A Pilot Study Investigating the Dynamics of Pigeon Circovirus Recombination in Domesticated Pigeons Housed in a Single Loft
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Pigeon Lofts and Sample Collection
2.3. Viral DNA Extraction and Recovery of PiCV Genomes
2.4. Bioinformatic Analyses of PiCV Genome Sequences
3. Results and Discussion
3.1. Characterization of PiCV Genotypes
3.2. Phylogenetic Analysis of PiCV Genomes
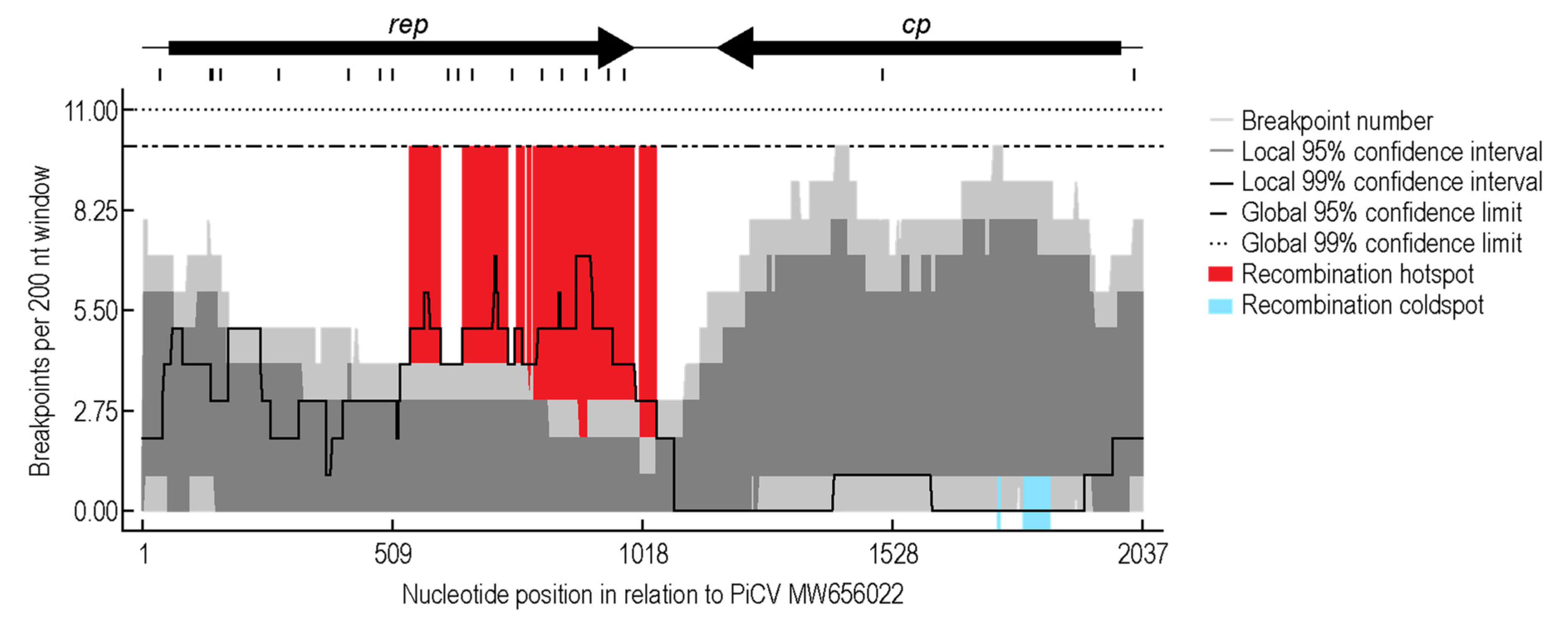
3.3. Recombination Analysis
4. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Haag-Wackernagel, D.; Moch, H. Health hazards posed by feral pigeons. J. Infect. 2004, 48, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Giunchi, D.; Mucci, N.; Bigi, D.; Mengoni, C.; Baldaccini, N.E. Feral pigeon populations: Their gene pool and links with local domestic breeds. Zoology (Jena) 2020, 142, 125817. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, G.; van Grouw, H.; Shapiro, M.D.; Gilbert, M.T.P.; Vieira, F.G. Darwin’s Fancy Revised: An Updated Understanding of the Genomic Constitution of Pigeon Breeds. Genome Biol. Evol. 2020, 12, 136–150. [Google Scholar] [CrossRef] [PubMed]
- Freick, M.; Muller, H.; Raue, R. Rapid detection of pigeon herpesvirus, fowl adenovirus and pigeon circovirus in young racing pigeons by multiplex PCR. J. Virol. Methods 2008, 148, 226–231. [Google Scholar] [CrossRef]
- Huang, Y.L.; Castaneda, O.A.; Thongchan, D.; Khatri-Chhetri, R.; Tsai, S.S.; Wu, H.Y. Pigeon circovirus infection in disqualified racing pigeons from Taiwan. Avian Pathol. 2017, 46, 359–366. [Google Scholar] [CrossRef]
- Omar, M.A.; Hassan, F.A.; Shahin, S.E. Economic Assessment of Various Levels of Protein and Energy in Pigeon Squabs Diet. Zagazig Vet. J. 2017, 45, 20–30. [Google Scholar] [CrossRef][Green Version]
- Proskura, W.S.; Kustosz, J.; Dybus, A.; Lanckriet, R. Polymorphism in dopamine receptor D4 gene is associated with pigeon racing performance. Anim. Genet. 2015, 46, 586–587. [Google Scholar] [CrossRef]
