Role of MicroRNAs in Bone Pathology during Chikungunya Virus Infection
Abstract
1. Background
2. Role of miRNAs during CHIKV Infection
2.1. Antiviral Role of miRNAs
2.2. Pro-Viral Role of miRNAs
2.3. Aberrant Expression of miRNAs in Mosquito Cells during CHIKV Infection
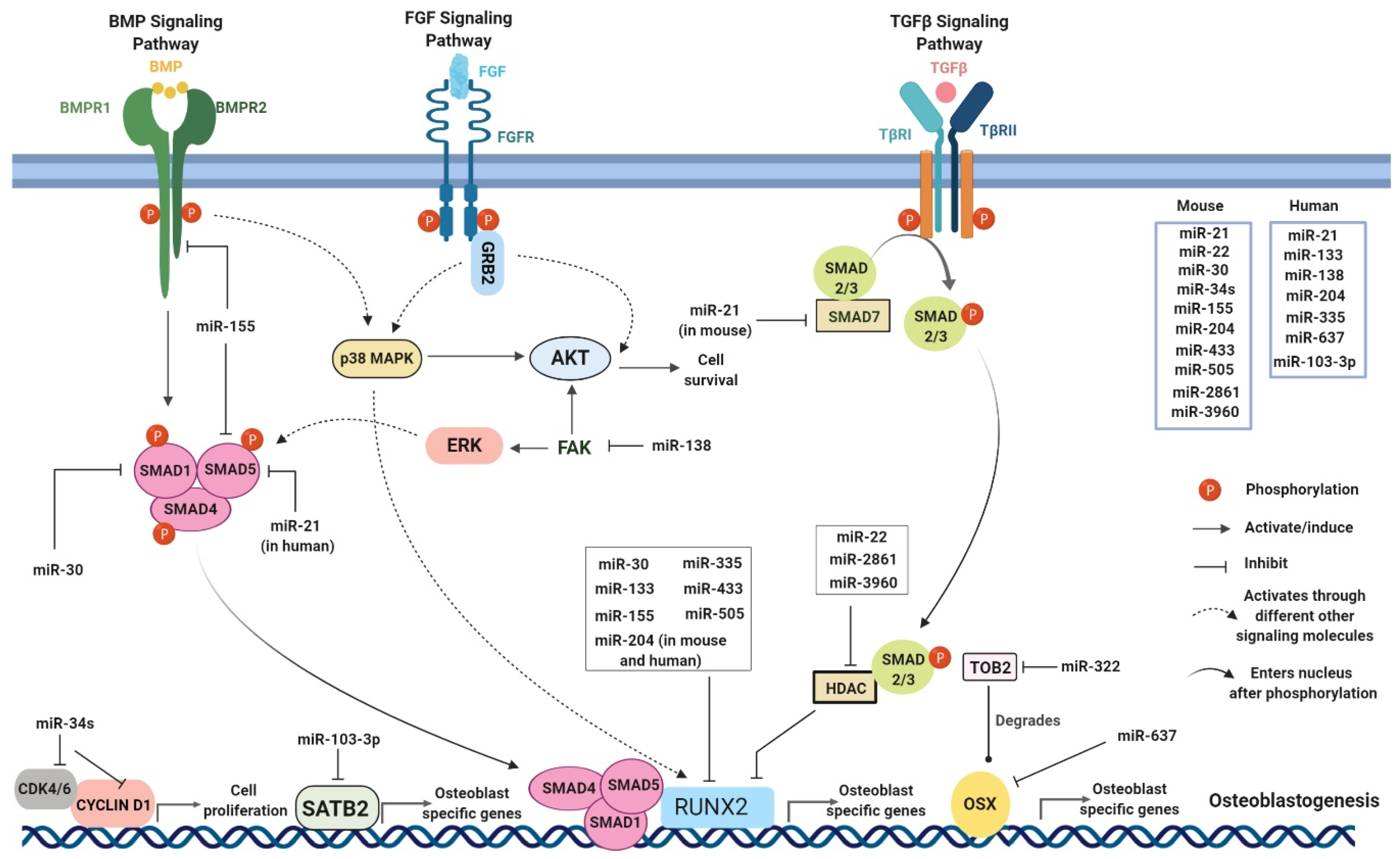
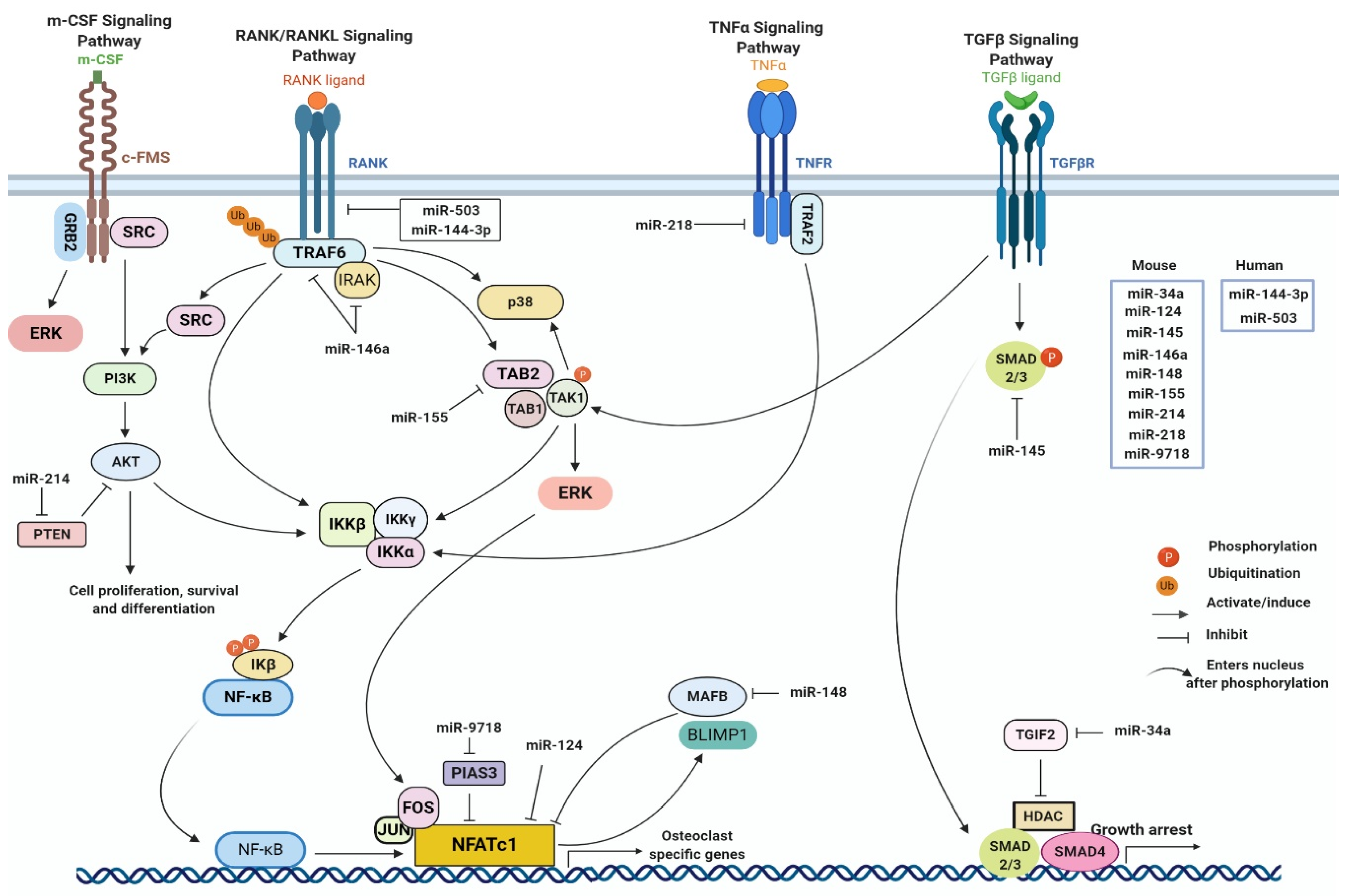
3. Possible Role of miRNAs in Bone Homeostasis in the Context of CHIKV Infection
4. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Morrison, C.R.; Plante, K.S.; Heise, M.T. Chikungunya Virus: Current Perspectives on a Reemerging Virus. Microbiol. Spectr. 2016, 4, 143–161. [Google Scholar]
- Robinson, M.C. An epidemic of virsus disease in Southern Province, Tanganyika Territory, in 1952–53, Clinical Features. Trans. R. Soc. Trop. Med. Hyg. 1955, 49, 28–32. [Google Scholar] [CrossRef]
- Lumsden, W.H.R. An epidemic of virus disease in Southern Province, Tanganyika Territory, in 1952–53. II. General description and epidemiology. Trans. R. Soc. Trop. Med. Hyg. 1955, 49, 33–57. [Google Scholar] [CrossRef]
- Zeller, H.; Van Bortel, W.; Sudre, B. Chikungunya: Its History in Africa and Asia and Its Spread to New Regions in 2013–2014. J. Infect. Dis. 2016, 214, S436–S440. [Google Scholar] [CrossRef]
- Weaver, S.C.; Forrester, N.L. Chikungunya: Evolutionary history and recent epidemic spread. Antivir. Res. 2015, 120, 32–39. [Google Scholar] [CrossRef]
- Petersen, L.R.; Powers, A.M. Chikungunya: Epidemiology. F1000 Res. 2016, 5. [Google Scholar] [CrossRef]
- Cunha, R.V.D.; Trinta, K.S. Chikungunya virus: Clinical aspects and treatment–A Review. Mem. Inst. Oswaldo Cruz. 2017, 112, 523–531. [Google Scholar] [CrossRef]
- Ganesan, V.K.; Duan, B.; Reid, S.P. Chikungunya Virus: Pathophysiology, Mechanism, and Modeling. Viruses 2017, 9, 368. [Google Scholar] [CrossRef]
- Rocha, V.F.D.; de Oliveira, A.H.P.; Bandeira, A.C.; Sardi, S.I.; Garcia, R.F.; Magalhães, S.A.; Sampaio, C.A.; Campos Soares, G. Chikungunya Virus Infection Associated with Encephalitis and Anterior Uveitis. Ocul. Immunol. Inflamm. 2018, 26, 677–679. [Google Scholar] [CrossRef]
- Mehta, R.; Gerardin, P.; Brito CAAd Soares, C.N.; Ferreira, M.L.B.; Solomon, T. The neurological complications of chikungunya virus: A systematic review. Rev. Med. Virol. 2018, 28, e1978. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Lee, I.; Ajay, S.S.; Yook, J.I.; Kim, H.S.; Hong, S.H.; Kim, N.H.; Dhanasekaran, S.M.; Chinnaiyan, A.M.; Athey, B.D. New class of microRNA targets containing simultaneous 5’-UTR and 3’-UTR interaction sites. Genome Res. 2009, 19, 1175–1183. [Google Scholar] [CrossRef]
- Liu, J.; Valencia-Sanchez, M.A.; Hannon, G.J.; Parker, R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005, 7, 719–723. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.H.; Kim, Y.K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Murchison, E.P.; Hannon, G.J. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr. Opin. Cell Biol. 2004, 16, 223–229. [Google Scholar] [CrossRef]
- Lund, E.; Dahlberg, J.E. Substrate Selectivity of Exportin 5 and Dicer in the Biogenesis of MicroRNAs. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 59–66. [Google Scholar] [CrossRef]
- Rana, T.M. Illuminating the silence: Understanding the structure and function of small RNAs. Nat. Rev. Mol. Cell Biol. 2007, 8, 23–36. [Google Scholar] [CrossRef]
- Girardi, E.; Lopez, P.; Pfeffer, S. On the Importance of Host MicroRNAs During Viral Infection. Front. Genet. 2018, 9, 439. [Google Scholar] [CrossRef] [PubMed]
- Bruscella, P.; Bottini, S.; Baudesson, C.; Pawlotsky, J.M.; Feray, C.; Trabucchi, M. Viruses and miRNAs: More Friends than Foes. Front. Microbiol. 2017, 8, 824. [Google Scholar] [CrossRef] [PubMed]
- Ardekani, A.M.; Naeini, M.M. The Role of MicroRNAs in Human Diseases. Avicenna J. Med. Biotechnol. 2010, 2, 161–179. [Google Scholar] [PubMed]
- Selvamani, S.P.; Mishra, R.; Singh, S.K. Chikungunya virus exploits miR-146a to regulate NF-kappaB pathway in human synovial fibroblasts. PLoS ONE 2014, 9, e103624. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, M.; Pandey, N.; Rastogi, M.; Dogra, S.; Singh, S.K. Chikungunya virus modulates the miRNA expression patterns in human synovial fibroblasts. J. Med. Virol. 2020, 92, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Parashar, D.; Paingankar, M.S.; More, A.; Patil, P.; Amdekar, S. Altered microRNA expression signature in Chikungunya-infected mammalian fibroblast cells. Virus Genes. 2018, 54, 502–513. [Google Scholar] [CrossRef] [PubMed]
- López, P.; Girardi, E.; Mounce, B.C.; Weiss, A.; Chane-Woon-Ming, B.; Messmer, M.; Kaukinen, P.; Kopp, A.; Bortolamiol-Becet, D.; Fendri, A.; et al. High-Throughput Fluorescence-Based Screen Identifies the Neuronal MicroRNA miR-124 as a Positive Regulator of Alphavirus Infection. J. Virol. 2020, 94, 2119–2145. [Google Scholar] [CrossRef]
- Saxena, T.; Tandon, B.; Sharma, S.; Chameettachal, S.; Ray, P.; Ray, A.R.; Kulshreshtha, R. Combined miRNA and mRNA signature identifies key molecular players and pathways involved in chikungunya virus infection in human cells. PLoS ONE 2013, 8, e79886. [Google Scholar] [CrossRef]
- Dubey, S.K.; Shrinet, J.; Sunil, S. Aedes aegypti microRNA, miR-2944b-5p interacts with 3’UTR of chikungunya virus and cellular target vps-13 to regulate viral replication. PLoS Negl. Trop Dis. 2019, 13, e0007429. [Google Scholar] [CrossRef]
