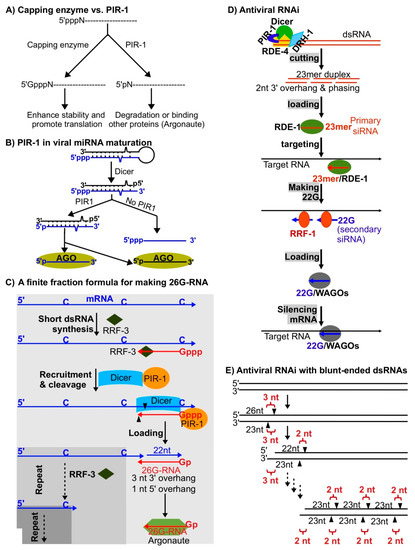
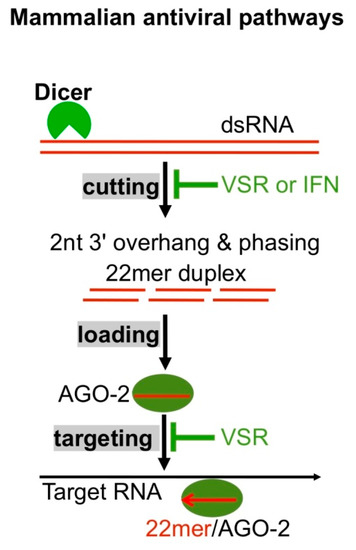
Abstract
Non-coding small RNAs play important roles in virus–host interactions. For hosts, small RNAs can serve as sensors in antiviral pathways including RNAi and CRISPR; for viruses, small RNAs can be involved in viral transcription and replication. This paper covers several recent discoveries on small RNA mediated virus–host interactions, and focuses on influenza virus cap-snatching and a few important virus sensors including PIR-1, RIG-I like protein DRH-1 and piRNAs. The paper also discusses recent advances in mammalian antiviral RNAi.
1. Introduction
The study of host–virus interaction is critical for developing effective antivirus strategies and cures. Its importance is fully exemplified in the current emergency of Coronavirus disease 2019 (COVID-19). Viruses interact with hosts using diverse mechanisms across infection stages. A typical mechanism involves protein–protein or protein–lipid interaction when viruses search target cells, such as in the cases of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and influenza virus infections [1,2,3,4]. Once entering cells, viruses hijack host transcription and translation machinery using viral suppressors including proteins and nucleic acids. In addition to protein factors, small RNAs play important roles in virus–host interactions. For example, recent studies demonstrate that influenza A virus (IAV) utilizes host capped non-coding small RNAs as primers to initiate viral mRNA synthesis [5,6]; host microRNAs (miRNA) can be invovled in regulating viral replication, transcription, and translation [7,8]; viral miRNAs can inhibit host antiviral mechanisms [8,9,10]; small interfering RNAs (siRNA), Piwi-interacting RNAs (piRNA), small nuclear RNAs (snRNAs), and CRISPR (clustered regularly interspaced short palindromic repeats) RNAs (crRNA) are all involved in virus–host interaction (Table 1).

Table 1.
The roles of small RNAs in virus–host interaction.
Host cells have developed multiple layers of antiviral mechanisms in the endless battles against viruses throughout evolution. In mammals, cellular immunity and humoral immunity based on T, B and other immune cells play critical roles. Host immunity can also be classified as innate and adaptive immunity. Innate immunity constitutes the first layer of immune responses and utilizes physical, chemical, cellular and molecular mechanisms to clear invading viruses while adaptive immunity involves the development of antibody and long term memory primarily involving T and B immune cells. In innate immunity, the interferon (IFN) I, II and III signaling pathways play critical roles in defending host cells against viruses. In these pathways, short double-stranded RNAs (dsRNA) are recognized by the Retinoic acid-inducible gene I (RIG-I) family proteins and stimulate the expression of interferons and downstream factors to clear viruses [22,23].