- Shao, Y.; Tian, H.Y.; Zhang, J.J.; Kharrati-Koopaee, H.; Guo, X.; Zhuang, X.L.; Li, M.L.; Nanaie, H.A.; Dehghani Tafti, E.; Shojaei, B.; et al. Genomic and Phenotypic Analyses Reveal Mechanisms Underlying Homing Ability in Pigeon. Mol. Biol. Evol. 2020, 37, 134–148. [Google Scholar] [CrossRef]
- Ye, M.; Xu, M.; Chen, C.; He, Y.; Ding, M.; Ding, X.; Wei, W.; Yang, S.; Zhou, B. Expression analyses of candidate genes related to meat quality traits in squabs from two breeds of meat-type pigeon. J. Anim. Physiol. Anim. Nutr. 2018, 102, 727–735. [Google Scholar] [CrossRef]
- Geraldy, K. Transport of racing pigeons and animal welfare. Dtsch. Tierarztl. Wochenschr. 2007, 114, 114–115. [Google Scholar]
- Kaleta, E.F.; Baldauf, C. Newcastle disease in free-living and pet birds. In Newcastle Disease; Springer: Berlin/Heidelberg, Germany, 1988; pp. 197–246. [Google Scholar]
- Phan, T.G.; Vo, N.P.; Boros, A.; Pankovics, P.; Reuter, G.; Li, O.T.; Wang, C.; Deng, X.; Poon, L.L.; Delwart, E. The viruses of wild pigeon droppings. PLoS ONE 2013, 8, e72787. [Google Scholar] [CrossRef]
- McCowan, C.; Crameri, S.; Kocak, A.; Shan, S.; Fegan, M.; Forshaw, D.; Rubbenstroth, D.; Chen, H.; Holmes, C.; Harper, J.; et al. A novel group A rotavirus associated with acute illness and hepatic necrosis in pigeons (Columba livia), in Australia. PLoS ONE 2018, 13, e0203853. [Google Scholar] [CrossRef]
- Zhang, Z.; Dai, W.; Dai, D. Molecular characterization of pigeon torque teno virus (PTTV) in Jiangsu province. Comput. Biol. Chem. 2017, 69, 10–18. [Google Scholar] [CrossRef]
- Agnihotri, K.; Smith, C.; Oakey, J.; Storie, G. Pigeon adenovirus and pigeon torque teno virus associated with acute multifocal hepatic necrosis in pigeons in Queensland, Australia. Arch. Virol. 2021. [Google Scholar] [CrossRef]