- Shrinet, J.; Jain, S.; Jain, J.; Bhatnagar, R.K.; Sunil, S. Next generation sequencing reveals regulation of distinct Aedes microRNAs during chikungunya virus development. PLoS Negl. Trop. Dis. 2014, 8, e2616. [Google Scholar] [CrossRef]
- Dubey, S.K.; Shrinet, J.; Jain, J.; Ali, S.; Sunil, S. Aedes aegypti microRNA miR-2b regulates ubiquitin-related modifier to control chikungunya virus replication. Sci. Rep. 2017, 7, 17666. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Balakathiresan, N.S.; Maheshwari, R.K. Chikungunya Virus Infection Alters Expression of MicroRNAs Involved in Cellular Proliferation, Immune Response and Apoptosis. Intervirology 2015, 58, 332–341. [Google Scholar] [CrossRef]
- Nan, Y.; Wu, C.; Zhang, Y.J. Interplay between Janus Kinase/Signal Transducer and Activator of Transcription Signaling Activated by Type I Interferons and Viral Antagonism. Front. Immunol. 2017, 8, 1758. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.S.; Webster, J.A.; Madzokere, E.T.; Stephenson, E.B.; Herrero, L.J. Mosquito antiviral defense mechanisms: A delicate balance between innate immunity and persistent viral infection. Parasit Vectors 2019, 12, 165. [Google Scholar] [CrossRef] [PubMed]
- Fros, J.J.; Liu, W.J.; Prow, N.A.; Geertsema, C.; Ligtenberg, M.; Vanlandingham, D.L.; Schnettler, E.; Vlak, J.M.; Suhrbier, A.; Khromykh, A.A.; et al. Chikungunya virus nonstructural protein 2 inhibits type I/II interferon-stimulated JAK-STAT signaling. J. Virol. 2010, 84, 10877–10887. [Google Scholar] [CrossRef]
- Varghese, F.S.; Thaa, B.; Amrun, S.N.; Simarmata, D.; Rausalu, K.; Nyman, T.A.; Merits, A.; McInerney, G.M.; Ng, L.F.P.; Ahola, T. The Antiviral Alkaloid Berberine Reduces Chikungunya Virus-Induced Mitogen-Activated Protein Kinase Signaling. J. Virol. 2016, 90, 9743–9757. [Google Scholar] [CrossRef]
- Smith, J.L.; Jeng, S.; McWeeney, S.K.; Hirsch, A.J. A MicroRNA Screen Identifies the Wnt Signaling Pathway as a Regulator of the Interferon Response during Flavivirus Infection. J. Virol. 2017, 91, e02388-16. [Google Scholar] [CrossRef]
- Li, G.; Qiu, Z. Deletion of miR-15 Protects Against Rheumatoid Arthritis via Deregulating its Target Gene BCL2L2 and Repressing NF-κB Pathway. Ann. Clin. Lab. Sci. 2019, 49, 581–589. [Google Scholar]
- Bhomia, M.; Sharma, A.; Gayen, M.; Gupta, P.; Maheshwari, R.K. Artificial microRNAs can effectively inhibit replication of Venezuelan equine encephalitis virus. Antivir. Res. 2013, 100, 429–434. [Google Scholar] [CrossRef]
- Gao, Z.; Dou, Y.; Chen, Y.; Zheng, Y. MicroRNA roles in the NF- kappaB signaling pathway during viral infections. Biomed. Res. Int. 2014, 2014, 436097. [Google Scholar] [CrossRef]
- Makkoch, J.; Poomipak, W.; Saengchoowong, S.; Khongnomnan, K.; Praianantathavorn, K.; Jinato, T.; Poovorawan, Y.; Payungporn, S. Human microRNAs profiling in response to influenza A viruses (subtypes pH1N1, H3N2, and H5N1). Exp. Biol. Med. 2016, 241, 409–420. [Google Scholar] [CrossRef]
- Hullinger, T.G.; Montgomery, R.L.; Seto, A.G.; Dickinson, B.A.; Semus, H.M.; Lynch, J.M.; Dalby, C.M.; Robinson, K.; Stack, C.; Latimer, P.A.; et al. Inhibition of miR-15 protects against cardiac ischemic injury. Circ. Res. 2012, 110, 71–81. [Google Scholar] [CrossRef]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef] [PubMed]
- Moran-Moguel, M.C.; Petarra-Del Rio, S.; Mayorquin-Galvan, E.E.; Zavala-Cerna, M.G. Rheumatoid Arthritis and miRNAs: A Critical Review through a Functional View. J. Immunol. Res. 2018, 2018, 2474529. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Chai, J.; Ma, L.; Duan, H.; Zhang, H. Downregulated microRNA-32 expression induced by high glucose inhibits cell cycle progression via PTEN upregulation and Akt inactivation in bone marrow-derived mesenchymal stem cells. Biochem. Biophys. Res. Commun. 2013, 433, 526–531. [Google Scholar] [CrossRef]
- Fabregat, I.; Caballero-Diaz, D. Transforming Growth Factor-beta-Induced Cell Plasticity in Liver Fibrosis and Hepatocarcinogenesis. Front. Oncol. 2018, 8, 357. [Google Scholar] [CrossRef]
- Mirzaei, H.; Faghihloo, E. Viruses as key modulators of the TGF-beta pathway; a double-edged sword involved in cancer. Rev. Med. Virol. 2018, 28, e1967. [Google Scholar] [CrossRef]
- Uhrlaub, J.L.; Pulko, V.; DeFilippis, V.R.; Broeckel, R.; Streblow, D.N.; Coleman, G.D.; Park, B.S.; Lindo, J.F.; Vickers, I.; Anzinger, J.J.; et al. Dysregulated TGF-beta Production Underlies the Age-Related Vulnerability to Chikungunya Virus. PLoS Pathog. 2016, 12, e1005891. [Google Scholar] [CrossRef]
- Nahand, J.S.; Karimzadeh, M.R.; Nezamnia, M.; Fatemipour, M.; Khatami, A.; Jamshidi, S.; Moghoofei, M.; Taghizadieh, M.; Hajighadimi, S.; Shafiee, A.; et al. The role of miR-146a in viral infection. IUBMB Life 2020, 72, 343–360. [Google Scholar] [CrossRef]
- Bartok, B.; Boyle, D.L.; Liu, Y.; Ren, P.; Ball, S.T.; Bugbee, W.D.; Rommel, C.; Firestein, G.S. PI3 kinase delta is a key regulator of synoviocyte function in rheumatoid arthritis. Am. J. Pathol. 2012, 180, 1906–1916. [Google Scholar] [CrossRef]
- Malemud, C.J. The PI3K/Akt/PTEN/mTOR pathway: A fruitful target for inducing cell death in rheumatoid arthritis? Future Med. Chem. 2015, 7, 1137–1147. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.V.; Lee, M.J.; Islam, A.S.; Rohrer, J.L.; Goldberg, V.M.; Beidelschies, M.A.; Greenfield, E.M. Inhibition of the PI3K-Akt signaling pathway reduces tumor necrosis factor-alpha production in response to titanium particles in vitro. J. Bone Joint Surg. Am. 2007, 89, 1019–1027. [Google Scholar] [PubMed]
