). The size of the genomes varied between 150,118–153,648 kbps, with the smallest genome belonging to the BC isolate (CAN/BC-10-1122) due to a 3563-nucleotide deletion on the 5′ end of its genome. Interestingly deletion was also present in TCO-like and TCO vaccine sequences. The 14 Canadian ILTV sequences were aligned with 36 ILTV whole-genome sequences representing various geographical areas available in the public domain, which included TCO and CEO ILT vaccine sequences (Supplementary Table S2). The 14 Canadian ILTV sequences were deposited in the GenBank, and their accession numbers are given in Table 2.

Table 2.
Canadian ILTV full genome sequences (n = 14).
3.3. Phylogenetic Analysis
Using the multiple sequence alignment of the 50 full genome sequences, a phylogenetic tree was generated. As shown in Figure 1, the first cluster mostly comprised wild-type ILTV isolates and virulent strains considered to be different from vaccines (genotype VI–IX), 3 of the 14 Canadian ILTV sequences clustered among this group (CAN/AB-S20, CAN/QC-2175807, CAN/AB-S63). The second cluster grouped ILTV isolates with CEO revertant viruses (genotype V), and 10 of the 14 Canadian ILTV sequences clustered with this group. The remaining Canadian ILTV sequence, represented by CAN/BC-10-1122, grouped within the cluster with the ILTV strains related to the TCO vaccines (genotype I, II, III). Additional information is shown in Supplementary Table S4. The last cluster included all the ILTV CEO commercial vaccine strains (Genotype IV), as well as European origin vaccine strain Serva, Chinese strains WG, K317, LJS09, Korean strain 40,798, US strains 63,140 and 3.26.90 and Australian strains CL9 and ACC78. None of the analyzed Canadian ILTV sequences clustered in this group.
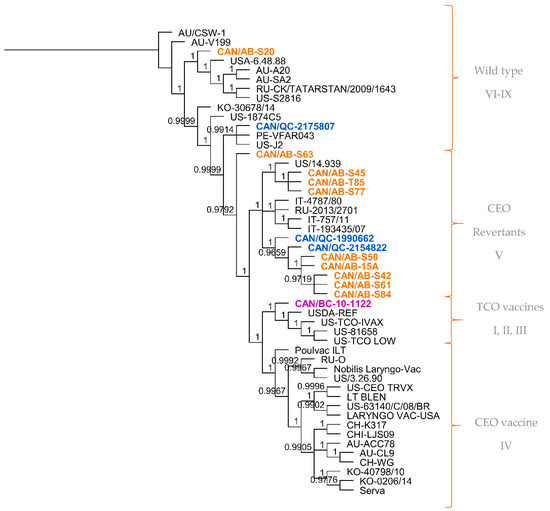
Figure 1.
Phylogenetic tree of the full genome sequences of 50 ILTV strains from different geographical regions. Tree was generated using the Bayesian inference method (MrBayes, Geneious software). Posterior probability values are indicated as branch labels in the tree. Sequences highlighted in orange represent the Alberta ILTV isolates; in blue, the Quebec ILTV isolates and in purple is the ILTV isolate originated in British Columbia. Brackets and labels to the right of the tree separate and indicate the suggested genotype according to the clustering of the sequences in the phylogenetic tree.
Of the 10 Canadian ILTV isolates clustered as CEO revertants following whole-genome sequence analysis, it was interesting to find a high percent nucleotide identity between nine of these sequences (CAN/QC-1990662, CAN/AB-S61, CAN/AB-S50, CAN/AB-S42, CAN/AB-T85, CAN/AB-S45, CAN/AB-S77, CAN/AB-15A, CAN/AB-S84) and three vaccine strains: European Serva, Nobilis Laringovac® (an attenuated ILTV Serva strain) and Poulvac ILT® (uses the ILTV Salisbury strain). A comparative analysis was made between these nine ILTV Canadian sequences and the European Serva. On average, these nine sequences shared 99.9% nucleotide identity with 34 SNPs inside the coding sequence in 36 different ORFs (Table 3), including a codon insertion in ORF C, resulting in a frameshift in the amino acid sequence. Fourteen of these SNPs are synonymous, and 20 non-synonymous. The rest of the Canadian sequences, CAN/QC-2175807, CAN/AB-S63, CAN/BC-10-1122, and CAN/AB-S20, did not appear to be as closely related to the Serva strain or any of the latter mentioned vaccine strains. Additional phylogenetic analysis was conducted using three separate portions of the ILTV genome, the UL, US and IR regions (Figure 2a–c, respectively). Overall, the analyzed Canadian ILTV isolates clustered similarly compared to Figure 1 results, with the exception of isolate CAN/QC-2154822, which clustered with CEO vaccines and a US virulent strain on the IR phylogenetic tree.