- Teske, L.; Rubbenstroth, D.; Meixner, M.; Liere, K.; Bartels, H.; Rautenschlein, S. Identification of a novel aviadenovirus, designated pigeon adenovirus 2 in domestic pigeons (Columba livia). Virus Res. 2017, 227, 15–22. [Google Scholar] [CrossRef]
- Wan, C.; Chen, C.; Cheng, L.; Shi, S.; Fu, G.; Liu, R.; Chen, H.; Fu, Q.; Huang, Y. Detection of novel adenovirus in sick pigeons. J. Vet. Med. Sci. 2018, 80, 1025–1028. [Google Scholar] [CrossRef]
- Rubbenstroth, D.; Peus, E.; Schramm, E.; Kottmann, D.; Bartels, H.; McCowan, C.; Schulze, C.; Akimkin, V.; Fischer, N.; Wylezich, C.; et al. Identification of a novel clade of group A rotaviruses in fatally diseased domestic pigeons in Europe. Transbound. Emerg. Dis. 2019, 66, 552–561. [Google Scholar] [CrossRef] [PubMed]
- Mankertz, A.; Hattermann, K.; Ehlers, B.; Soike, D. Cloning and sequencing of columbid circovirus (coCV), a new circovirus from pigeons. Arch. Virol. 2000, 145, 2469–2479. [Google Scholar] [CrossRef]
- Todd, D.; Fringuelli, E.; Scott, A.N.; Borghmans, B.J.; Duchatel, J.P.; Shivaprasad, H.L.; Raidal, S.R.; Abadie, J.X.; Franciosini, M.P.; Smyth, J.A. Sequence comparison of pigeon circoviruses. Res. Vet. Sci. 2008, 84, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Todd, D.; Weston, J.H.; Soike, D.; Smyth, J.A. Genome sequence determinations and analyses of novel circoviruses from goose and pigeon. Virology 2001, 286, 354–362. [Google Scholar] [CrossRef]
- Coletti, M.; Franciosini, M.P.; Asdrubali, G.; Passamonti, F. Atrophy of the primary lymphoid organs of meat pigeons in Italy associated with circoviruslike particles in the bursa of Fabricius. Avian Dis. 2000, 44, 454–459. [Google Scholar] [CrossRef]
- Woods, L.W.; Latimer, K.S.; Niagro, F.D.; Riddell, C.; Crowley, A.M.; Anderson, M.L.; Daft, B.M.; Moore, J.D.; Campagnoli, R.P.; Nordhausen, R.W. A retrospective study of circovirus infection in pigeons: Nine cases (1986–1993). J. Vet. Diagn. Investig. 1994, 6, 156–164. [Google Scholar] [CrossRef]