- Thaa, B.; Biasiotto, R.; Eng, K.; Neuvonen, M.; Götte, B.; Rheinemann, L.; Mutso, M.; Utt, A.; Varghese, F.; Balistreri, G.; et al. Differential Phosphatidylinositol-3-Kinase-Akt-mTOR Activation by Semliki Forest and Chikungunya Viruses Is Dependent on nsP3 and Connected to Replication Complex Internalization. J. Virol. 2015, 89, 11420–11437. [Google Scholar] [CrossRef]
- Rouse, B.T.; Sehrawat, S. Immunity and immunopathology to viruses: What decides the outcome? Nat. Rev. Immunol. 2010, 10, 514–526. [Google Scholar] [CrossRef]
- Yang, R.; Xu, X.; Li, H.; Chen, J.; Xiang, X.; Dong, Z.; Zhang, D. p53 induces miR199a-3p to suppress SOCS7 for STAT3 activation and renal fibrosis in UUO. Sci. Rep. 2017, 7, 43409. [Google Scholar] [CrossRef]
- Kuchipudi, S.V. The Complex Role of STAT3 in Viral Infections. J. Immunol. Res. 2015, 2015, 272359. [Google Scholar] [CrossRef]
- Gao, G.; Luo, H. The ubiquitin-proteasome pathway in viral infections. Can. J. Physiol. Pharmacol. 2006, 84, 5–14. [Google Scholar] [CrossRef]
- Calistri, A.; Munegato, D.; Carli, I.; Parolin, C.; Palu, G. The ubiquitin-conjugating system: Multiple roles in viral replication and infection. Cells 2014, 3, 386–417. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhao, W.; Zhao, K.; Zhang, L.; Gao, C. TRIM26 negatively regulates interferon-beta production and antiviral response through polyubiquitination and degradation of nuclear IRF3. PLoS Pathog. 2015, 11, e1004726. [Google Scholar] [CrossRef]
- Liu, J.; Huang, X.; Hao, S.; Wang, Y.; Liu, M.; Xu, J.; Zhang, X.; Yu, T.; Gan, S.; Dai, D.; et al. Peli1 negatively regulates noncanonical NF-kappaB signaling to restrain systemic lupus erythematosus. Nat. Commun. 2018, 9, 1136. [Google Scholar] [CrossRef] [PubMed]
- Slota, J.A.; Booth, S.A. MicroRNAs in Neuroinflammation: Implications in Disease Pathogenesis, Biomarker Discovery and Therapeutic Applications. Noncoding RNA 2019, 5, 35. [Google Scholar] [CrossRef]
- Li, J.; Song, Q.; Shao, L.; Zhang, L.L.; Guo, X.H.; Mao, Y.J. MiR-124a inhibits proliferation and invasion of rheumatoid arthritis synovial fibroblasts. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 4581–4588. [Google Scholar] [PubMed]
- Tchankouo-Nguetcheu, S.; Bourguet, E.; Lenormand, P.; Rousselle, J.C.; Namane, A.; Choumet, V. Infection by chikungunya virus modulates the expression of several proteins in Aedes aegypti salivary glands. Parasit Vectors 2012, 5, 1–11. [Google Scholar] [CrossRef]
- Reshi, L.; Hong, J. Mitochondria as a Favourite Organelle for Invading Viruses. Mol. Biol. 2017, 6, 1–12. [Google Scholar] [CrossRef]
- Maharaj, P.D.; Widen, S.G.; Huang, J.; Wood, T.G.; Thangamani, S. Discovery of mosquito saliva microRNAs during CHIKV infection. PLoS Negl. Trop. Dis. 2015, 9, e0003386. [Google Scholar] [CrossRef]
- Yen, P.S.; Chen, C.H.; Sreenu, V.; Kohl, A.; Failloux, A.B. Assessing the Potential Interactions between Cellular miRNA and Arboviral Genomic RNA in the Yellow Fever Mosquito, Aedes aegypti. Viruses 2019, 11, 540. [Google Scholar] [CrossRef]
- Goupil, B.A.; McNulty, M.A.; Martin, M.J.; McCracken, M.K.; Christofferson, R.C.; Mores, C.N. Novel Lesions of Bones and Joints Associated with Chikungunya Virus Infection in Two Mouse Models of Disease: New Insights into Disease Pathogenesis. PLoS ONE 2016, 11, e0155243. [Google Scholar] [CrossRef] [PubMed]
- Manimunda, S.P.; Vijayachari, P.; Uppoor, R.; Sugunan, A.P.; Singh, S.S.; Rai, S.K.; Sudeep, A.B.; Muruganandam, N.; Chaitanya, I.K.; Guruprasad, D.R. Clinical progression of chikungunya fever during acute and chronic arthritic stages and the changes in joint morphology as revealed by imaging. Trans. R. Soc. Trop. Med. Hyg. 2010, 104, 392–399. [Google Scholar] [CrossRef]
- Sims, N.A.; Gooi, J.H. Bone remodeling: Multiple cellular interactions required for coupling of bone formation and resorption. Semin. Cell Dev. Biol. 2008, 19, 444–451. [Google Scholar] [CrossRef]
- Friedenstein, A.J.; Petrakova, K.V.; Kurolesova, A.I.; Frolova, G.P. Heterotopic of bone marrow. Analysis of precursor cells for osteogenic and hematopoietic tissues. Transplantation 1968, 6, 230–247. [Google Scholar] [CrossRef]
- Parfitt, A.M. Osteonal and Hemi-Osteonal Remodeling: The Spatial and Temporal Framework for Signal Traffic in Adult Human Bone. J. Cell Biochem. 1994, 55, 273–286. [Google Scholar] [CrossRef]
- James, A.W. Review of Signaling Pathways Governing MSC Osteogenic and Adipogenic Differentiation. Scientifica 2013, 2013, 684736. [Google Scholar] [CrossRef] [PubMed]
- Shahi, M.; Peymani, A.; Sahmani, M. Regulation of bone metabolism. Rep. Biochem. Mol. Biol. 2017, 5, 73–82. [Google Scholar] [PubMed]
- Xu, J.; Li, Z.; Hou, Y.; Fang, W. Potential mechanisms underlying the Runx2 induced osteogenesis of bone marrow mesenchymal stem cells. Am. J. Transl. Res. 2015, 7, 2527–2535. [Google Scholar] [PubMed]
- Wang, C.; Liao, H.; Cao, Z. Role of Osterix and MicroRNAs in Bone Formation and Tooth Development. Med. Sci. Monit. 2016, 22, 2934–2942. [Google Scholar] [CrossRef]
- Komori, T. Regulation of osteoblast differentiation by transcription factors. J. Cell. Biochem. 2006, 99, 1233–1239. [Google Scholar] [CrossRef]