Table 3.
Single nucleotide polymorphisms (SNPs) of the nine Canadian ILTV sequences from the Alberta (CAN/AB-S61, CAN/AB-S50, CAN/AB-S42, CAN/AB-T85, CAN/AB-S45, CAN/AB-S77, CAN/AB-15A, CAN/AB-S84) and Quebec (CAN/QC-1990662) provinces using the vaccine strain Serva as a reference sequence. Asterisks (*) represent all nine Canadian sequences. RDR = ribonucleoside-diphosphate reductase, DUTN = deoxyuridine 5′-triphosphate nucleotidohydrolase.
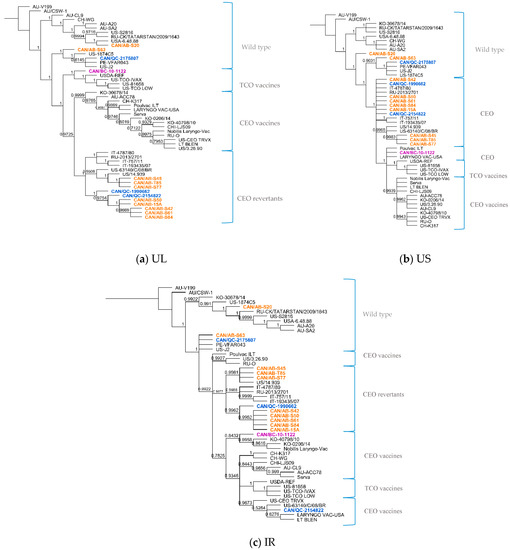
Figure 2.
Phylogenetic analysis of 50 ILTV sequences using the unique long region (a), unique short region (b) and the internal repeat regions (c) using the Bayesian inference method (MrBayes, Geneious software). Sequences highlighted in orange represent the Alberta ILTV isolates; in blue, the Quebec ILTV isolates and in purple is the ILTV isolate originated in British Columbia. Brackets and labels to the right of the trees separate and indicate the suggested genotype according to the clustering of the sequences in the phylogenetic trees. Posterior probability values are indicated as branch labels in the tree.
The position within trees of sequence CAN/QC-2175807 remained constant along with VFAR043, J2 and Canadian CAN/AB-S63. A more detailed analysis of these sequences showed a close relationship among them. Between the first two mentioned strains, only 13 SNPs inside coding sequences differed between them, 7 of them synonymous and 6 non-synonymous. Between CAN/QC-2175807 and J2, only 10 SNPs inside coding sequences differed between them, 5 synonymous and 5 non-synonymous. The same comparison was made among sequences CAN/AB-S63, VFAR043 and J2. Between the first two sequences, 37 SNPs inside of coding sequences could be detected, 19 synonymous and 18 non-synonymous and between CAN/AB-63 and US strain J2, only 34 SNPs, 17 synonymous and 17 non-synonymous in 13 different coding sequences (UL2, US4, UL5, US5, UL6, UL9, UL23, UL25, UL27, UL32, UL38, UL39, UL41). CAN/QC-2154822 ILTV sequence, grouped with CEO vaccine strains (LT BLEN, LARYNGO-VAC and CEO TRVX) and a US strain 63140/C/08/BR in the IR region phylogenetic tree (Figure 2c), this Canadian isolate seemed to have more discrepancies with the rest of the sequences in the alignment. Hundred and forty-four (144) SNPs were identified inside coding sequences when compared to Serva, 21 SNPs synonymous and 123 non-synonymous, including 2 insertions, one in UL27, the other one in ORFC, and a total of 57 deletions, 36 of them concluding on frameshifts in the amino acid sequences in 21 coding regions. However, it was with Serva and Serva-like sequences that it was more closely related to. The Canadian ILTV isolate CAN/BC-10-1122, which clustered among TCO vaccines in the phylogenetic tree following the whole-genome sequence analysis, was found to cluster along with CEO vaccines in the phylogenetic trees of the US and IR regions (Figure 2b,c), showing a nucleotide percent identity of 100% with vaccine Poulvac ILT and Laryngo-vac vaccine sequences in the US multiple sequence alignment, and a 99.8% with Serva and Serva-like vaccine sequences in the IR region.