- Stenzel, T.A.; Pestka, D.; Tykalowski, B.; Smialek, M.; Koncicki, A. Epidemiological investigation of selected pigeon viral infections in Poland. Vet. Rec. 2012, 171, 562. [Google Scholar] [CrossRef]
- Ledwoń, A.; Bailey, T.; O’Donovan, D.; Mckeown, S.; Lloyd, C.; Więckowski, T.; Kinne, J.; Silvanose, C.; Szeleszczuk, P.; Wernery, U. Prevalence of circovirus and adenovirus in pigeons in Dubai. Med. Weter. 2011, 67, 752–756. [Google Scholar]
- Loiko, M.R.; Junqueira, D.M.; Varela, A.P.M.; Tochetto, C.; Scheffer, C.M.; Lima, D.A.; Morel, A.P.; Cerva, C.; Paim, W.P.; Mayer, F.Q.; et al. Columbid circoviruses detected in free ranging pigeons from Southern Brazil: Insights on PiCV evolution. Arch. Virol. 2018, 163, 3083–3090. [Google Scholar] [CrossRef]
- Sarker, S.; Das, S.; Ghorashi, S.A.; Forwood, J.K.; Raidal, S.R. Pigeon circoviruses from feral pigeons in Australia demonstrate extensive recombination and genetic admixture with other circoviruses. Avian Pathol. 2019, 48, 512–520. [Google Scholar] [CrossRef]
- Stenzel, T.; Pestka, D. Occurrence and genetic diversity of pigeon circovirus strains in Poland. Acta Vet. Hung. 2014, 62, 274–283. [Google Scholar] [CrossRef]
- Wang, K.C.; Zhuang, Q.Y.; Qiu, Y.; Wang, T.; Chen, J.M. Genome sequence characterization of pigeon circoviruses in China. Virus Res. 2017, 233, 1–7. [Google Scholar] [CrossRef]
- Duchatel, J.P.; Todd, D.; Curry, A.; Smyth, J.A.; Bustin, J.C.; Vindevogel, H. New data on the transmission of pigeon circovirus. Vet. Rec. 2005, 157, 413–415. [Google Scholar] [CrossRef]
- Duchatel, J.P.; Todd, D.; Smyth, J.A.; Bustin, J.C.; Vindevogel, H. Observations on detection, excretion and transmission of pigeon circovirus in adult, young and embryonic pigeons. Avian Pathol. 2006, 35, 30–34. [Google Scholar] [CrossRef]