The recent discoveries in RNA interference (RNAi) and CRISPR demonstrate a novel but long awaited mechanism to clear viruses, i.e., using small RNA sensors to recognize viral nucleic acids and using enzymes to destroy them [21,24]. In RNAi, dsRNAs are processed into 20 to 30-nt siRNAs by Dicer, an RNase III like enzyme [25,26]. Some small RNAs, such as piRNAs and crRNAs, are generated in Dicer-independent manners [21,27,28,29,30,31]. Regardless, small RNAs are primarily used as sensors to monitor target RNAs. Unlike anybody-based target recognition, which involves three-dimensional structures, small RNA-based target recognition only requires primary sequence (one-dimensional), greatly simplifying sensor design. This beauty and simplicity allow quick adaptation of these discoveries to bio-engineering tools for manipulating nucleic acids in all kingdoms of life.
Here, we discuss recent advances in host–virus interactions via small RNAs, primarily focusing on IAV cap-snatching, virus sensors, and mammallian antiviral small RNAs.
2. Capped Small RNAs Play Important Roles in IAV mRNA Synthesis
The IAV genome is composed of eight negative-sense viral RNAs (vRNA) for generating mRNAs encoding polymerase basic protein 1 (PB1), polymerase basic protein 2 (PB2), polymerase acidic protein (PA), hemagglutinin (HA), nucleoprotein (NP), neuraminidase (NA), matrix protein (M including M1 and M2), and nonstructural protein (NS including NS1 and NS2). PA, PB1 and PB2 constitute the IAV RNA-dependent RNA polymerase (RdRP) complex responsible for RNA transcription and replication; glycoprotein HA and NA on the IAV virion surface are responsible for target cell interaction (virion entry to and release from host cells, respectively); M plays roles in virion assembly and budding; NP binds/protects vRNAs in the virion; NS1 inhibits host RNAi and interferon-mediated innate immunity, and NS2, also called nuclear export protein (NEP), is involved in IAV virion export from host cell nuclei [32,33,34]. In the early stage of infection, IAV RdRP utilizes template vRNAs to generate positive-sense mRNAs, each of which contains a coding frame flanked by a 5′ and 3′ non-coding regions (NCR). Interestingly, a poly(A) tail is added using a stuttering mechanism, which repeatedly utilizes a short poly(U) sequence on template vRNAs to add/extend the poly(A) tail on IAV mRNAs (Figure 1A). Since the poly(U) sequences are not at the 5′ end of template vRNAs, IAV mRNAs are shorter than vRNAs if the poly(A) tail is not counted [32,33,34]. In the late stage of infection, IAV utilizes RdRP to generate complementary RNA (cRNA) using template vRNA and vRNA using template cRNA. vRNA and cRNA are exactly reverse complementary and both bear 5′ triphosphate (ppp) without a poly(A) tail (Figure 1A) [32,33,34].
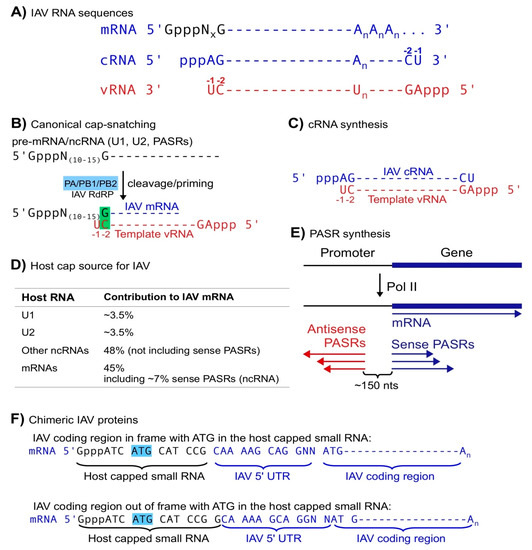
Figure 1.
Canonical cap-snatching. (A) IAV mRNA, cRNA and vRNA sequences (An represents a poly(A) tail and GpppNx represents a host capped small RNA with x equal to 10 to 15 nts). (B) the canonical cap-snatching mechanism: IAV RdRP cleaves host capped RNAs at positions 10–15 nts downstream of the 5′ caps and utilizes the last nt (G) of the resulting host capped small RNAs to anneal with the penultimate (-2, C) nt of template vRNAs, initiating mRNA synthesis. (C) Synthesis of IAV cRNA starts with the last nt (-1 U) of template vRNA in a primer-independent manner. (D) Host cap sources of IAV mRNAs. (E) PASR biogenesis: sense PASRs start at the same transcription start sites as the annotated ”gene” and antisense PASRs are mapped ~150 nts upstream of sense PASRs but on the opposite strand. (F) Schemes of chimeric IAV proteins utilizing a start codon ATG in the host capped small RNAs.