3.4. Recombination Analysis
Of the 14 Canadian ILTV whole-genome sequences examined for potential recombination events, CAN/BC-10-1122 was found to be a recombinant virus with vaccine strains as possible parental strains (Table 4). The first suggested parent is TCO vaccine LT-IVAX®, which shared an identity of 96.3% with the Canadian BC ILTV sequence. The analysis suggested two minor potential parents (i.e., sequences with a minor contribution to the genome of the suggested recombinant), US-CEO origin vaccine Poulvac ILT®, which shared 97.7% identity with the sequence CAN/BC-10-1122. CEO origin vaccine Nobilis Laryngo-vac®, which shared 97.4% identity with the Canadian isolate. Both CEO vaccine strains were almost identical, with 99.9% identity and only 19 nucleotide differences between them in the coding sequence. As shown in Table 4, the detection of recombination in the CAN/BC-10-1122 isolate using various methods was highly significant with very low p values.

Table 4.
Recombination signals involving Canadian ILTV isolates, recombination analysis carried out on Recombination Detection Program (RDP4) software.
A second recombination event was detected in sequence 6.48.88 of US origin. In this suggested recombination event, sequence CAN/AB-S20 was indicated as a minor parent (sequence with a minor contribution to the genome of the suggested recombinant), and vaccine SA2 was suggested as a potential major parent in this event (Table 4). As shown in Table 4, the detection of recombination in 6.48.88 ILTV isolate using various methods was highly significant with p values < 0.05.
Interestingly, RDP4 software pointed to another recombination event with sequence CAN/QC-2154822 as a major parent for Australian CL9 and minor parent A20. This recombination event had been previously described with Serva as a major parent for the CL9 isolate. After considering the background information on this isolate, belonging to Australia where vaccination with Serva is done with regularity, comparing and doing a visual verification of the alignment including the QC sequence, Serva and the rest of the isolates involved, this event was no longer taken into consideration.
Both recombination events were originally revealed using RDP4 software, confirmed by RDP, GENCONV, MaxChi, Chimaera, SiScan and 3Seq methods [37,38,39,40,41,42]. To further confirm the RDP4 analysis results, a Bootscan analysis using the full genome sequence alignment done with MAFFT of the parental vaccine strains, suggested by RDP4 software, TCO LT-IVAX® and Poulvac ILT® was carried out. In this analysis, the US 1874C5ILTV strain was used as a control and CAN/BC10-1122 as the query sequence (Figure 3). Table 5 summarizes the percentage nucleotide identity of the BC ILTV isolate (BC-10-1122) with CEO Poulvac ILT® and TCO LT-IVAX® in different segments of the ILTV genome (1–15,393; 15,393–37,509; 37,509–113,748; 113,748–end) as indicated by bootscan analysis. It is noteworthy to mention that the same plot was obtained when using Nobilis Laryngo-vac® as a minor parent in the Bootscan analysis.
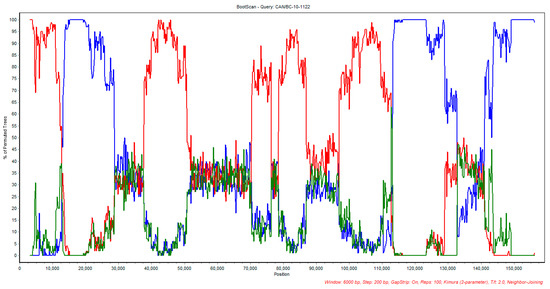
Figure 3.
Bootscan analysis plot of CAN/BC-10-1122 with the suggested parental vaccine strain TCO LT-IVAX® (red) and Poulvac ILT® strain (blue). US 1874C5 strain was used as control (green).

Table 5.
Percentages of nucleotide identity of vaccines CEO-Poulvac ILT® and TCO-LT-IVAX®with British Columbia ILTV isolate (BC-10-1122) indicated by bootscan.
The bootscan analysis also corroborated the recombination event involving CAN/AB-S20 ILTV and Australian SA2 as potential parental strains of US 6.48.88, as suggested by the RDP4 analysis; the results are illustrated in Figure 4. In this analysis, the US-TCO strain was used as a control and US 6.48.88 was used as the query sequence.
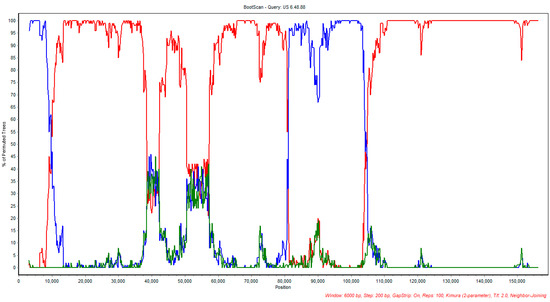
Figure 4.