- Franciosini, M.P.; Fringuelli, E.; Tarhuni, O.; Guelfi, G.; Todd, D.; Casagrande Proietti, P.; Falocci, N.; Asdrubali, G. Development of a polymerase chain reaction-based in vivo method in the diagnosis of subclinical pigeon circovirus infection. Avian Dis. 2005, 49, 340–343. [Google Scholar] [CrossRef]
- Abadie, J.; Nguyen, F.; Groizeleau, C.; Amenna, N.; Fernandez, B.; Guereaud, C.; Guigand, L.; Robart, P.; Lefebvre, B.; Wyers, M. Pigeon circovirus infection: Pathological observations and suggested pathogenesis. Avian Pathol. 2001, 30, 149–158. [Google Scholar] [CrossRef]
- Stenzel, T.; Dziewulska, D.; Tykalowski, B.; Koncicki, A. The Clinical Infection with Pigeon Circovirus (PiCV) Leads to Lymphocyte B Apoptosis But Has No Effect on Lymphocyte T Subpopulation. Pathogens 2020, 9, 632. [Google Scholar] [CrossRef]
- Stenzel, T.; Piasecki, T.; Chrząstek, K.; Julian, L.; Muhire, B.M.; Golden, M.; Martin, D.P.; Varsani, A. Pigeon circoviruses display patterns of recombination, genomic secondary structure and selection similar to those of beak and feather disease viruses. J. Gen. Virol. 2014, 95, 1338–1351. [Google Scholar] [CrossRef] [PubMed]
- Khalifeh, A.; Kraberger, S.; Dziewulska, D.; Stenzel, T.; Varsani, A. Complete Genome Sequence of a Phapecoctavirus Isolated from a Pigeon Cloacal Swab Sample. Microbiol. Resour. Announc. 2021, 10. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Stover, B.C.; Muller, K.F. TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinform. 2010, 11, 7. [Google Scholar] [CrossRef]
- Muhire, B.M.; Varsani, A.; Martin, D.P. SDT: A virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 2014, 9, e108277. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Varsani, A.; Roumagnac, P.; Botha, G.; Maslamoney, S.; Schwab, T.; Kelz, Z.; Kumar, V.; Murrell, B. RDP5: A computer program for analyzing recombination in, and removing signals of recombination from, nucleotide sequence datasets. Virus Evol. 2021, 7, veaa087. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
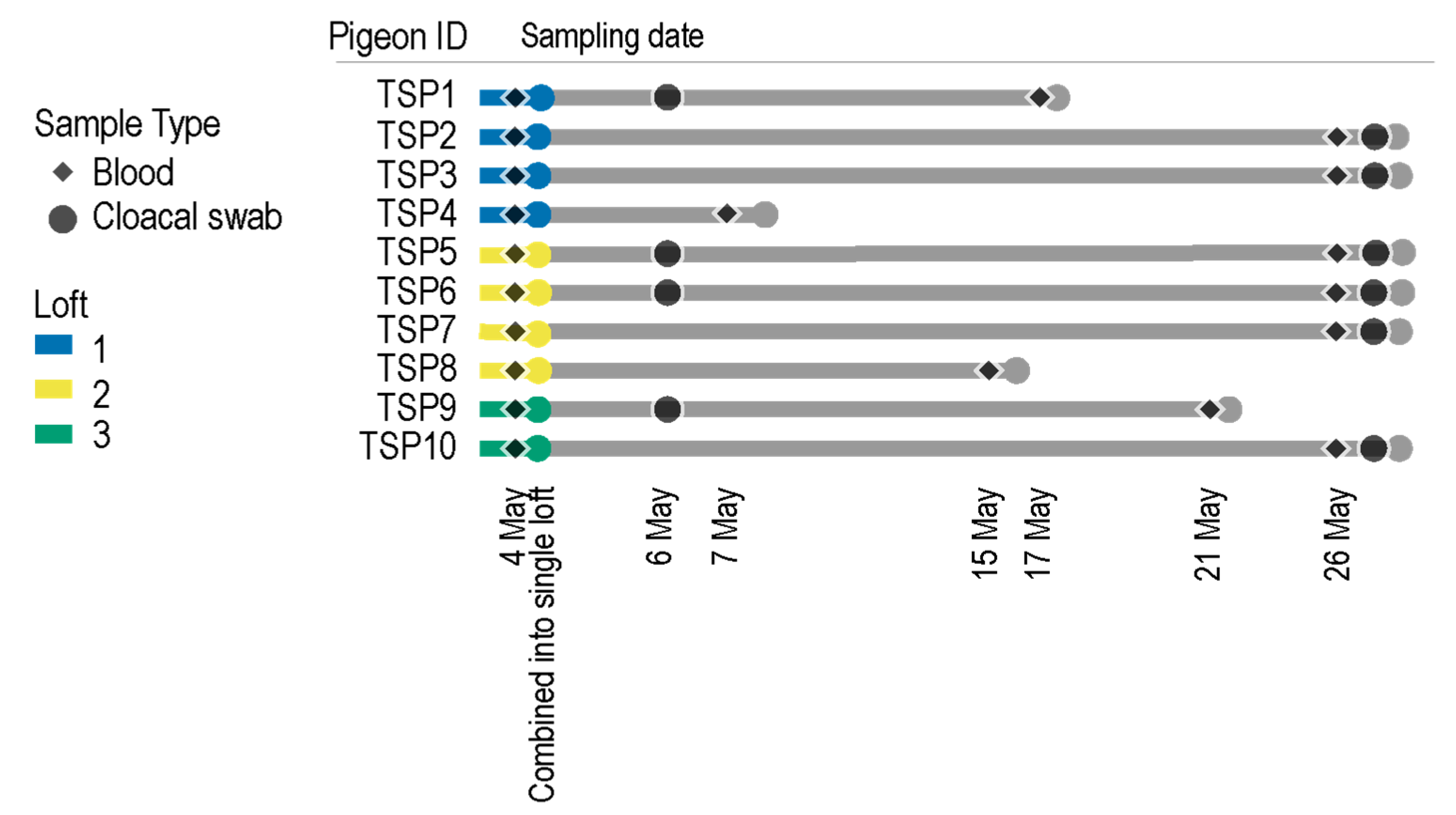