To make viral proteins, most viruses hijack host translation systems, which usually utilize capped RNAs, i.e., mRNAs, as substrates. To make capped viral mRNAs, viruses can utilize host capping enzymes, which reside in nuclei. However, many RNA viruses have to encode their own RNA capping enzymes since they perform transcription only in cytoplasm, which lacks host RNA capping enzymes [35]. Influenza virus utilizes a special process, cap-snatching, to generate viral mRNAs composed of a host capped small RNA and a virus-encoded RNA [36,37,38]. To generate such mRNAs, the IAV RdRP utilizes PB2 to bind host capped RNAs, PA to cleave at positions 10–15 nucleotides (nt) downstream of the 5′ cap, the last nt, usually G, of the resulting capped small RNAs to anneal with the penultimate nt (−2; always C) of template vRNAs, and the polymerase activity to synthesize viral mRNAs based on vRNA templates (Figure 1A,B; [5,6,11,12,39,40,41,42,43]). Thus, IAV utilizes cap-snatching to obtain caps for its mRNAs from host capped RNAs, indirectly utilizing host capping enzymes. In cap-snatching, host capped small RNAs may appear to serve as primers [6,12]. However, if the last nt (G) of the host capped small RNA is treated as an nt 5′ modified with a capped RNA oligo (Figure 1B), this single-basepair-mediated priming mechanism is actually equivalent to that in de novo RNA synthesis without any primer, the initiation mode utilized by most RNA polymerases including for IAV complementary RNA (cRNA) synthesis (Figure 1C). In summary, cap-snatching allows IAV to obtain caps while maintaining the authenticity of viral RNA sequences.
Host non-coding (nc) small RNAs likely serve as the major cap donors in IAV cap-snatching. For decades, host ncRNAs have not been within the radar range of cap-snatching studies. Based on limited sequencing data and annotations, host mRNAs were initially identified as the major cap source [36,37,38]. However, the short sequences of host capped small RNAs on IAV mRNAs basically allow almost all of them to match host mRNA sequences even if matching was restricted to mRNA transcription start site regions, since most mRNAs utilize multiple transcription start sites. Given that mature mRNAs are not localized in nuclei where cap-snatching occurs, pre-mRNAs were proposed as the authentic donors despite lack of sequencing evidence such as intron-containing sequences [36,37,38]. Several groups have utilized high-throughput sequencing to analyze the cap donor profile of IAV cap-snatching [6,11,12]. Among them, Koppstein et al. and Gu et al. identified U1 and U2 snRNAs as the top cap donors while Sikora et al. did not include snRNAs in their search despite the existence of such sequences (Figure 1D) [6,11,12]. Although mature snRNAs are localized in nuclei, Koppstein et al. speculated that only pre-snRNAs, which share the same sequences with mature snRNAs but bear a 7-methyl Guanosine (m7G) cap instead of a 2,2,7 trimethyl Guanosine (m2,2,7G) cap (mature snRNAs), are the authentic donors. Their speculation was based on two previous observations: (1) the m2,2,7G cap on mRNA has a lower affinity to translation factor eIF4E than m7G cap; and (2) the ratio of U1 snRNA cap to U2 cap on IAV mRNAs corresponds well with the transcription rate (representing pre-RNA levels) of U1 and U2 snRNAs rather than the steady-state levels (primarily representing mature RNA levels) [44,45,46]. These arguments may bear flaws since (1) IAV mRNA translation may not require eIF4E [47]; (2) m2,2,7G caps may help virus-specific translation such as in human immunodeficiency virus (HIV) translation [48]; and (3) the paper did not examine the U1 and U2 snRNA levels in the IAV-infected cells but only used those in the non-infected cells published previously to correlate pre-snRNA levels to cap usage [45,46]. Actually, Gu et al. demonstrated that U2 contributed a similar number rather than 3-folds of caps as U1 at two post-infection time points [6,12]. This discrepancy can be caused by different experimental designs including cell lines used, infection stages, cloning methods, etc. Regardless, further experiments, for example, an analysis of viral caps using m2,2,7G immunoprecipitation, can address whether cap-snatching utilizes mature U1 and U2 snRNAs or only pre-U1 and U2.