Bootscan analysis plot of CAN/AB-S20 with the suggested parental vaccine strain CAN/AB-S20 (blue) and SA2 (red). US 1874C5 strain was used as control (green).
4. Discussion
The aim of the current study was to genetically characterize the ILTV isolates linked to ILT-positive cases in Canada using WGS. Among the highlights of this research was finding that most of the obtained Canadian sequences (n = 10) are genetically related to CEO vaccines. Our observation of CEO vaccine-related ILTV circulation in Canada is consistent with observations in other studies [1,21,43], and more recently, coinciding with findings on a study on the molecular characterization of circulating Western Canadian ILTV using Iltovirus specific genes, ORF a and b [26], where 84% ILTV isolates were characterized as genotype V, CEO revertants. It is noteworthy that some of the ILTV isolates belonging to AB and BC used in this study were also included in the previously mentioned study [26]. However, molecular characterization using partial ORF a and b genes did not classify any of the examined Canadian ILTV isolates as TCO vaccine-related. Instead, using partial ORF a and b genes, classified one of the isolates in this study, BC-10-1122 (considered to be TCO vaccine-related using WGS) as genotype IV, with the CEO vaccines, along with other two BC isolates [26].
In 1971, the BC poultry industry used three CEO attenuated vaccines for the vaccination of its flocks against ILT. A few years later, an attenuated vaccine (TCO origin) was also introduced to be used via eyedrop. During 1971–1973, the BC poultry industry encountered an ILT outbreak related to attenuated vaccines that involved 25–50% of the vaccinated flocks [3]. The introduction of live attenuated vaccines probably led to the circulation of the CEO vaccine virus in the province. The current practice of inadequate vaccination with the TCO vaccine (for example, administering the vaccine via drinking water, when it is indicated by the manufacturers for eyedrop administration only) may facilitate TCO vaccine virus reversion to virulence [12] and its circulation among the flocks in BC. The concurrent circulation of vaccine-related ILTV strains and wild-type ILTV probably aided in the recombination event found in the BC ILTV isolate.
In the case of AB, vaccination of commercial flocks is not a common practice, but it is done among backyard flocks. Based on flock history data (Table 1 and Supplementary Table S3), all Alberta-derived ILT samples in this study originated from noncommercial small back yard flocks. These flocks generally comprise different susceptible avian species of varying ages, and new birds are frequently introduced to the flock without adhering to proper biosecurity measures. Contact of these birds with other wild avian species and insects [44], which could be carriers of the disease, is also likely. Along with the transportation of show birds to various exhibitions and competitions, these set of conditions create an ideal situation for ILTV transmission and maintenance since biosecurity in such flocks cannot be maintained effectively [25,45].
It was also clear that Canada is not exempt from ILTV recombination events, as two of the 14 Canadian ILTV isolates (CAN/BC-10-1122 and CAN/AB-S20) appeared to be associated with recombination, and in both cases involving the presence of CEO or TCO live attenuated vaccine viruses, although vaccination with CEO attenuated vaccines is no longer recommended to be used in the Western part of Canada.
Highlighted by the results from the recombination event involving Australian vaccine strain SA2 and Canadian isolate CAN/AB-20, the results of this work are also in agreement with previous work suggesting a US origin for the Australian vaccine strain SA2, which could have later diversified and spread into different geographical areas [15]. This could explain its involvement in the later mentioned recombination event with the Canadian isolate. This view is based on sequence similarity of Canadian ILTV sequence CAN/AB-S20 with the Australian vaccine strains SA2 and A20, US field strains 6.48.88 and S2 816 and Russian strain RU/CK/TATARSTAN/2009/1043 and pointed out by the phylogenetic analysis results [15]. A US origin could also be the case for the Canadian isolate CAN/AB-S20. This event could have been facilitated too by the presence of wild species of migratory birds that could have played an important role in the spread and diversity of the viruses [7].