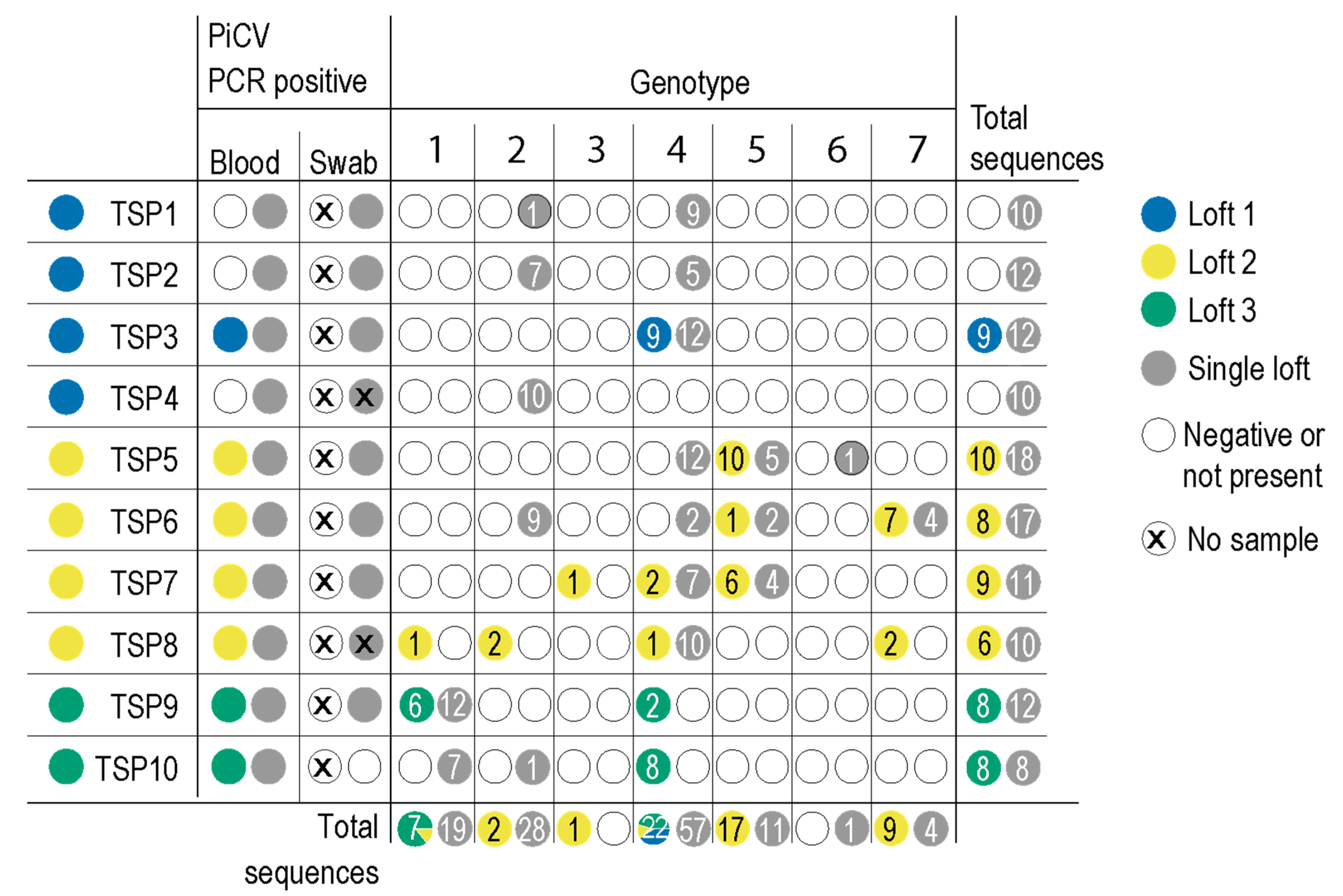
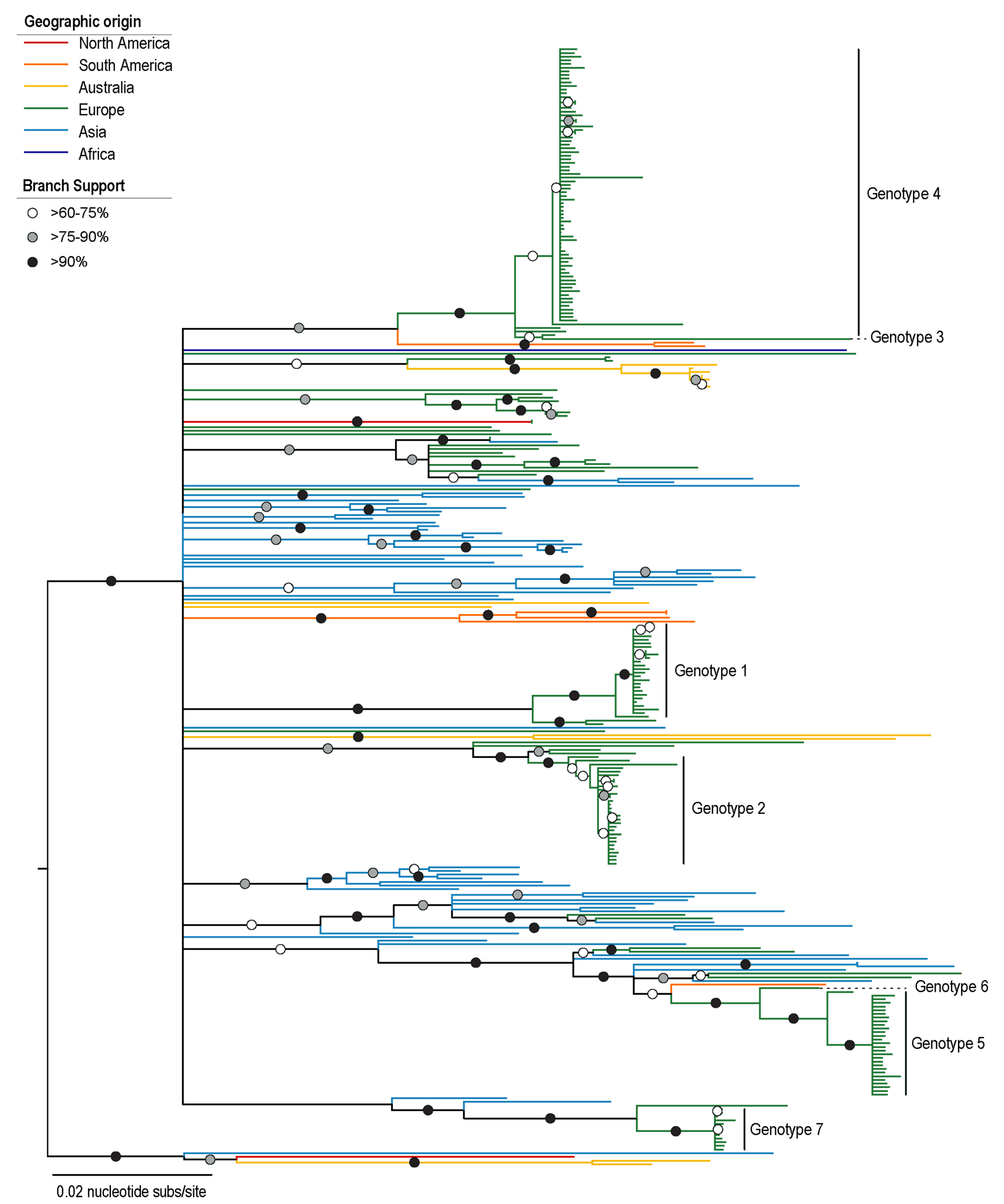
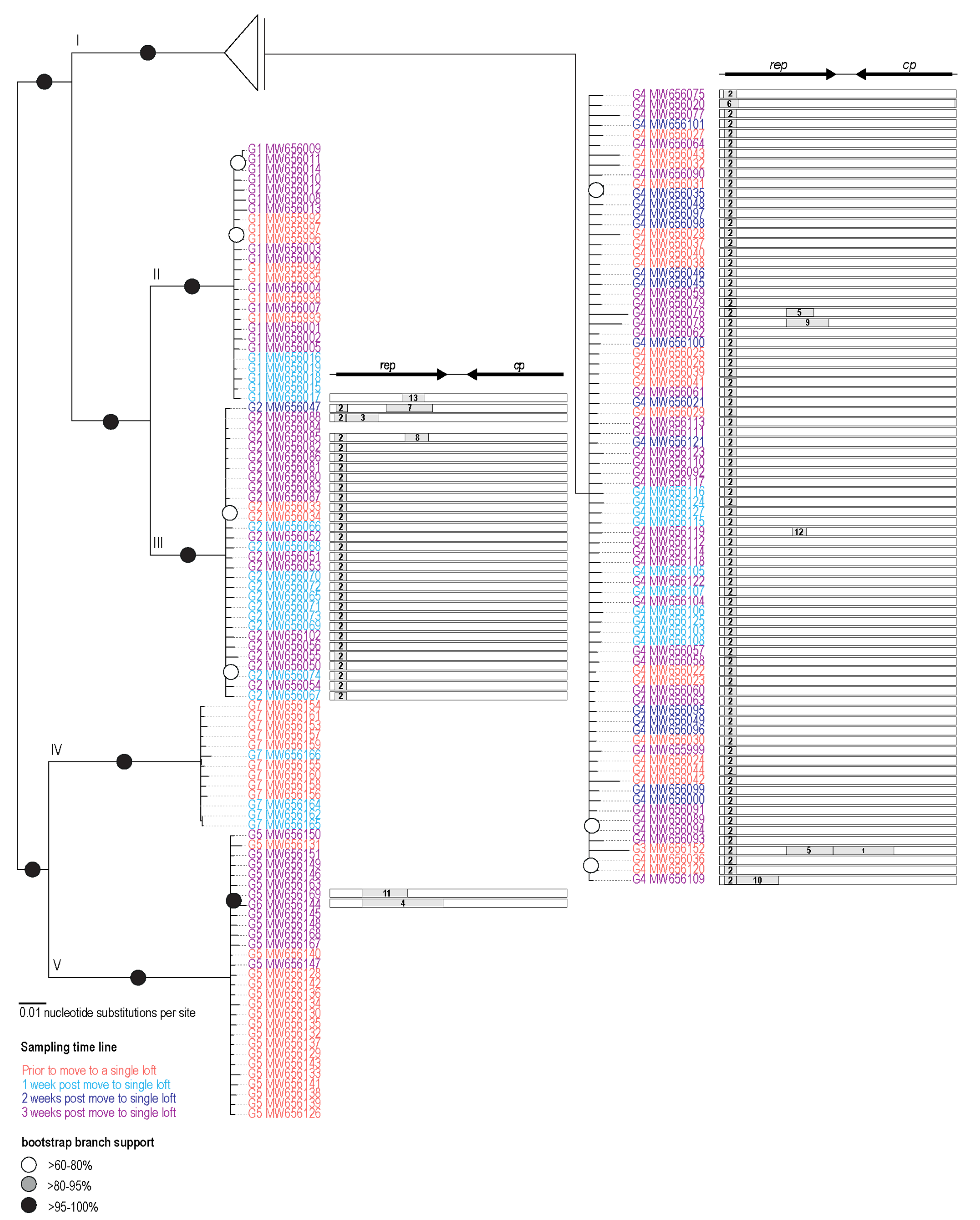
| Pigeon | Date | Blood | Cloacal Swab | Accession #s |
|---|---|---|---|---|
| TSP1 | 4 May 2020 | x | negative | |
| 13 May 2020 | x | MW656045, MW656046, MW656047, MW656048, MW656049 | ||
| 6 May 2020 | x | MW656103, MW656106, MW656107, MW656108, MW656125 | ||
| TSP2 | 4 May 2020 | x | negative | |
| 26 May 2020 | x | MW656050, MW656051, MW656052, MW656053, MW656054, MW656055, MW656056 | ||
| 26 May 2020 | x | MW656109, MW656110, MW656111, MW656104, MW656123 | ||
| TSP3 | 4 May 2020 | x | MW656022, MW656023, MW656024, MW656025, MW656026, MW656027, MW656028, MW656029, MW656030 | |
| 26 May 2020 | x | MW656057, MW656058, MW656059, MW656060, MW656061, MW656062, MW656063, MW656064 | ||
| 26 May 2020 | x | MW656112, MW656113, MW656114, MW656122 | ||
| TSP4 | 4 May 2020 | x | negative | |
| 7 May 2020 | x | MW656065, MW656066, MW656067, MW656068, MW656069, MW656070, MW656071, MW656072, MW656073, MW656074 | ||
| TSP5 | 04 May 2020 | x | MW656128, MW656129, MW656130, MW656131, MW656132, MW656133, MW656134, MW656126, MW656135, MW656136 | |
| 6 May 2020 | x | MW656105, MW656115, MW656116, MW656124, MW656127 | ||
| 26 May 2020 | x | MW655999, MW656079, MW656075, MW656076, MW656077, MW656078, MW656020, MW656144 | ||
| 26 May 2020 | x | MW656145, MW656149, MW656150, MW656151, MW656168 | ||
| TSP6 | 4 May 2020 | x | MW656153, MW656154, MW656159, MW656137, MW656155, MW656156, MW656157, MW656158 | |
| 6 May 2020 | x | MW656162, MW656164, MW656165, MW656166 | ||
| 26 May 2020 | x | MW656080, MW656081, MW656082, MW656083, MW656084, MW656085, MW656086, MW656087, MW656088 | ||
| 26 May 2020 | x | MW656118, MW656119, MW656146, MW656169 | ||
| TSP7 | 4 May 2020 | x | MW656138, MW656143, MW656152, MW656139, MW656140, MW656031, MW656141, MW656120, MW656142 | |
| 26 May 2020 | x | MW656089, MW656090, MW656091, MW656092, MW656093, MW656094 | ||