Cap-snatching may prefer host capped ncRNAs as substrates. Unlike the other two groups, which only sequenced viral capped RNAs, Gu et al. simultaneously obtained host and viral capped RNAs, allowing them to obtain unique matches to host capped RNAs (substrate) for the capped small RNA parts on IAV mRNAs (product) in the same samples and to obtain cap-snatching rates (product/(product + substrate)). U1/U2 snRNAs combined provided ~7% caps on IAV mRNAs; all known ncRNAs including U1/U2, other snRNAs and snoRNAs provided at least 55% caps; pre-mRNAs provided less than 45% including ~7% snatched from sense promoter-associated small RNAs (PASR), a class of small ncRNAs associated with Pol II transcription initiation (Figure 1D,E; see below) [6]. Although host ncRNAs have a higher snatching rate than mRNAs, Gu et al. did not distinguish pre-RNAs from mature RNAs due to the limitations of their experimental strategy. A transcription rate analysis using global run-on sequencing (GRO-seq) and other methods may answer whether pre-ncRNAs are preferred over pre-mRNAs as cap-snatching substrates [49,50].
PASRs, another type of capped small ncRNAs usually with sizes of less than 200 nts, serve as a significant cap source for IAV mRNAs [6]. Transcription initiation by Pol II usually generates sense PASRs starting exactly at the transcription start sites of annotated mRNAs and other RNAs transcribed by Pol II, and antisense PASRs mapped ~150 nts upstream on the antisense strands (relative to annotated genes; Figure 1E) [49,51,52,53,54,55]. In other words, transcription initiation by Pol II is usually bidirectional or divergent and may pause or fail, generating PASRs, while elongation is usually unidirectional. PASRs serve as piRNA precursors in C. elegans and can be processed into miRNAs in animals [13,14]. In theory, sense PASRs bear the same sequences as annotated host mRNAs and other Pol II products. Therefore, the 45% caps on IAV mRNAs thought to be derived from host pre-mRNAs could represent an alternative source, i.e., sense PASRs. Gu et al. found that ~7% IAV mRNA caps were explicitly derived from antisense PASRs, which were mapped to genomic regions without any known annotations [6]. Based on PASR symmetry, they also proposed that sense PASRs may also contribute a similar amount. Therefore, among the 45% IAV mRNA caps assigned to host pre-mRNAs, at least 7% can be traced to sense PASRs, further reducing the contribution of host pre-mRNAs as an IAV mRNA cap source. Othmar et al. showed that IAV RdRP interacts with host Pol II via Pol II C terminal domain (CTD) [56], thereby likely regulating Pol II initiation and elongation. Since PASRs are likely generated by abortive Pol II transcription, it is tempting to propose that IAV RdRP pause Pol II, thus promoting the biogenesis of PASRs for cap-snatching while inhibiting host mRNA elongation, a double jeopardy game to promote virus infection and inhibit host transcription. Further studies are needed to examine this hypothesis.
The chimeric feature of IAV mRNAs allows IAV to generate chimeric proteins. Yuin Ho et al. recently demonstrated that IAV mRNAs can utilize ATG in host capped small RNAs to initiate translation, generating two types of novel proteins containing a few amino acids encoded by host capped small RNAs and the IAV 5′ untranslated region (UTR) or NCRs: one attached to in-framed IAV-encoded proteins and the other attached to out-of-frame IAV-coded “novel proteins” (Figure 1F). Although both types of proteins are expressed at very low levels in the IAV infected cells, they contributed to virulence and were able to initiate host immune response via T cells [57].