Similar to other viral families, recombination has been documented among different members of the Herpesviridae family, with many of the recombination events involving the use of attenuated vaccines [46,47,48,49,50]. In the case of ILTV, the attenuated CEO vaccines are usually involved in recombination events with more frequency than the TCO vaccines, which could be influenced by the higher degree of attenuation of the TCO vaccine strains. Recombination between attenuated ILT vaccines (either CEO or TCO) depends on a number of factors including, but not limited to, the quantity and ratio of the vaccines used [17]. However, naturally occurring ILTV recombination events are common, and our results are in agreement with these studies [16,22,23]. However, we have not investigated the consequence of these recombinant field ILTV isolates yet; it is important to know the frequency of these recombination events and whether they lead to increased virulence, as was recorded for other members of the Herpesviridae family such as the varicella-zoster virus (VZV) [50], HSV-1 and HSV-2) [50], and other ILTV recombinants [16,22,23]. Future work will be required to evaluate the pathogenicity and transmission potential of the Canadian recombinant ILTV isolates.
5. Conclusions
Using a WGS approach, we obtained the full genomes of 14 ILTVs of Canadian origin and detected naturally occurring recombination. The CEO vaccine viruses that circulate actively within chicken flocks in Canada added to the inadequate vaccination practices with live attenuated vaccines in some of the provinces, could favor recombination events between vaccine and wild-type viruses, as has been observed in the current study. Our findings add to the knowledge of ILTV evolution and challenges the practical implications of ILT control using live attenuated vaccines.
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4915/12/11/1302/s1, Table S1. Depth variation along the genome of the 14 ILTV Canadian sequences, Table S2. The representative ILTV whole genome sequences (n = 36) that were obtained from public domain and their genome length and Genbank accession numbers for the purpose of aligning with the Canadian ILTV whole genome sequences, Table S3. Clinicopathological history of the ILTV infected chickens where the 14 Canadian ILTV samples that yielded full genome sequences originated, Table S4. Proposed genotype for the 14 Canadian ILTV sequences adressed in this study, and average percent identity with the rest of the sequences belonging in the corresponding groups.
Author Contributions
Conceptualization, M.F.A.-C., T.J., R.K., M.R., D.P., C.A.G., S.C. and D.O.; methodology, M.F.A.-C., T.J., C.P., R.K., M.R., D.P., S.C., D.O., F.v.d.M., C.A.G. and K.F.; software, A.P.C., C.P. and F.v.d.M.; formal analysis, A.P.C., C.P. and F.v.d.M.; investigation, A.P.C., C.B., C.A.G., C.P. and M.F.A.-C.; resources, T.J., R.K., C.P., M.R., D.P., C.A.G., D.O. and M.F.A.-C.; writing—original draft preparation, A.P.C. and M.F.A.-C.; writing—review and editing, A.P.C., T.J., R.K., M.R., D.P., C.A.G., S.C., D.O., M.F.A.-C. and F.v.d.M.; supervision, M.F.A.-C., K.P. and F.v.d.M.; project administration, M.F.A.-C. and S.C.; funding acquisition, M.F.A.-C., T.J., R.K., M.R., D.P., C.A.G., S.C., F.v.d.M. and D.O. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Alberta Agriculture and Forestry (AAF, project number 10020202) and Egg Farmers of Alberta (EFA, Grant Number 10022788).
Acknowledgments