| 26 May 2020 | x | MW656147, MW656148, MW656117, MW656163, MW656167 | ||
| TSP8 | 4 May 2020 | x | MW655992, MW656032, MW656033, MW656034, MW656160, MW656161, | |
| 15 May 2020 | x | MW656000, MW656021, MW656095, MW656096, MW656097, MW656098, MW656099, MW656100, MW656101, MW656121 | ||
| TSP9 | 4 May 2020 | x | MW655993, MW655994, MW655995, MW655996, MW655997, MW656036, MW655998, MW656035 | |
| 06 May 2020 | x | MW656015, MW656016, MW656017, MW656018, MW656019 | ||
| 21 May 2020 | x | MW656001, MW656002, MW656003, MW656004, MW656005, MW656006, MW656007 | ||
| TSP10 | 4 May 2020 | x | MW656037, MW656038, MW656039, MW656040, MW656041, MW656042, MW656043, MW656044 | |
| 26 May 2020 | x | MW656008, MW656009, MW656010, MW656011, MW656102, MW656012, MW656013, MW656014 |
| Event # | Recombinant Region | Minor Parental Sequence(s) | Major Parental Sequence(s) | Method | p-Value | ||
|---|---|---|---|---|---|---|---|
| Recombinant Sequence(s) | Begin | End | |||||
| 1 | Genotype 3 | 983 | 1512 | Genotype 6 (all) Genotype 5 (MW656131, MW65613, MW656150, MW656169) | Genotype 4 (all) | GBMCST | 8.88 × 10−47 |
| 2 | Genotype 4 (all) Genotype 2 (all) Genotype 3 | 38 | 140 | Genotype 5 (all) | Genotype 1 (all) | GBMCS | 6.04 × 10−20 |
| 3 | Genotype 2 (MW656088) | 142 | 418 | Genotype 7 (all) Genotype 5 (MW656159) | Genotype 2 (expect MW656088) | GMCST | 2.77 × 10−17 |
| 4 | Genotype 6 | 275 | 983 | Genotype 4 (MW656020) | Genotype 5 (all) | GMCST | 4.69 × 10−16 |
| 5 | Genotype 4 (MW656076) Genotype 3 (MW656152)[P] | 577 * | 816 | Genotype 5 (all) | Genotype 4 (MW656020, MW656093, MW656106, MW656109, MW656120) | GMCST | 1.52 × 10−13 |
| 6 | Genotype 4 (MW656020) | 2018 | 158 | Genotype 1 (all) | Genotype 2 (all) | GMCST | 2.46 × 10−13 |
| 6 | Genotype 4 (MW656020) | 2018 | 158 | Genotype 1 (all) | Genotype 2 (all) | GMCST | 2.46 × 10−13 |