Li and Hui et al. recently reported that IAV utilizes non-canonical cap-snatching to diversify its mRNAs and ncRNAs [5]. Canonical IAV mRNA synthesis starts using the basepairing of the last nt (usually G) of snatched capped small RNAs and the penultimate (-2) nt (always C) of template vRNAs via cap-snatching (Figure 1B). However, non-canonical cap-snatching occurs primarily in two types of regions. In the first type, named as mRNA 3′ clusters, IAV mRNA synthesis utilizes the basepairing of the last nt (G) of snatched capped small RNAs and an internal C nt on template vRNAs, generating mRNAs or ncRNAs usually covering the last ~300 (up to 1000) nts of normal IAV mRNAs (Figure 2A). In the second type, named as vRNA 5′ regions (Figure 2B), cap-snatching primarily occurs at the second position of IAV vRNA, i.e., using the basepairing of the last nt (G) of snatched capped small RNAs and the -2 nt (C) of template cRNAs to synthesize capped vRNAs (normal vRNAs contain a 5′ triphosphate group and start at the first position, which corresponds to -1 nt of template cRNA). This constitutes a perfect symmetric transcription pattern in which IAV mRNAs primarily start using the template vRNA -2 nt and non-capped cRNAs usually utilize the −1 nt, while capped vRNAs start using the template cRNA -2 nt and non-capped vRNAs predominantly utilize -1 (Figure 2A,B). However, the transcription (capped RNA)/replication (non-capped RNA) activities are different since IAV mRNAs (capped) are expressed at much higher levels than non-capped cRNAs, while the opposite, i.e., much more non-capped RNAs, occurs on vRNA strands. Regardless, like canonical cap-snatching, non-canonical cap-snatching also generates host-tagged (a few amino acids) IAV proteins, host-tagged novel proteins, and many ncRNAs. Although most IAV mRNAs (~98%) are generated using the canonical initiation sites (-2 C of template vRNAs), ~9% NA mRNAs utilize the non-canonical sites (mRNA 3′ cluster). Since NA protein plays critical roles in defining the antigenicity of IAV and is used as the major drug target, it is tempting to assume that these non-canonical cap-snatching events could lead to novel NA proteins, which may contribute to IAV virulence, initiate host immune response and affect drug efficacy.
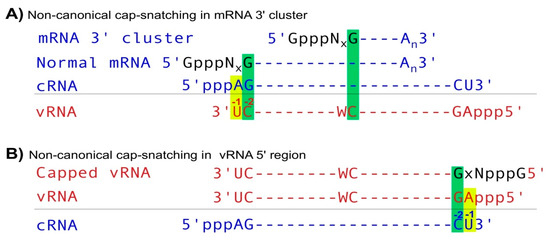
Figure 2.
Noncanonical cap-snatching. (A) In mRNA 3′ clusters, IAV mRNA transcription starts using the basepairing of between the -1 nt (G) of host capped small RNAs and an internal nt C embedded in a 5′CW (W represents A or U) sequence on template vRNAs. (B) In vRNA 5′ regions, IAV RdRP utilizes the -1 nt (G) of host capped small RNAs to anneal with the -2 C of template cRNAs, initiating capped vRNA synthesis.
5. piRNAs Serve as Virus Sensors
In animal germlines, piRNA, a class of 20 to 30 nts long ncRNAs play critical roles in silencing transposons, maintaining germline genome integrity [27,28,29,31]. Piwi Argonautes utilize piRNAs to recognize target RNAs including viral RNAs and cleave them. Interestingly, most piRNAs (21U-RNAs with size 21 nts and preferentially starting with U) in C. elegans do not target transposons. Instead, at least some of them target endogenous RNAs via basepairing with mismatches (Figure 4) [18,19,89,90,91,92]. It is assumed that the ~30,000 piRNAs constitute a repertoire of sensors that recognize any foreign RNAs including viral RNAs via imperfect basepairing (Figure 4) [13,18,89]. Since the relaxed basepairing mechanism may cause self-targeting, many mRNAs expressed in C. elegans germline cells are protected via a CSR-1 (Argonaute)/22G-RNA mediated mechanism [87]. This piRNA-targeting/CSR-1 protection mechanism has been tested using germline transgenes [93]. However, since there is no natural virus that can infect C. elegans germline cells (Orsay virus only infects intestine cells), the model has not been tested with any live viruses. It is also interesting to see whether the piRNAs of low abundance can protect hosts from viruses since there is no evidence suggesting that the expression of those piRNAs can be induced in response to foreign RNA invasion.