We acknowledge the help of M. Sarjoon Abdul-Cader and Upasama De Silva Senapathi for providing help in processing some of the samples at the University of Calgary. We would like to acknowledge Mohammad Mostafa Nazari Zanjani of the University of Calgary for his insight in the use of bioinformatics tools and Victor Palomino-Tapia for his insight on poultry vaccination practices in the province of BC.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Menendez, K.R.; García, M.; Spatz, S.; Tablante, N.L. Molecular epidemiology of infectious laryngotracheitis: A review. Avian Pathol. 2014, 43, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Ou, S.C.; Giambrone, J.J. Infectious laryngotracheitis virus in chickens. World J. Virol. 2012, 1, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Hayles, L.B.; Macdonald, K.R.; Newby, W.C.; Wood, C.W.; Gilchrist, E.W.; MacNeill, A.C. Epizootiology of infectious laryngotracheitis in British Columbia 1971–1973. Can. Vet. J. 1976, 17, 101–108. [Google Scholar] [PubMed]
- Fuchs, W.; Veits, J.; Helferich, D.; Granzow, H.; Teifke, J.P.; Mettenleiter, T.C. Molecular biology of avian infectious laryngotracheitis virus. Vet. Res. 2007, 38, 261–279. [Google Scholar] [CrossRef]
- Bagust, T. Laryngotracheitis (gallid-1) herpesvirus infection in the chicken 4. Latency establishment by wild and vaccine strains of ILT virus. Avian Pathol. 1986, 15, 581–595. [Google Scholar] [CrossRef]
- García, M.; Volkening, J.; Riblet, S.; Spatz, S. Genomic sequence analysis of the United States infectious laryngotracheitis vaccine strains chicken embryo origin (CEO) and tissue culture origin (TCO). Virology 2013, 440, 64–74. [Google Scholar] [CrossRef]
- Bagust, T.J.; Johnson, M.A. Avian infectious laryngotracheitis: Virus-host interactions in relation to prospects for eradication. Avian Pathol. 1995, 24, 373–391. [Google Scholar] [CrossRef]
- García, M.; Zavala, G. Commercial Vaccines and Vaccination Strategies Against Infectious Laryngotracheitis: What We Have Learned and Knowledge Gaps That Remain. Avian Dis. 2019, 63, 325–334. [Google Scholar] [CrossRef]
- Oldoni, I.; Garcia, M. Characterization of infectious laryngotracheitis virus isolates from the US by polymerase chain reaction and restriction fragment length polymorphism of multiple genome regions. Avian Pathol. 2007, 36, 167–176. [Google Scholar] [CrossRef]
- Groves, P.J.; Williamson, S.L.; Sharpe, S.M.; Gerber, P.F.; Gao, Y.K.; Hirn, T.J.; Walkden-Brown, S. Uptake and spread of infectious laryngotracheitis vaccine virus within meat chicken flocks following drinking water vaccination. Vaccine 2019, 37, 5035–5043. [Google Scholar] [CrossRef]
- Maekawa, D.; Beltrán, G.; Riblet, S.M.; García, M. Protection Efficacy of a Recombinant Herpesvirus of Turkey Vaccine against Infectious Laryngotracheitis Virus Administered In Ovo to Broilers at Three Standardized Doses. Avian Dis. 2019, 63, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Guy, J.S.; Barnes, H.J.; Smith, L. Increased virulence of modified live laryngotracheitis vaccine virus folliwing bird to bird passage. Avian Dis. 1991, 35, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Muylkens, B.; Farnir, F.; Meurens, F.; Schynts, F.; Vanderplasschen, A.; Georges, M.; Thiry, E. Coinfection with Two Closely Related Alphaherpesviruses Results in a Highly Diversified Recombination Mosaic Displaying Negative Genetic Interference. J. Virol. 2009, 83, 3127–3137. [Google Scholar] [CrossRef] [PubMed]
- Loncoman, C.A.; Vaz, P.K.; Coppo, M.J.C.; Hartley, C.A.; Morera, F.J.; Browning, G.F.; Devlin, J.M. Natural recombination in alphaherpesviruses: Insights into viral evolution through full genome sequencing and sequence analysis. Infect. Genet. Evol. 2017, 49, 174–185. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-W.; Devlin, J.M.; Markham, J.; Noormohammadi, A.H.; Browning, G.F.; Ficorilli, N.P.; Hartley, C.A.; Markham, P.F. Phylogenetic and Molecular Epidemiological Studies Reveal Evidence of Multiple Past Recombination Events between Infectious Laryngotracheitis Viruses. PLoS ONE 2013, 8, e55121. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-W.; Markham, P.F.; Coppo, M.J.C.; Legione, A.R.; Markham, J.F.; Noormohammadi, A.H.; Browning, G.F.; Ficorilli, N.; Hartley, C.A.; Devlin, J.M. Attenuated Vaccines Can Recombine to Form Virulent Field Viruses. Science 2012, 337, 188. [Google Scholar] [CrossRef] [PubMed]
