High Rate of Circulating MERS-CoV in Dromedary Camels at Slaughterhouses in Riyadh, 2019
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Rapid MERS-CoV Ag Assay
2.3. Reverse Transcriptase Polymerase Chain Reaction (RT-PCR)
2.4. ELISA
2.5. MERS Pseudotyped Viral Particles (MERSpp) Neutralization Assay
2.6. Sequencing and Phylogenic Analysis
2.7. Ethical Approval
2.8. Statistical Analysis
3. Results
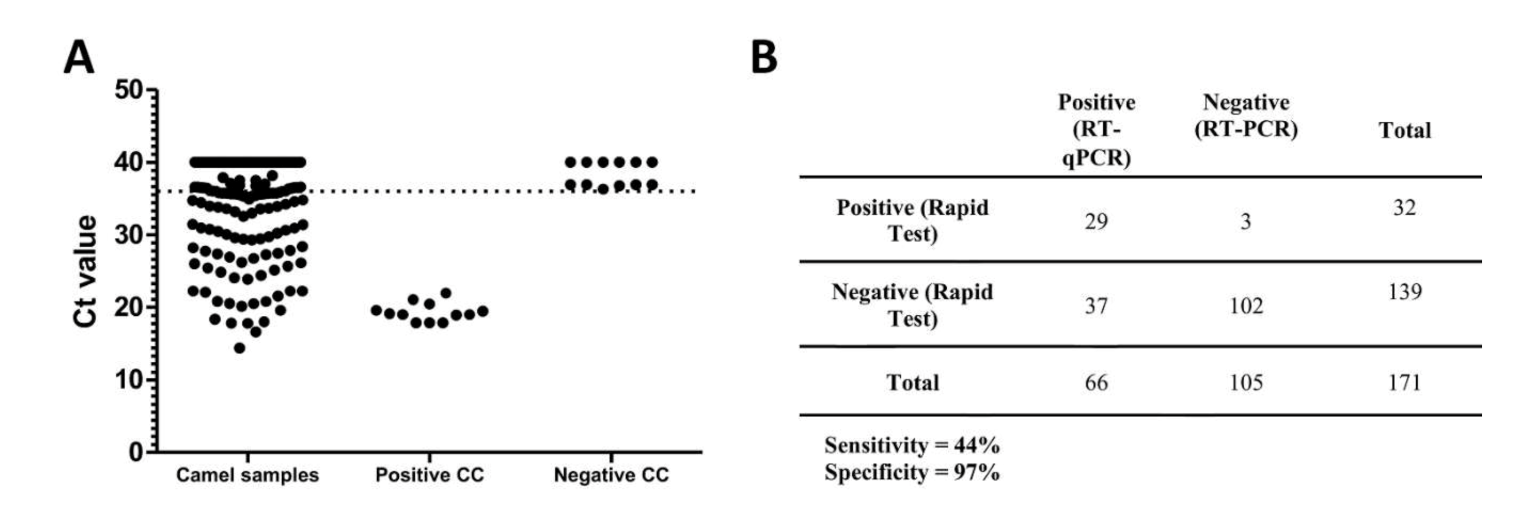
3.1. Detection of MERS-CoV in Slaughterhouse Camels in Riyadh
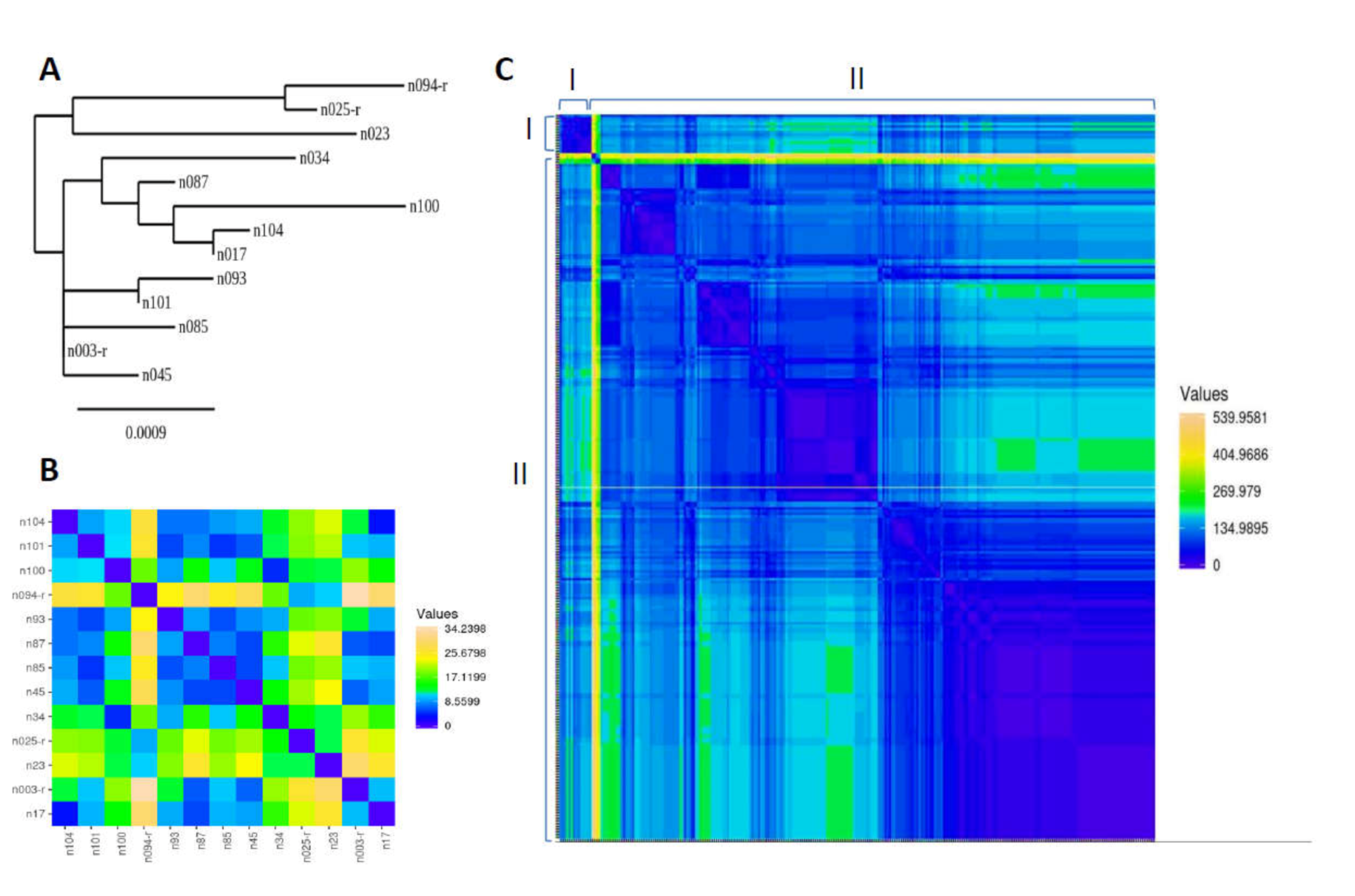
3.2. Genetic Sequence of MERS-CoV Circulating in Slaughterhouse Camels in Riyadh
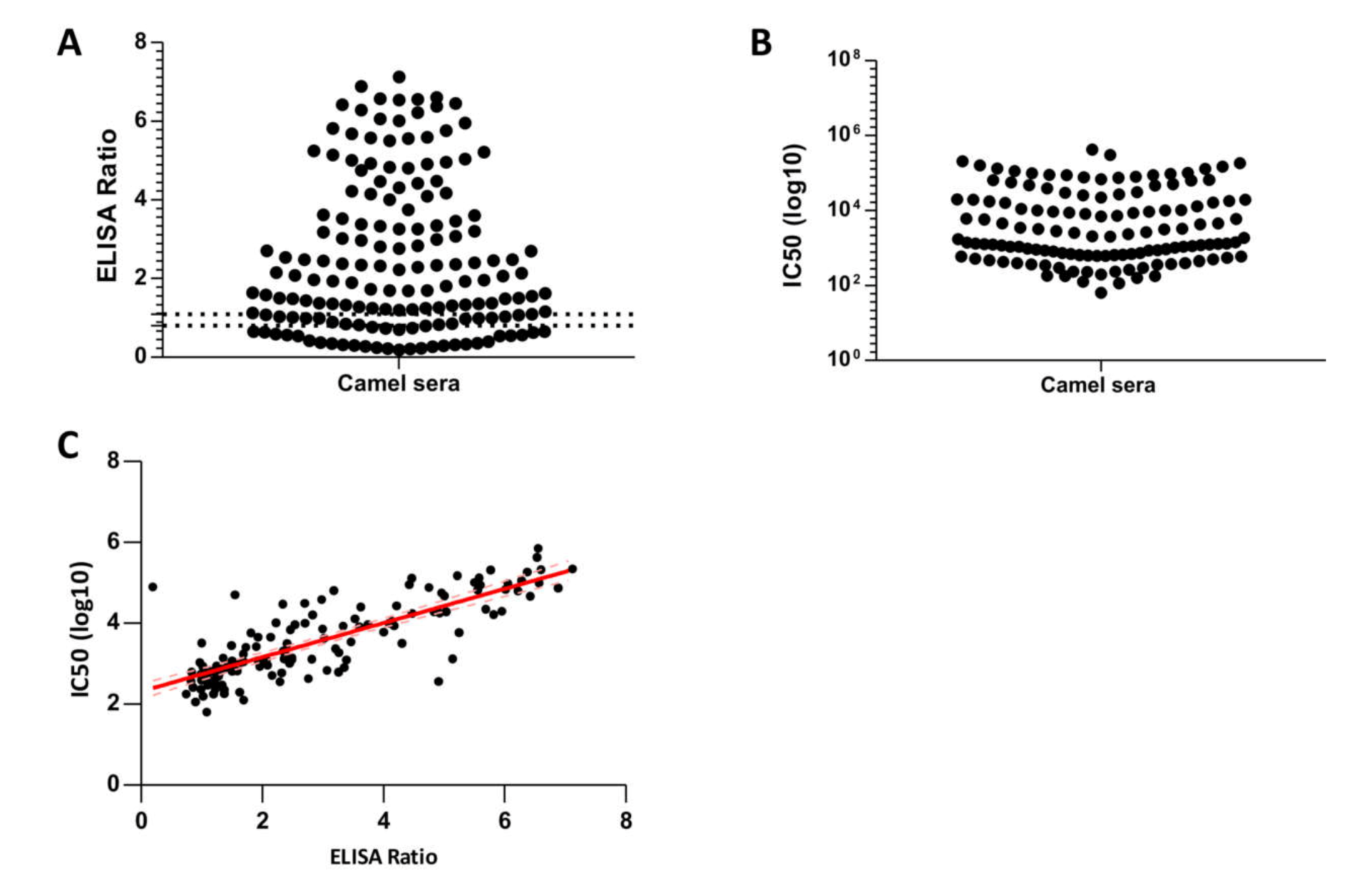
3.3. Seroprevalence of MERS-CoV in Slaughterhouse Camels in Riyadh
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- WHO. WHO Middle EAST Respiratory Syndrome Coronavirus (MERS-CoV); World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Assiri, A.; Al-Tawfiq, J.A.; A Al-Rabeeah, A.; A Al-Rabiah, F.; Al-Hajjar, S.; Al-Barrak, A.; Flemban, H.; Al-Nassir, W.N.; Balkhy, H.H.; Al-Hakeem, R.F.; et al. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: A descriptive study. Lancet Infect. Dis. 2013, 13, 752–761. [Google Scholar] [CrossRef]
- Zaki, A. (Ali); Van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A. (Albert); Fouchier, R. Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia. New Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef] [PubMed]