- Almalki, S.G.; Agrawal, D.K. Key transcription factors in the differentiation of mesenchymal stem cells. Differentiation 2016, 92, 41–51. [Google Scholar] [CrossRef]
- Hanna, H.; Mir, L.M.; Andre, F.M. In vitro osteoblastic differentiation of mesenchymal stem cells generates cell layers with distinct properties. Stem Cell Res. Ther. 2018, 9, 203. [Google Scholar] [CrossRef]
- Winslow, M.M.; Pan, M.; Starbuck, M.; Gallo, E.M.; Deng, L.; Karsenty, G.; Crabtree, G.R. Calcineurin/NFAT signaling in osteoblasts regulates bone mass. Dev. Cell. 2006, 10, 771–782. [Google Scholar] [CrossRef]
- Kim, K.; Lee, S.H.; Ha Kim, J.; Choi, Y.; Kim, N. NFATc1 induces osteoclast fusion via up-regulation of Atp6v0d2 and the dendritic cell-specific transmembrane protein (DC-STAMP). Mol. Endocrinol. 2008, 22, 176–185. [Google Scholar] [CrossRef]
- Macián, F.; García-Rodríguez, C.; Rao, A. Gene expression elicited by NFAT in the presence or absence of cooperative recruitment of Fos and Jun. EMBO J. 2000, 19, 4783–4795. [Google Scholar] [CrossRef] [PubMed]
- Takayanagi, H.; Kim, S.; Koga, T.; Nishina, H.; Isshiki, M.; Yoshida, H.; Saiura, A.; Isobe, M.; Yokochi, T.; Inoue, J.; et al. Induction and Activation of the Transcription Factor NFATc1 (NFAT2) Integrate RANKL Signaling in Terminal Differentiation of Osteoclasts. Dev. Cell 2002, 3, 889–901. [Google Scholar] [CrossRef]
- Matsuo, K.; Galson, D.L.; Zhao, C.; Peng, L.; Laplace, C.; Wang, K.Z.; Bachler, M.A.; Amano, H.; Aburatani, H.; Ishikawa, H.; et al. Nuclear factor of activated T-cells (NFAT) rescues osteoclastogenesis in precursors lacking c-Fos. J. Biol. Chem. 2004, 279, 26475–26480. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Kim, J.H.; Lee, J.; Jin, H.M.; Lee, S.H.; Fisher, D.E.; Kook, H.; Kim, K.K.; Choi, Y.; Kim, N. Nuclear factor of activated T cells c1 induces osteoclast-associated receptor gene expression during tumor necrosis factor-related activation-induced cytokine-mediated osteoclastogenesis. J. Biol. Chem. 2005, 280, 35209–35216. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, M.; Kogawa, M.; Wada, S.; Takayanagi, H.; Tsujimoto, M.; Katayama, S.; Hisatake, K.; Nogi, Y. Essential role of p38 mitogen-activated protein kinase in cathepsin K gene expression during osteoclastogenesis through association of NFATc1 and PU.1. J. Biol. Chem. 2004, 279, 45969–45979. [Google Scholar] [CrossRef]
- Yasuda, H.; Shima, N.; Nakagawa, N.; Yamaguchi, K.; Kinosaki, M.; Mochizuki, S.; Tomoyasu, A.; Yano, K.; Goto, M.; Murakami, A.; et al. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc. Natl. Acad. Sci. USA 1998, 95, 3597–3602. [Google Scholar] [CrossRef] [PubMed]
- Lacey, D.L.; Timms, E.; Tan, H.L.; Kelley, M.J.; Dunstan, C.R.; Burgess, T.; Elliott, R.; Colombero, A.; Elliott, G.; Scully, S.; et al. Osteoprotegerin Ligand Is a Cytokine that Regulates Osteoclast Differentiation and Activation. Cell 1998, 93, 165–176. [Google Scholar] [CrossRef]
- Kim, K.; Kim, J.H.; Lee, J.; Jin, H.M.; Kook, H.; Kim, K.K.; Lee, S.Y.; Kim, N. MafB negatively regulates RANKL-mediated osteoclast differentiation. Blood 2007, 109, 3253–3259. [Google Scholar] [CrossRef]
- Miyauchi, Y.; Ninomiya, K.; Miyamoto, H.; Sakamoto, A.; Iwasaki, R.; Hoshi, H.; Miyamoto, K.; Hao, W.; Yoshida, S.; Morioka, H.; et al. The Blimp1-Bcl6 axis is critical to regulate osteoclast differentiation and bone homeostasis. J. Exp. Med. 2010, 207, 751–762. [Google Scholar] [CrossRef]
- Zhao, B.; Takami, M.; Yamada, A.; Wang, X.; Koga, T.; Hu, X.; Tamura, T.; Ozato, K.; Choi, Y.; Ivashkiv, L.B.; et al. Interferon regulatory factor-8 regulates bone metabolism by suppressing osteoclastogenesis. Nat. Med. 2009, 15, 1066–1071. [Google Scholar] [CrossRef]
- Kameda, T.; Mano, H.; Yuasa, T.; Mori, Y.; Miyazawa, K.; Shiokawa, M.; Nakamaru, Y.; Hiroi, E.; Hiura, K.; Kameda, A.; et al. Estrogen Inhibits Bone Resorption by Directly Inducing Apoptosis of the Bone-resorbing Osteoclasts. J. Exp. Med. 1997, 186, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Jing, D.; Hao, J.; Shen, Y.; Tang, G.; Li, M.L.; Huang, S.H.; Zhao, Z.H. The role of microRNAs in bone remodeling. Int. J. Oral Sci. 2015, 7, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Fan, J.; Fan, L.; Li, T.; Yang, Y.; Xu, H.; Deng, L.; Li, J.; Li, T.; Weng, X.; et al. MiRNA-10b Reciprocally Stimulates Osteogenesis and Inhibits Adipogenesis Partly through the TGF-beta/SMAD2 Signaling Pathway. Aging Dis. 2018, 9, 1058–1073. [Google Scholar] [CrossRef]
- Wei, F.; Yang, S.; Guo, Q.; Zhang, X.; Ren, D.; Lv, T.; Xu, X. MicroRNA-21 regulates Osteogenic Differentiation of Periodontal Ligament Stem Cells by targeting Smad5. Sci. Rep. 2017, 7, 16608. [Google Scholar] [CrossRef]
- Li, H.; Yang, F.; Wang, Z.; Fu, Q.; Liang, A. MicroRNA-21 promotes osteogenic differentiation by targeting small mothers against decapentaplegic 7. Mol. Med. Rep. 2015, 12, 1561–1567. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Wang, S.; Bian, C.; Yang, Z.; Zhou, H.; Zeng, Y.; Li, H.; Han, Q.; Zhao, R.C. Upregulation of miR-22 promotes osteogenic differentiation and inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells by repressing HDAC6 protein expression. Stem. Cells Dev. 2012, 21, 2531–2540. [Google Scholar] [CrossRef]
- Kapinas, K.; Kessler, C.B.; Delany, A.M. miR-29 suppression of osteonectin in osteoblasts: Regulation during differentiation and by canonical Wnt signaling. J. Cell Biochem. 2009, 108, 216–224. [Google Scholar] [CrossRef]