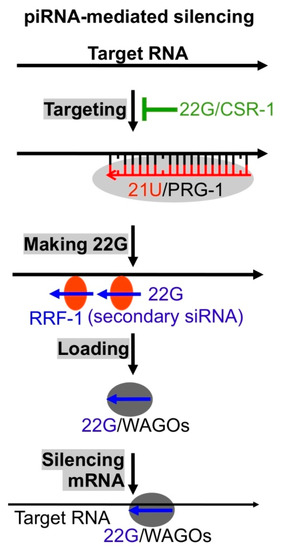
Figure 4.
The antivirus sensor piRNA. C. elegans 21U-RNA (21U, piRNA)/Argonaute Piwi related gene 1 (PRG-1) complex binds target RNAs with up to 3 mismatches, RdRP RRF-1 is recruited to generate secondary siRNAs (22Gs) using target RNAs as templates, 22Gs are loaded to WAGOs to form secondary RISCs, and these RISCs directly silence target RNAs.
7. Conclusions
Small RNAs play important antiviral roles, as exemplified by the CRISPR Nobel Prize this year and RNAi Nobel prize in 2006. Ideally, the small RNA based sensors are more specific and easy to design than conventional chemical and antibody drugs. However, viral RNAs are usually protected by proteins and may not be accessible. It is still challenging to design more target-specific and efficient strategies for delivering small RNA based drugs. Since many RNA viruses encode RNAi inhibitors, small molecule drugs targeting these inhibitors have potential in inhibiting virus infection. IAV strictly utilizes a WG (W represents A or U) motif to initiate mRNA transcription [5]. This motif can be used to develop nucleotide analog drugs, which may be more efficient and specific than the currently available drugs.
Author Contributions
H.D. and W.G. co-wrote this review. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by NIH grant GM124349.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bristow, M.R.; Zisman, L.S.; Altman, N.L.; Gilbert, E.M.; Lowes, B.D.; Minobe, W.A.; Slavov, D.; Schwisow, J.A.; Rodriguez, E.M.; Carroll, I.A.; et al. Dynamic Regulation of SARS-Cov-2 Binding and Cell Entry Mechanisms in Remodeled Human Ventricular Myocardium. JACC Basic Transl. Sci. 2020, 5, 871–883. [Google Scholar] [CrossRef] [PubMed]
- Seyedpour, S.; Khodaei, B.; Loghman, A.H.; Seyedpour, N.; Kisomi, M.F.; Balibegloo, M.; Nezamabadi, S.S.; Gholami, B.; Saghazadeh, A.; Rezaei, N. Targeted therapy strategies against SARS-CoV-2 cell entry mechanisms: A systematic review of in vitro and in vivo studies. J. Cell Physiol. 2020, 1–29. [Google Scholar] [CrossRef]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef] [PubMed]
- Ramos, I.; Fernandez-Sesma, A. Cell receptors for influenza a viruses and the innate immune response. Front. Microbiol. 2012, 3, 117. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Dai, H.; Nguyen, A.P.; Hai, R.; Gu, W. Influenza A virus utilizes noncanonical cap-snatching to diversify its mRNA/ncRNA. RNA 2020, 26, 1170–1183. [Google Scholar] [CrossRef]
- Gu, W.; Gallagher, G.R.; Dai, W.; Liu, P.; Li, R.; Trombly, M.I.; Gammon, D.B.; Mello, C.C.; Wang, J.P.; Finberg, R.W. Influenza A virus preferentially snatches noncoding RNA caps. RNA 2015, 21, 2067–2075. [Google Scholar] [CrossRef]
- Jangra, R.K.; Yi, M.; Lemon, S.M. Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. J. Virol. 2010, 84, 6615–6625. [Google Scholar] [CrossRef]
- Trobaugh, D.W.; Klimstra, W.B. MicroRNA Regulation of RNA Virus Replication and Pathogenesis. Trends Mol. Med. 2017, 23, 80–93. [Google Scholar] [CrossRef] [PubMed]
- McFadden, M.J.; Gokhale, N.S.; Horner, S.M. Protect this house: Cytosolic sensing of viruses. Curr. Opin. Virol. 2017, 22, 36–43. [Google Scholar] [CrossRef]
- Garcia-Sastre, A. Ten Strategies of Interferon Evasion by Viruses. Cell Host Microbe 2017, 22, 176–184. [Google Scholar] [CrossRef]