- Reusken, C.B.; Ababneh, M.; Raj, V.S.; Meyer, B.; Eljarah, A.; Abutarbush, S.; Godeke, G.J.; Bestebroer, T.M.; Zutt, I.; A Müller, M.; et al. Middle East Respiratory Syndrome coronavirus (MERS-CoV) serology in major livestock species in an affected region in Jordan, June to September 2013. Eurosurveillance 2013, 18, 20662. [Google Scholar] [CrossRef] [PubMed]
- Meyer, B.; Müller, M.A.; Corman, V.M.; Reusken, C.B.E.M.; Ritz, D.; Godeke, G.; Erik, L.; Stephan, K.; Artem, S.; van Beek, J.; et al. Antibodies against MERS Coronavirus in Dromedaries, United Arab Emirates, 2003 and 2013. Emerg. Infect Dis. 2014, 20, 552–559. [Google Scholar] [CrossRef]
- Kasem, S.; Qasim, I.; Al-Doweriej, A.; Hashim, O.; Alkarar, A.; Abu-Obeida, A.; Saleh, M.; Al-Hofufi, A.; Al-Ghadier, H.; Hussien, R.; et al. The prevalence of Middle East respiratory Syndrome coronavirus (MERS-CoV) infection in livestock and temporal relation to locations and seasons. J. Infect. Public Health 2018, 11, 884–888. [Google Scholar] [CrossRef]
- Khalafalla, A.I.; Lu, X.; Al-Mubarak, A.I.; Dalab, A.H.S.; Al-Busadah, K.A.; Erdman, D.D. MERS-CoV in Upper Respiratory Tract and Lungs of Dromedary Camels, Saudi Arabia, 2013–2014. Emerg. Infect. Dis. 2015, 21, 1153–1158. [Google Scholar] [CrossRef]
- Farag, E.A.B.A.; Reusken, C.B.; Haagmans, B.L.; Mohran, K.A.; Raj, V.S.; Pas, S.D.; Voermans, J.; Smits, S.L.; Godeke, G.-J.; Al-Hajri, M.M.; et al. High proportion of MERS-CoV shedding dromedaries at slaughterhouse with a potential epidemiological link to human cases, Qatar 2014. Infect. Ecol. Epidemiol. 2015, 5, 28305. [Google Scholar] [CrossRef]
- Wernery, U.; Corman, V.M.; Wong, E.Y.; Tsang, A.K.; Muth, D.; Lau, S.K.P.; Khazanehdari, K.; Zirkel, F.; Ali, M.; Nagy, P.; et al. Acute Middle East Respiratory Syndrome Coronavirus Infection in Livestock Dromedaries, Dubai, 2014. Emerg. Infect. Dis. 2015, 21, 1019–1022. [Google Scholar] [CrossRef]