- Wu, T.; Zhou, H.; Hong, Y.; Li, J.; Jiang, X.; Huang, H. miR-30 family members negatively regulate osteoblast differentiation. J. Biol. Chem. 2012, 287, 7503–7511. [Google Scholar] [CrossRef]
- Wang, J.; Guan, X.; Guo, F.; Zhou, J.; Chang, A.; Sun, B.; Cai, Y.; Ma, Z.; Dai, C.; Li, X.; et al. miR-30e reciprocally regulates the differentiation of adipocytes and osteoblasts by directly targeting low-density lipoprotein receptor-related protein 6. Cell Death Dis. 2013, 4, e845. [Google Scholar] [CrossRef]
- Chen, L.; Holmstrøm, K.; Qiu, W.; Ditzel, N.; Shi, K.; Hokland, L.; Kassem, M. MicroRNA-34a inhibits osteoblast differentiation and in vivo bone formation of human stromal stem cells. Stem Cells 2014, 32, 902–912. [Google Scholar] [CrossRef]
- Bae, Y.; Yang, T.; Zeng, H.C.; Campeau, P.M.; Chen, Y.; Bertin, T.; Dawson, B.C.; Munivez, E.; Tao, J.; Lee, B.H. miRNA-34c regulates Notch signaling during bone development. Hum. Mol. Genet. 2012, 21, 2991–3000. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Shi, Y.; Zheng, L.; Zhou, B.; Inose, H.; Wang, J.; Guo, X.E.; Grosschedl, R.; Karsenty, G. miR-34s inhibit osteoblast proliferation and differentiation in the mouse by targeting SATB2. J. Cell Biol. 2012, 197, 509–521. [Google Scholar] [CrossRef]
- Shen, H.; Lu, C.; Shi, J.; Li, H.; Si, J.; Shen, G. Satb2 expression in Foxc1-promoted osteogenic differentiation of MC3T3-E1 cells is negatively regulated by microRNA-103–3p. Acta Biochim. Biophys. Sin. 2019, 51, 588–597. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Lu, S.L.; Bai, Y.; Fang, X.; Huang, H.; Zhuang, X.Q. MiR-133a inhibits fracture healing via targeting RUNX2/BMP2. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 2519–2526. [Google Scholar]
- Jia, J.; Tian, Q.; Ling, S.; Liu, Y.; Yang, S.; Shao, Z. miR-145 suppresses osteogenic differentiation by targeting Sp7. FEBS Lett. 2013, 587, 3027–3031. [Google Scholar] [CrossRef]
- Gu, Y.; Ma, L.; Song, L.; Li, X.; Chen, D.; Bai, X. miR-155 Inhibits Mouse Osteoblast Differentiation by Suppressing SMAD5 Expression. Biomed. Res. Int. 2017, 2017, 1893520. [Google Scholar] [CrossRef]
- Liu, H.; Zhong, L.; Yuan, T.; Chen, S.; Zhou, Y.; An, L.; Guo, Y.; Fan, M.; Li, Y.; Sun, Y.; et al. MicroRNA-155 inhibits the osteogenic differentiation of mesenchymal stem cells induced by BMP9 via downregulation of BMP signaling pathway. Int. J. Mol. Med. 2018, 41, 3379–3393. [Google Scholar] [CrossRef]
- Davis, C.; Dukes, A.; Drewry, M.; Helwa, I.; Johnson, M.H.; Isales, C.M.; Hill, W.D.; Liu, Y.; Shi, X.; Fulzele, S.; et al. MicroRNA-183–5p Increases with Age in Bone-Derived Extracellular Vesicles, Suppresses Bone Marrow Stromal (Stem) Cell Proliferation, and Induces Stem Cell Senescence. Tissue Eng. Part A 2017, 23, 1231–1240. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, L.; Xing, L.; Chen, D. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells 2010, 28, 357–364. [Google Scholar] [CrossRef]
- Ito, K.; Tomoki, R.; Ogura, N.; Takahashi, K.; Eda, T.; Yamazaki, F.; Kato, Y.; Goss, A.; Kondoh, T. MicroRNA-204 regulates osteogenic induction in dental follicle cells. J. Dent. Sci. 2020. [Google Scholar] [CrossRef]
- Zheng, X.; Dai, J.; Zhang, H.; Ge, Z. MicroRNA-221 promotes cell proliferation, migration, and differentiation by regulation of ZFPM2 in osteoblasts. Braz. J. Med. Biol. Res. 2018, 51, e7574. [Google Scholar] [CrossRef] [PubMed]
- Gamez, B.; Rodriguez-Carballo, E.; Bartrons, R.; Rosa, J.L.; Ventura, F. MicroRNA-322 (miR-322) and its target protein Tob2 modulate Osterix (Osx) mRNA stability. J. Biol. Chem. 2013, 288, 14264–14275. [Google Scholar] [CrossRef] [PubMed]
- Tomé, M.; López-Romero, P.; Albo, C.; Sepúlveda, J.C.; Fernández-Gutiérrez, B.; Dopazo, A.; Bernad, A.; González, M.A. miR-335 orchestrates cell proliferation, migration and differentiation in human mesenchymal stem cells. Cell Death Differ. 2011, 18, 985–995. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Tu, Q.; Bonewald, L.F.; He, X.; Stein, G.; Lian, J.; Chen, J. Effects of miR-335–5p in modulating osteogenic differentiation by specifically downregulating Wnt antagonist DKK1. J. Bone Miner. Res. 2011, 26, 1953–1963. [Google Scholar] [CrossRef] [PubMed]
- Long, H.; Zhu, Y.; Lin, Z.; Wan, J.; Cheng, L.; Zeng, M.; Tang, Y.; Zhao, R. miR-381 modulates human bone mesenchymal stromal cells (BMSCs) osteogenesis via suppressing Wnt signaling pathway during atrophic nonunion development. Cell Death Dis. 2019, 10, 470. [Google Scholar] [CrossRef]
- Kim, E.J.; Kang, I.H.; Lee, J.W.; Jang, W.G.; Koh, J.T. MiR-433 mediates ERRgamma-suppressed osteoblast differentiation via direct targeting to Runx2 mRNA in C3H10T1/2 cells. Life Sci. 2013, 92, 562–568. [Google Scholar] [CrossRef]
- Zhu, L.; Lin, Z.W.; Wang, G.; Zhang, H.; Liu, B.; Xu, Q.J. MicroRNA-495 downregulates AQP1 and facilitates proliferation and differentiation of osteoblasts in mice with tibial fracture through activation of p38 MAPK signaling pathway. Sci. Rep. 2019, 9, 16171. [Google Scholar] [CrossRef]
- Li, W.; Chen, Z.; Cai, C.; Li, G.; Wang, X.; Shi, Z. MicroRNA-505 is involved in the regulation of osteogenic differentiation of MC3T3-E1 cells partially by targeting RUNX2. J. Orthop. Surg. Res. 2020, 15, 143. [Google Scholar] [CrossRef]