| 7 | Genotype 2 (MW656047) | 484 | 904 | Genotype 4 (MW656020) | Genotype 2 (MW656085) | GBMCST | 2.59 × 10−13 |
| 8 | Genotype 2 (MW656085) | 644 | 856 | Genotype 7 (all) | Genotype 2 (except MW656085) | GBMCST | 4.99 × 10−12 |
| 9 | Genotype 4 (MW656078) | 577* | 948 | Genotype 1 (all) | Genotype 4 (except MW656078) | GBMCST | 5.69 × 10−11 |
| 10 | Genotype 4 (MW656109) | 142* | 508 | Genotype 5 (all) | Genotype 4 (MW656093, MW656119, MW656020) Genotype 3 | GBMCST | 8.88 × 10−22 |
| 11 | Genotype 5 (MW656169) | 277 | 676 | Genotype 4 (all) | Genotype 5 (except MW656169) | GBMCST | 3.20 × 10−17 |
| 12 | Genotype 4 (MW656119) | 624 | 752 | Genotype 5 (all) | Genotype 4 (except MW656119) Genotype 2 (all) | GBT | 2.32 × 10−9 |
| 13 | Genotype 1 (MW656017) | 621 | 813 | Genotype 5 (all) | Genotype 1 (except MW656017) | GBT | 1.29 × 10−8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khalifeh, A.; Kraberger, S.; Dziewulska, D.; Varsani, A.; Stenzel, T. A Pilot Study Investigating the Dynamics of Pigeon Circovirus Recombination in Domesticated Pigeons Housed in a Single Loft. Viruses 2021, 13, 964. https://doi.org/10.3390/v13060964
Khalifeh A, Kraberger S, Dziewulska D, Varsani A, Stenzel T. A Pilot Study Investigating the Dynamics of Pigeon Circovirus Recombination in Domesticated Pigeons Housed in a Single Loft. Viruses. 2021; 13(6):964. https://doi.org/10.3390/v13060964
Chicago/Turabian StyleKhalifeh, Anthony, Simona Kraberger, Daria Dziewulska, Arvind Varsani, and Tomasz Stenzel. 2021. "A Pilot Study Investigating the Dynamics of Pigeon Circovirus Recombination in Domesticated Pigeons Housed in a Single Loft" Viruses 13, no. 6: 964. https://doi.org/10.3390/v13060964
APA StyleKhalifeh, A., Kraberger, S., Dziewulska, D., Varsani, A., & Stenzel, T. (2021). A Pilot Study Investigating the Dynamics of Pigeon Circovirus Recombination in Domesticated Pigeons Housed in a Single Loft. Viruses, 13(6), 964. https://doi.org/10.3390/v13060964