- Chu, D.K.; Poon, L.L.; Gomaa, M.M.; Shehata, M.M.; Perera, R.A.; Abu Zeid, D.; El Rifay, A.S.; Siu, L.Y.; Guan, Y.; Webby, R.J.; et al. MERS Coronaviruses in Dromedary Camels, Egypt. Emerg. Infect. Dis. 2014, 20, 1049–1053. [Google Scholar] [CrossRef]
- Omrani, A.S.; Al-Tawfiq, J.A.; A Memish, Z. Middle East respiratory syndrome coronavirus (MERS-CoV): Animal to human interaction. Pathog. Glob. Heal. 2015, 109, 354–362. [Google Scholar] [CrossRef]
- Kasem, S.; Qasim, I.; Al-Hufofi, A.; Hashim, O.; Alkarar, A.; Abu-Obeida, A.; Gaafer, A.; Elfadil, A.; Zaki, A.; Al-Romaihi, A.; et al. Cross-sectional study of MERS-CoV-specific RNA and antibodies in animals that have had contact with MERS patients in Saudi Arabia. J. Infect. Public Heal. 2018, 11, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Reusken, C.B.; Haagmans, B.L.; A Müller, M.; Gutierrez, C.; Godeke, G.-J.; Meyer, B.; Muth, D.; Raj, V.S.; Vries, L.S.-D.; Corman, V.M.; et al. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: A comparative serological study. Lancet Infect. Dis. 2013, 13, 859–866. [Google Scholar] [CrossRef]
- Goldman, D.L.; Khine, H.; Abadi, J.; Lindenberg, D.J.; Pirofski, L.; Niang, R. Serologic Evidence for MERS-CoV Infection in Dromedary Camels, Punjab, Pakistan, 2012–2015. Emerg. Infect Dis. 2001, 107, 1–8. [Google Scholar]