- Zhang, J.F.; Fu, W.M.; He, M.L.; Wang, H.; Wang, W.M.; Yu, S.C.; Bian, X.W.; Zhou, J.; Lin, M.C.; Lu, G.; et al. MiR-637 maintains the balance between adipocytes and osteoblasts by directly targeting Osterix. Mol. Biol. Cell. 2011, 22, 3955–3961. [Google Scholar] [CrossRef]
- Hu, R.; Liu, W.; Li, H.; Yang, L.; Chen, C.; Xia, Z.Y.; Guo, L.J.; Xie, H.; Zhou, H.D.; Wu, X.P.; et al. A Runx2/miR-3960/miR-2861 regulatory feedback loop during mouse osteoblast differentiation. J. Biol. Chem. 2011, 286, 12328–12339. [Google Scholar] [CrossRef]
- Kim, K.; Kim, J.H.; Kim, I.; Lee, J.; Seong, S.; Park, Y.W.; Kim, N. MicroRNA-26a regulates RANKL-induced osteoclast formation. Mol. Cells 2015, 38, 75–80. [Google Scholar] [PubMed]
- Franceschetti, T.; Kessler, C.B.; Lee, S.K.; Delany, A.M. miR-29 promotes murine osteoclastogenesis by regulating osteoclast commitment and migration. J. Biol. Chem. 2013, 288, 33347–33360. [Google Scholar] [CrossRef]
- Lian, W.S.; Ko, J.Y.; Chen, Y.S.; Ke, H.J.; Hsieh, C.K.; Kuo, C.W.; Wang, S.Y.; Huang, B.W.; Tseng, J.G.; Wang, F.S. MicroRNA-29a represses osteoclast formation and protects against osteoporosis by regulating PCAF-mediated RANKL and CXCL12. Cell Death Dis. 2019, 10, 705. [Google Scholar] [CrossRef] [PubMed]
- Sul, O.J.; Rajasekaran, M.; Park, H.J.; Suh, J.H.; Choi, H.S. MicroRNA-29b Enhances Osteoclast Survival by Targeting BCL-2-Modifying Factor after Lipopolysaccharide Stimulation. Oxid. Med. Cell Longev. 2019, 2019, 6018180. [Google Scholar] [CrossRef] [PubMed]
- Mizoguchi, F.; Murakami, Y.; Saito, T.; Miyasaka, N.; Kohsaka, H. miR-31 controls osteoclast formation and bone resorption by targeting RhoA. Arthritis Res. Ther. 2013, 15, R102. [Google Scholar] [CrossRef] [PubMed]
- Krzeszinski, J.Y.; Wei, W.; Huynh, H.; Jin, Z.; Wang, X.; Chang, T.C.; Xie, X.J.; He, L.; Mangala, L.S.; Lopez-Berestein, G.; et al. miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature 2014, 512, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Cong, F.; Wu, N.; Tian, X.; Fan, J.; Liu, J.; Song, T.; Fu, H. MicroRNA-34c promotes osteoclast differentiation through targeting LGR4. Gene 2017, 610, 1–8. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, H.J.; Park, C.K.; Kim, Y.G.; Lee, H.J.; Kim, J.Y.; Kim, H.H. MicroRNA-124 regulates osteoclast differentiation. Bone 2013, 56, 383–389. [Google Scholar] [CrossRef]
- Tang, L.; Yin, Y.; Liu, J.; Li, Z.; Lu, X. MiR-124 Attenuates Osteoclastogenic Differentiation of Bone Marrow Monocytes Via Targeting Rab27a. Cell Physiol. Biochem. 2017, 43, 1663–1672. [Google Scholar] [CrossRef]
- Yang, S.; Zhang, W.; Cai, M.; Zhang, Y.; Jin, F.; Yan, S.; Baloch, Z.; Fang, Z.; Xue, S.; Tang, R.; et al. Suppression of Bone Resorption by miR-141 in Aged Rhesus Monkeys. J. Bone Miner. Res. 2018, 33, 1799–1812. [Google Scholar] [CrossRef]
- Wang, C.; He, H.; Wang, L.; Jiang, Y.; Xu, Y. Reduced miR-144–3p expression in serum and bone mediates osteoporosis pathogenesis by targeting RANK. Biochem. Cell Biol. 2018, 96, 627–635. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Liu, L.; Liu, Y.; Luo, S.; Song, Y.; Fang, B. miR-144–3p Suppresses Osteogenic Differentiation of BMSCs from Patients with Aplastic Anemia through Repression of TET2. Mol. Ther. Nucleic Acids. 2020, 19, 619–626. [Google Scholar] [CrossRef]
- Yu, F.Y.; Xie, C.Q.; Sun, J.T.; Peng, W.; Huang, X.W. Overexpressed miR-145 inhibits osteoclastogenesis in RANKL-induced bone marrow-derived macrophages and ovariectomized mice by regulation of Smad3. Life Sci. 2018, 202, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Nakasa, T.; Shibuya, H.; Nagata, Y.; Niimoto, T.; Ochi, M. The inhibitory effect of microRNA-146a expression on bone destruction in collagen-induced arthritis. Arthritis Rheum. 2011, 63, 1582–1590. [Google Scholar] [CrossRef]
- Cheng, P.; Chen, C.; He, H.B.; Hu, R.; Zhou, H.D.; Xie, H.; Zhu, W.; Dai, R.C.; Wu, X.P.; Liao, E.Y.; et al. miR-148a regulates osteoclastogenesis by targeting V-maf musculoaponeurotic fibrosarcoma oncogene homolog B. J. Bone Miner Res. 2013, 28, 1180–1190. [Google Scholar] [CrossRef]
- Sul, O.J.; Sung, Y.B.; Rajasekaran, M.; Ke, K.; Yu, R.; Back, S.H.; Choi, H.S. MicroRNA-155 induces autophagy in osteoclasts by targeting transforming growth factor beta-activated kinase 1-binding protein 2 upon lipopolysaccharide stimulation. Bone 2018, 116, 279–289. [Google Scholar] [CrossRef]
- Zhao, C.; Sun, W.; Zhang, P.; Ling, S.; Li, Y.; Zhao, D.; Peng, J.; Wang, A.; Li, Q.; Song, J.; et al. miR-214 promotes osteoclastogenesis by targeting Pten/PI3k/Akt pathway. RNA Biol. 2015, 12, 343–353. [Google Scholar] [CrossRef]
- Liu, J.; Li, D.; Dang, L.; Liang, C.; Guo, B.; Lu, C.; He, X.; Cheung, H.Y.; He, B.; Liu, B.; et al. Osteoclastic miR-214 targets TRAF3 to contribute to osteolytic bone metastasis of breast cancer. Sci. Rep. 2017, 7, 40487. [Google Scholar] [CrossRef]
- Wang, W.; Yang, L.; Zhang, D.; Gao, C.; Wu, J.; Zhu, Y.; Zhang, H. MicroRNA-218 Negatively Regulates Osteoclastogenic Differentiation by Repressing the Nuclear Factor-kappaB Signaling Pathway and Targeting Tumor Necrosis Factor Receptor 1. Cell Physiol. Biochem. 2018, 48, 339–347. [Google Scholar] [CrossRef]