- Fakhri, O.; Devlin, J.M.; Browning, G.F.; Vaz, P.K.; Thilakarathne, D.; Lee, S.-W.; Hartley, C.A. Genomic recombination between infectious laryngotracheitis vaccine strains occurs under a broad range of infection conditions in vitro and in ovo. PLoS ONE 2020, 15, e0229082. [Google Scholar] [CrossRef]
- Sanjuán, R.; Domingo, P. Genetic Diversity and Evolution of Viral Populations, in Reference Module in Life Sciences; Elsevier: Valencia, Spain, 2019. [Google Scholar]
- Loncoman, C.A.; Hartley, C.A.; Coppo, M.J.C.; Vaz, P.K.; Diaz-Méndez, A.; Browning, G.F.; García, M.; Spatz, S.; Devlin, J.M. Genetic Diversity of Infectious Laryngotracheitis Virus during In Vivo Coinfection Parallels Viral Replication and Arises from Recombination Hot Spots within the Genome. Appl. Environ. Microbiol. 2017, 83, e01532-17. [Google Scholar] [CrossRef]
- Piccirillo, A.; Lavezzo, E.; Niero, G.; Moreno, A.; Massi, P.; Franchin, E.; Toppo, S.; Salata, C.; Palù, G. Full Genome Sequence-Based Comparative Study of Wild-Type and Vaccine Strains of Infectious Laryngotracheitis Virus from Italy. PLoS ONE 2016, 11, e0149529. [Google Scholar] [CrossRef]
- Blacker, H.P.; Kirkpatrick, N.C.; Rubite, A.; O’Rourke, D.; Noormohammadi, A.H. Epidemiology of recent outbreaks of infectious laryngotracheitis in poultry in Australia. Aust. Vet. J. 2011, 89, 89–94. [Google Scholar] [CrossRef]
- Zhao, Y.; Kong, C.; Wang, Y. Multiple Comparison Analysis of Two New Genomic Sequences of ILTV Strains from China with Other Strains from Different Geographic Regions. PLoS ONE 2015, 10, e0132747. [Google Scholar] [CrossRef] [PubMed]
- La, T.-M.; Choi, E.-J.; Lee, J.-B.; Park, S.-Y.; Song, C.-S.; Choi, I.-S.; Lee, S.-W. Comparative genome analysis of Korean field strains of infectious laryngotracheitis virus. PLoS ONE 2019, 14, e0211158. [Google Scholar] [CrossRef]
- Norberg, P.; Depledge, D.P.; Kundu, S.; Atkinson, C.; Brown, J.; Haque, T.; Hussaini, Y.; MacMahon, E.; Molyneaux, P.; Papaevangelou, V.; et al. Recombination of Globally Circulating Varicella-Zoster Virus. J. Virol. 2015, 89, 7133–7146. [Google Scholar] [CrossRef]
- Morris, S. The Early History of Infectious Laryngotracheitis. Avian Dis. 1996, 40, 494–500. [Google Scholar]
- Barboza-Solis, C.; Contreras, A.P.; Palomino-Tapia, V.A.; Joseph, T.; King, R.; Ravi, M.; Peters, D.; Fonseca, K.; Gagnon, C.A.; Van Der Meer, F.; et al. Genotyping of Infectious Laryngotracheitis Virus (ILTV) Isolates from Western Canadian Provinces of Alberta and British Columbia Based on Partial Open Reading Frame (ORF) a and b. Animals 2020, 10, 1634. [Google Scholar] [CrossRef] [PubMed]
- Ojkic, D.; Swinton, J.; Vallieres, M.; Martin, E.; Shapiro, J.; Sanei, B.; Binnington, B. Characterization of field isolates of infectious laryngotracheitis virus from Ontario. Avian Pathol. 2006, 35, 286–292. [Google Scholar] [CrossRef] [PubMed]
- Callison, S.; Riblet, S.; Oldoni, I.; Sun, S.; Zavala, G.; Williams, S.; Resurreccion, R.; Spackman, E.; García, M. Development and validation of a real-time Taqman® PCR assay for the detection and quantitation of infectious laryngotracheitis virus in poultry. J. Virol. Methods 2007, 139, 31–38. [Google Scholar] [CrossRef]
- Cunningham, C.H. A Laboratory Guide in Virology, 7th ed.; University of Georgia: Athens, GA, USA, 1973. [Google Scholar]
- Thapa, S.; Nagy, E.; Abdul-Careem, M. In ovo delivery of toll-like receptor 2 ligand, lipoteichoic acid induces pro-inflammatory mediators reducing post-hatch infectious laryngotracheitis virus infection. Vet. Immunol. Immunopathol. 2015, 164, 170–178. [Google Scholar] [CrossRef]
- Abdul-Cader, M.S.; Amarasinghe, A.; Palomino-Tapia, V.; Ahmed-Hassan, H.; Bakhtawar, K.; Nagy, E.; Sharif, S.; Gomis, S.; Abdul-Careem, M.F. In ovo CpG DNA delivery increases innate and adaptive immune cells in respiratory, gastrointestinal and immune systems post-hatch correlating with lower infectious laryngotracheitis virus infection. PLoS ONE 2018, 13, e0193964. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Lemey, P.; Lott, M.; Moulton, V.; Posada, D.; Lefeuvre, P. RDP3: A flexible and fast computer program for analyzing recombination. Bioinformatics 2010, 26, 2462–2463. [Google Scholar] [CrossRef] [PubMed]