- A Mueller, M.; Corman, V.M.; Jores, J.; Meyer, B.; Younan, M.; Liljander, A.; Bosch, B.-J.; Lattwein, E.; Hilali, M.; Musa, B.E.; et al. MERS Coronavirus Neutralizing Antibodies in Camels, Eastern Africa, 1983–1997. Emerg. Infect. Dis. 2014, 20, 2093–2095. [Google Scholar]
- Hemida, M.G.; Perera, R.; A Al Jassim, R.; Kayali, G.; Siu, L.Y.; Wang, P.; Chu, K.W.; Perlman, S.; A Ali, M.; Alnaeem, A.; et al. Seroepidemiology of Middle East respiratory syndrome (MERS) coronavirus in Saudi Arabia (1993) and Australia (2014) and characterisation of assay specificity. Eurosurveillance 2014, 19, 20828. [Google Scholar] [CrossRef] [PubMed]
- Hemida, M.G.; Chu, D.K.W.; Poon, L.L.M.; Perera, R.A.P.M.; Alhammadi, M.A.; Ng, H. MERS Coronavirus in Dromedary Camel Herd, Saudi Arabia. Emerg. Infect. Dis. 2014, 20, 1231–1234. [Google Scholar]
- Hemida, M.G.; Alnaeem, A.; Chu, D.K.; Perera, R.A.; Chan, S.M.; Almathen, F.; Emily, Y.; Brian, C.Y.; Richard, J.; Leo, L.M.P.; et al. Longitudinal study of Middle East Respiratory Syndrome coronavirus infection in dromedary camel herds in Saudi Arabia, 2014-2015. Emerg. Microbes Infect. 2017, 6, e56. [Google Scholar] [CrossRef]
- Killerby, M.E.; Biggs, H.M.; Midgley, C.M.; Gerber, S.I.; Watson, J.T. Middle East Respiratory Syndrome Coronavirus Transmission. Emerg. Infect. Dis. 2020, 26, 191–198. [Google Scholar] [CrossRef]
- Conzade, R.; Grant, R.; Malik, M.R.; Elkholy, A.; Elhakim, M.; Samhouri, D.; Ben Embarek, P.K.; Van Kerkhove, M.D. Reported Direct and Indirect Contact with Dromedary Camels among Laboratory-Confirmed MERS-CoV Cases. Viruses 2018, 10, 425. [Google Scholar] [CrossRef]