- Chen, C.; Cheng, P.; Xie, H.; Zhou, H.D.; Wu, X.P.; Liao, E.Y.; Luo, X.H. MiR-503 regulates osteoclastogenesis via targeting RANK. J. Bone Miner. Res. 2014, 29, 338–347. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Qin, A.P.; Liao, B.; Shao, H.G.; Guo, L.J.; Xie, G.Q.; Yang, L.; Jiang, T.J. A novel microRNA regulates osteoclast differentiation via targeting protein inhibitor of activated STAT3 (PIAS3). Bone 2014, 67, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Leong, K.-Y.B.; Chan, Y.-H.; Abdullah, W.M.A.N.W.; Lim, E.; Lai, K.-S. MicroRNAs in Bone Diseases: Progress and Prospects. In Transcriptional and Post-Transcriptional Regulation; IntechOpen: London, UK, 2018. [Google Scholar]
- Sun, Y.; Xu, L.; Huang, S.; Hou, Y.; Liu, Y.; Chan, K.M.; Pan, X.H.; Li, G. mir-21 overexpressing mesenchymal stem cells accelerate fracture healing in a rat closed femur fracture model. Biomed. Res. Int. 2015, 2015, 412327. [Google Scholar] [CrossRef] [PubMed]
- Salasznyk, R.M.; Klees, R.F.; Williams, W.A.; Boskey, A.; Plopper, G.E. Focal adhesion kinase signaling pathways regulate the osteogenic differentiation of human mesenchymal stem cells. Exp. Cell Res. 2007, 313, 22–37. [Google Scholar] [CrossRef] [PubMed]
- Eskildsen, T.; Taipaleenmäki, H.; Stenvang, J.; Abdallah, B.M.; Ditzel, N.; Nossent, A.Y.; Bak, M.; Kauppinen, S.; Kassem, M. MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc. Natl. Acad. Sci. USA 2011, 108, 6139–6144. [Google Scholar] [CrossRef]
- Rodriguez-Carballo, E.; Gamez, B.; Ventura, F. p38 MAPK Signaling in Osteoblast Differentiation. Front. Cell Dev. Biol. 2016, 4, 40. [Google Scholar] [CrossRef] [PubMed]
- Pathak, H.; Mohan, M.C.; Ravindran, V. Chikungunya arthritis. Clin. Med. 2019, 19, 381–385. [Google Scholar] [CrossRef]
- Birlik, M.; Koçak, A.; Harmanci, D. Role of MicroRNAs in Rheumatoid Arthritis. In New Developments in the Pathogenesis of Rheumatoid Arthritis; IntechOpen: London, UK, 2017. [Google Scholar]
- Ordas, A.; Kanwal, Z.; Lindenberg, V.; Rougeot, J.; Mink, M.; Spaink, H.P.; Meijer, A.H. MicroRNA-146 function in the innate immune transcriptome response of zebrafish embryos to Salmonella typhimurium infection. BMC Genom. 2013, 14, 696. [Google Scholar] [CrossRef]
- Lim, J.; Byun, J.; Guk, K.; Hwang, S.G.; Bae, P.K.; Jung, J.; Kang, T.; Lim, E.K. Highly Sensitive in Vitro Diagnostic System of Pandemic Influenza A (H1N1) Virus Infection with Specific MicroRNA as a Biomarker. ACS Omega 2019, 4, 14560–14568. [Google Scholar] [CrossRef]
- Janssen, H.L.; Reesink, H.W.; Lawitz, E.J.; Zeuzem, S.; Rodriguez-Torres, M.; Patel, K.; van der Meer, A.J.; Patick, A.K.; Chen, A.; Zhou, Y.; et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013, 368, 1685–1694. [Google Scholar] [CrossRef]
- Stelma, F.; van der Ree, M.H.; Sinnige, M.J.; Brown, A.; Swadling, L.; de Vree, J.M.L.; Willemse, S.B.; van der Valk, M.; Grint, P.; Neben, S.; et al. A single dose of anti-miR-122, RG-101, in CHC patients results in NK cell normalization with no effect on HCV-specific CD8+ T cell function. Hepatology 2017, 66, 57–68. [Google Scholar] [CrossRef]
| MicroRNAs | Target mRNA | Expression Status (Upregulated/ Downregulated) | References |
|---|---|---|---|
| aae-miR-2b | 3′ UTR of ubiquitin related modifier (URM), ubiquitin, and 3′ UTR of CHIKV | Up | [31] |
| hsa-miR-21-5p | B-cell lymphoma 2 (BCL2), chemokine ligand 1 (CCL1), FASLG, pellino E3 ubiquitin protein ligase 1 (PELI1), interleukin 12 A (IL12A), transforming growth factor beta (TGFβ1) | Up | [25] |
| hsa-miR-138-2-3p | Tripartite motif containing 26 (TRIM5), TGF-Beta activated kinase 1 (MAP3K7) binding protein 3 (TAB3), tumor necrosis factor receptor superfamily (TNFRSF19), mitogen-activated protein kinase 13 (MAPK13), Apoptotic protease activating factor 1 (APAF1), Forkhead Box O3 (FOXO3) | Up | [25] |
| hsa-miR-146 | Tumor necrosis factor receptor (TNFR)-associated factor 6 (TRAF6), interleukin-1 receptor-associated kinase 1 (IRAK1/2) | Up | [24] |
| hsa-miR-216a-5p | Cluster of Differentiation 6 (CD6), Janus kinase 2 (JAK2) | Up | [25] |
| aal-miR-305-3p | ECM receptor interaction, endocytosis, and SNARE interactions in vesicular transport | Up | [30] |
| hsa-miR-382-3p | Beta-transducin repeat containing E3 ubiquitin protein ligase (BTRC), gap junction protein alpha 1 (GJA1), TRIM8, ubiquitin-conjugating enzyme E2 D2 (UBE2D2) | Up | [25] |
| hsa-miR-409-3p | DNA topoisomerase 2-beta (TOP2B) | Up | [32] |
| hsa-miR-491-3p | UBE2B | Up | [25] |
| hsa-miR-921 | nicotinic acid uptake protein (NIAP) | Up | [25] |
| aal-miR-927 | Soluble NSF attachment proteins receptor (SNARE) interactions in vesicular transport | Up | [30] |
| aae-miR-989 | sh2/sh3 adaptor and vacuolar ATP synthase | Down | [31] |
| hsa-miR-1260a | BCL2 antagonist/killer 1 (BAK1), activating transcription factor 6 beta (ATF6B) | Up | [25] |
| hsa-miR-1260b | Receptor interacting serine/threonine kinase 1 (RIPK1), E3 ubiquitin-protein ligase NRDP1 (RNF41), suppressor of cytokine signaling 6 (SOCS6), NLR family CARD domain containing 5 (NLRC5), caspase 10 (CASP10) | Up | [25] |
| hsa-miR-1264 | TRIM26, bone morphogenetic protein 2 (BMP2), baculoviral IAP repeat containing 6 (BIRC6), interleukin 6 signal transducer (IL6ST), listerin E3 ubiquitin protein ligase 1 (LTN1), itchy E3 ubiquitin protein ligase (ITCH), MAPK8, SOCS5, UBE2D3 | Up | [25] |
| hsa-miR-3074 | Integrin alpha-V (ITGAV), TNF receptor associated factor 3 (TRAF3) | Up | [25] |
| hsa-miR-4286 | Interferon alpha and beta receptor subunit 1 (IFNAR2), interleukin 13 receptor, alpha 1 (IL13RA1), interferon regulatory factor 1 (IRF1), ubiquitin conjugating enzyme E2 Z (UBE2Z), IL6R | Up | [25] |