- Lole, K.S.; Bollinger, R.C.; Paranjape, R.S.; Gadkari, D.; Kulkarni, S.S.; Novak, N.G.; Ingersoll, R.; Sheppard, H.W.; Ray, S.C. Full-Length Human Immunodeficiency Virus Type 1 Genomes from Subtype C-Infected Seroconverters in India, with Evidence of Intersubtype Recombination. J. Virol. 1999, 73, 152–160. [Google Scholar] [CrossRef]
- Martin, D.; Rybicki, E. DP: Detection of recombination amongst aligned sequences. Bioinformatics 2000, 16, 562–563. [Google Scholar] [CrossRef] [PubMed]
- Padidam, M.; Sawyer, S.; Fauquet, C.M. Possible emergence of new geminiviruses by frequent recombination. Virology 1999, 265, 218–225. [Google Scholar] [CrossRef]
- Maynard Smith, J. Analyzing the mosaic structure of genes. J. Mol. Evol. 1992, 34, 126–129. [Google Scholar]
- Posada, D.; Crandall, K.A. Evaluation of methods for detecting recombination from DNA sequences: Computer simulations. Proc. Natl. Acad. Sci. USA 2001, 98, 13757–13762. [Google Scholar] [CrossRef]
- Gibbs, M.J.; Armstrong, J.S.; Gibbs, A.J. Sister-scanning: A Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 2000, 16, 573–582. [Google Scholar] [CrossRef]
- Boni, M.F.; Posada, D.; Feldman, M.W. An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics 2007, 176, 1035–1047. [Google Scholar] [CrossRef]
- Chacón, J.L.; Núñez, L.F.N.; Vejarano, M.P.; Parra, S.H.S.; Astolfi-Ferreira, C.S.; Ferreira, A.J.P. Persistence and spreading of field and vaccine strains of infectious laryngotracheitis virus (ILTV) in vaccinated and unvaccinated geographic regions, in Brazil. Trop. Anim. Health Prod. 2015, 47, 1101–1108. [Google Scholar] [CrossRef]
- Ou, S.-C.; Giambrone, J.J.; Macklin, K.S. Detection of infectious laryngotracheitis virus from darkling beetles and their immature stage (lesser mealworms) by quantitative polymerase chain reaction and virus isolation. J. Appl. Poult. Res. 2012, 21, 33–38. [Google Scholar] [CrossRef]
- Johnson, Y.J.; Colby, M.M.; Tablante, N.L.; Hegngi, F.N.; Salem, M.; Gedamu, M.; Pope, C. Application of commercial and backyard poultry geographic information system databases for the identification of risk factors for clinical Infectious laryngotracheitis in a cluster of cases on Delmarva Peninsula. Int. J. Poult. Sci. 2004, 3, 201–205. [Google Scholar]
- Previdelli, R.L.; Bertzbach, L.D.; Wight, D.J.; Vychodil, T.; You, Y.; Arndt, S.; Kaufer, B.B. The Role of Marek’s Disease Virus UL12 and UL29 in DNA Recombination and the Virus Lifecycle. Viruses 2019, 11, 111. [Google Scholar] [CrossRef]
- He, L.; Li, J.; Peng, P.; Nie, J.; Luo, J.; Cao, Y.; Xue, C. Genomic analysis of a Chinese MDV strain derived from vaccine strain CVI988 through recombination. Infect. Genet. Evol. 2020, 78, 104045. [Google Scholar] [CrossRef]
- Casto, A.M.; Roychoudhury, P.; Xie, H.; Selke, S.; Perchetti, G.A.; Wofford, H.; Huang, M.-L.; Verjans, G.M.G.M.; Gottlieb, G.S.; Wald, A.; et al. Large, Stable, Contemporary Interspecies Recombination Events in Circulating Human Herpes Simplex Viruses. J. Infect. Dis. 2019, 221, 1271–1279. [Google Scholar] [CrossRef]
- Cudini, J.; Roy, S.; Houldcroft, C.J.; Bryant, J.M.; Depledge, D.P.; Tutill, H.; Veys, P.; Williams, R.; Worth, A.J.J.; Tamuri, A.U.; et al. Human cytomegalovirus haplotype reconstruction reveals high diversity due to superinfection and evidence of within-host recombination. Proc. Natl. Acad. Sci. USA 2019, 116, 5693–5698. [Google Scholar] [CrossRef]
- Norberg, P.; Kasubi, M.J.; Haarr, L.; Bergström, T.; Liljeqvist, J.-A. Divergence and Recombination of Clinical Herpes Simplex Virus Type 2 Isolates. J. Virol. 2007, 81, 13158–13167. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).