- Alshukairi, A.N.; Zheng, J.; Zhao, J.; Nehdi, A.; Baharoon, S.A.; Layqah, L.; Bokhari, A.; Al Johani, S.M.; Samman, N.; Boudjelal, M.; et al. High Prevalence of MERS-CoV Infection in Camel Workers in Saudi Arabia. mBio 2018, 9, 1–10. [Google Scholar] [CrossRef]
- Alharbi, N.K.; Ibrahim, O.H.; Alhafufi, A.; Kasem, S.; Aldowerij, A.; Albrahim, R.; Abu-Obaidah, A.; Alkarar, A.; Bayoumi, F.A.; Almansour, A.M.; et al. Challenge infection model for MERS-CoV based on naturally infected camels. Virol. J. 2020, 17, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Grehan, K.; Ferrara, F.; Temperton, N.J. An optimised method for the production of MERS-CoV spike expressing viral pseudotypes. MethodsX 2015, 2, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Almasaud, A.; Alharbi, N.K.; Hashem, A.M. Generation of MERS-CoV pseudotyped viral particles for the evaluation of neutralizing antibodies in mammalian sera. In Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Alharbi, N.K.; Qasim, I.; Almasoud, A.; Aljami, H.A.; Alenazi, M.W.; Alhafufi, A.; Aldibasi, O.S.; Hashem, A.M.; Kasem, S.; Albrahim, R.; et al. Humoral Immunogenicity and Efficacy of a Single Dose of ChAdOx1 MERS Vaccine Candidate in Dromedary Camels. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef]
- Nowotny, N.; Kolodziejek, J. Middle East respiratory syndrome coronavirus (MERS-CoV) in dromedary camels, Oman 2013. Eurosurveillance 2014, 19, 20781. [Google Scholar] [CrossRef]
- Tolah, A.M.; Al Masaudi, S.B.; El-Kafrawy, S.A.; Mirza, A.A.; Harakeh, S.M.; Hassan, A.M.; Alsaadi, M.A.; Alzahrani, A.A.; Alsaaidi, G.A.; Amor, N.M.S.; et al. Cross-sectional prevalence study of MERS-CoV in local and imported dromedary camels in Saudi Arabia, 2016-2018. PLoS ONE 2020, 15, 2016–2018. [Google Scholar] [CrossRef]