| hsa-miR-4299 | SOCS7, signal transducer and activator of transcription 5 (STAT5B), TRIM28, mitogen-activated protein kinase kinase kinase 7 (MAP3K7), TAB1, AKT serine/threonine kinase 1 (AKT1), MAP3K11, MAP kinase-activated protein kinase 3 (MAPKAPK3), CAMP responsive element binding protein 1 (CREB1) | Up | [25] |
| hsa-miR-4443 | Interferon regulatory factor 3 (IRF3), mitogen-activated protein kinase kinase kinase 8 (MAP3K8), receptor interacting serine/threonine kinase 3 (RIPK3) | Up | [25] |
| hsa-miR-4695-3p | CASP8, chemokine C-X-C motif ligand 2 (CXCL2) | Up | [25] |
| hsa-miR-4717-3p | AKT3, UBE2M, sortilin (SORT1), ring finger protein 213 (RNF213), nerve growth factor receptor-associated protein 1 (NGFRAP1), MAPK10, IL11RA, C-C motif chemokine ligand 4 like 2 (CCL4L2), IFNAR1, IL7R, mitochondrial ubiquitin E3 ligase (MARCH5) | Up | [25] |
| hsa-miR-4762-5p | Nemo like kinase (NLK) | Up | [25] |
| hsa-miR-4775 | TNFRSF10A, toll-like receptors (TLR1), CASP3, ATF2, TAB2, glycogen synthase kinase 3 beta (GSK3B) | Up | [25] |
| hsa-miR-4794-5p | UBE2S, STAT1, TRAF5, RIPK1, protein inhibitor of activated STAT 1 (PIAS1), AKT1 | Up | [25] |
| hsa-miR-4878-3p | Diablo IAP-Binding Mitochondrial Protein (DIABLO) | Up | [25] |
| hsa-miR-5100 | UBE2J1, axis inhibition protein 2 (AXIN2), IRAK4 | Up | [25] |
| hsa-miR-5581-3p | MAPK6, IL4R, MAP3K1, CCL18, death inducer-obliterator 1 (DIDO1) | Up | [25] |
| miRNA | Target mRNA | Effect on Osteoblastogenesis | References |
|---|---|---|---|
| hsa-miR-10b | SMAD2 | Enhances | [93] |
| hsa-miR-21, mmu-miR-21 | SMAD5 (in human), SMAD7 (in mouse) | Inhibits (in human), Enhances (in mouse) | [94,95] |
| mmu-miR-22 | Histone deacetylase 6 (HDAC-6) | Enhances | [96] |
| mmu-miR-29 | ON | Inhibits | [97] |
| mmu-miR-30 | RUNX2, SMAD1, LDL receptor related protein 6 (LRP6) | Inhibits | [98,99] |
| hsa-miR-34a | Notch1, Notch2, and Jagged-1 (JAG1) | Inhibits | [100] |
| mmu-miR-34c | Notch1, Notch2, and JAG1 | Inhibits | [101] |
| mmu-miR-34s | CYCLIN D1, CDK4, CDK6, special AT-rich sequence-binding protein 2 (SATB2) | Inhibits | [102] |
| mmu-miR-103-3p | SATB2 | Inhibits | [103] |
| hsa-miR-133 | RUNX2 | Inhibits | [104] |
| hsa-miR-138 | Focal adhesion kinase (FAK) | Inhibits | |
| mmu-miR-145 | OSX gene (SP7) | Inhibits | [105] |
| mmu-miR-155 | SMAD5, RUNX2, and bone morphogenetic protein receptor type II (BMPR2) | Inhibits | [106,107] |
| mmu-miR-183 | Heme oxygenase 1 (HMOX-1) | Inhibits | [108] |
| mmu-miR-204 and hsa-miR-204 | RUNX2 (both in mouse and human), ALP (in human), ON (in human) | Inhibits | [109,110] |
| mmu-miR-221 | Zinc finger protein FOG family member 2 (ZFPM2) | Enhances | [111] |
| mmu-miR-322 | Transducer of ERBB2,2 (TOB2) | Enhances | [112] |
| hsa-miR-335 | RUNX2 | Inhibits | [113] |
| mmu-miR-335-5p | Dickkopf WNT signaling pathway inhibitor 1 (DKK1) | Enhances | [114] |
| hsa-miR-381 | WNT5A, frizzled class receptor 3 (FZD3) | Inhibits | [115] |
| mmu-mir-433 | RUNX2 | Inhibits | [116] |
| mmu-miR-495 | Aquaporin 1 (AQP1) | Enhances | [117] |
| mmu-miR-505 | RUNX2 | Inhibits | [118] |
| hsa-miR-637 | OSX | Inhibits | [119] |
| mmu-miR-2861 | HDAC-5 and homeobox A2 (HOXA2) | Enhances | [120] |
| mmu-miR-3960 | HDAC-5 and HOXA2 | Enhances | [120] |
| miRNA | Target mRNA | Effect on Osteoclastogenesis | References |
|---|---|---|---|
| mmu-miR-26a | Connective tissue growth factor (CTGF) | Inhibits | [121] |
| mmu-miR-29 family | G protein-coupled receptor 85 (GPR85), CD93, nuclear factor I A (NFIA) | Enhances | [122] |
| mmu-miR-29a | RANKL, CXCL12 | Inhibits | [123] |
| mmu-miR-29b | Bcl-2-modifying factor (BMF) | Enhances | [124] |
| mmu-miR-31 | Rhodopsin (RHOA) and the GTPases of the RHO family (Ras-related C3 botulinum toxin substrate 1 (RAC1), RAC2, CDC42, RHOA, and RHOU) | Inhibits | [125] |
| mmu-miR-34a | TGFB induced factor homeobox 2 (TGIF2) | Inhibits | [126] |
| mmu-miR-34c | Leucine rich repeat containing G protein-coupled receptor 4 (LGR4) | Enhances | [127] |
| mmu-miR-124 | NFATC1, ras-related protein 27a (RAB27a) | Inhibits | [128,129] |
| mml-miR-141 | EPH receptor A2 (EPHA2) | Inhibits | [130] |
| hsa-miR-144-3p | RANK, tet methylcytosine dioxygenase 2 (TET2) | Inhibits | [131,132] |
| mmu-miR-145 | SMAD3 | Inhibits | [133] |
| mmu-miR-146a | TRAF6 and IRAK-1 | Inhibits | [134] |
| mmu-miR-148 | MAF BZIP transcription factor B (MAFB) | Enhances | [135] |
| mmu-miR-155 | TAB2 | Inhibits | [136] |
| mmu-miR-214 | PTEN | Enhances | [137] |
| mmu-miR-214-3p | TRAF3 | Enhances | [138] |
| mmu-miR-218 | TNFR1 | Inhibits | [139] |
| hsa-miR-503 | RANK | Inhibits | [140] |
| mmu-miR-9718 | PIAS3 | Enhances | [141] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roy, E.; Byrareddy, S.N.; Reid, S.P. Role of MicroRNAs in Bone Pathology during Chikungunya Virus Infection. Viruses 2020, 12, 1207. https://doi.org/10.3390/v12111207
Roy E, Byrareddy SN, Reid SP. Role of MicroRNAs in Bone Pathology during Chikungunya Virus Infection. Viruses. 2020; 12(11):1207. https://doi.org/10.3390/v12111207
Chicago/Turabian StyleRoy, Enakshi, Siddappa N. Byrareddy, and St Patrick Reid. 2020. "Role of MicroRNAs in Bone Pathology during Chikungunya Virus Infection" Viruses 12, no. 11: 1207. https://doi.org/10.3390/v12111207
APA StyleRoy, E., Byrareddy, S. N., & Reid, S. P. (2020). Role of MicroRNAs in Bone Pathology during Chikungunya Virus Infection. Viruses, 12(11), 1207. https://doi.org/10.3390/v12111207