- Sohrab, S.S.; Azhar, E.I. Genetic diversity of MERS-CoV spike protein gene in Saudi Arabia. J. Infect. Public Heal. 2020, 13, 709–717. [Google Scholar] [CrossRef]
- Kim, D.-W.; Kim, Y.-J.; Park, S.H.; Yun, M.-R.; Yang, J.-S.; Kang, H.J.; Han, Y.W.; Lee, H.S.; Kim, H.M.; Kim, H.; et al. Variations in Spike Glycoprotein Gene of MERS-CoV, South Korea, 2015. Emerg. Infect. Dis. 2016, 22, 100–104. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aljasim, T.A.; Almasoud, A.; Aljami, H.A.; Alenazi, M.W.; Alsagaby, S.A.; Alsaleh, A.N.; Alharbi, N.K. High Rate of Circulating MERS-CoV in Dromedary Camels at Slaughterhouses in Riyadh, 2019. Viruses 2020, 12, 1215. https://doi.org/10.3390/v12111215
Aljasim TA, Almasoud A, Aljami HA, Alenazi MW, Alsagaby SA, Alsaleh AN, Alharbi NK. High Rate of Circulating MERS-CoV in Dromedary Camels at Slaughterhouses in Riyadh, 2019. Viruses. 2020; 12(11):1215. https://doi.org/10.3390/v12111215
Chicago/Turabian StyleAljasim, Taibah A., Abdulrahman Almasoud, Haya A. Aljami, Mohamed W. Alenazi, Suliman A. Alsagaby, Asma N. Alsaleh, and Naif Khalaf Alharbi. 2020. "High Rate of Circulating MERS-CoV in Dromedary Camels at Slaughterhouses in Riyadh, 2019" Viruses 12, no. 11: 1215. https://doi.org/10.3390/v12111215
APA StyleAljasim, T. A., Almasoud, A., Aljami, H. A., Alenazi, M. W., Alsagaby, S. A., Alsaleh, A. N., & Alharbi, N. K. (2020). High Rate of Circulating MERS-CoV in Dromedary Camels at Slaughterhouses in Riyadh, 2019. Viruses, 12(11), 1215. https://doi.org/10.3390/v12111215