- Sikora, D.; Rocheleau, L.; Brown, E.G.; Pelchat, M. Deep sequencing reveals the eight facets of the influenza A/HongKong/1/1968 (H3N2) virus cap-snatching process. Sci. Rep. 2014, 4, 6181. [Google Scholar] [CrossRef] [PubMed]
- Koppstein, D.; Ashour, J.; Bartel, D.P. Sequencing the cap-snatching repertoire of H1N1 influenza provides insight into the mechanism of viral transcription initiation. Nucleic Acids Res. 2015, 43, 5052–5064. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Lee, H.C.; Chaves, D.; Youngman, E.M.; Pazour, G.J.; Conte, D., Jr.; Mello, C.C. CapSeq and CIP-TAP identify Pol II start sites and reveal capped small RNAs as C. elegans piRNA precursors. Cell 2012, 151, 1488–1500. [Google Scholar] [CrossRef]
- Zamudio, J.R.; Kelly, T.J.; Sharp, P.A. Argonaute-bound small RNAs from promoter-proximal RNA polymerase II. Cell 2014, 156, 920–934. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Yigit, E.; Li, W.X.; Ding, S.W. An RIG-I-Like RNA helicase mediates antiviral RNAi downstream of viral siRNA biogenesis in Caenorhabditis elegans. PLoS Pathog. 2009, 5, e1000286. [Google Scholar] [CrossRef] [PubMed]
- Maillard, P.V.; van der Veen, A.G.; Poirier, E.Z.; Reis e Sousa, C. Slicing and dicing viruses: Antiviral RNA interference in mammals. EMBO J. 2019, 38, e100941. [Google Scholar] [CrossRef]
- Han, Q.; Chen, G.; Wang, J.; Jee, D.; Li, W.X.; Lai, E.C.; Ding, S.W. Mechanism and Function of Antiviral RNA Interference in Mice. mBio 2020, 11. [Google Scholar] [CrossRef]
- Lee, H.C.; Gu, W.; Shirayama, M.; Youngman, E.; Conte, D.; Mello, C.C. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell 2012, 150, 78–87. [Google Scholar] [CrossRef]
- Shirayama, M.; Seth, M.; Lee, H.C.; Gu, W.; Ishidate, T.; Conte, D.; Mello, C.C. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 2012, 150, 65–77. [Google Scholar] [CrossRef]
- Gasiunas, G.; Barrangou, R.; Horvath, P.; Siksnys, V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, E2579–E2586. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Kato, H.; Sato, S.; Yoneyama, M.; Yamamoto, M.; Uematsu, S.; Matsui, K.; Tsujimura, T.; Takeda, K.; Fujita, T.; Takeuchi, O.; et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity 2005, 23, 19–28. [Google Scholar] [CrossRef]
- Yoneyama, M.; Kikuchi, M.; Natsukawa, T.; Shinobu, N.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Akira, S.; Fujita, T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004, 5, 730–737. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef]
- Zamore, P.D.; Tuschl, T.; Sharp, P.A.; Bartel, D.P. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 2000, 101, 25–33. [Google Scholar] [CrossRef]
- Aravin, A.; Gaidatzis, D.; Pfeffer, S.; Lagos-Quintana, M.; Landgraf, P.; Iovino, N.; Morris, P.; Brownstein, M.J.; Kuramochi-Miyagawa, S.; Nakano, T.; et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 2006, 442, 203–207. [Google Scholar] [CrossRef]
- Girard, A.; Sachidanandam, R.; Hannon, G.J.; Carmell, M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 2006, 442, 199–202. [Google Scholar] [CrossRef] [PubMed]
- Grivna, S.T.; Beyret, E.; Wang, Z.; Lin, H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006, 20, 1709–1714. [Google Scholar] [CrossRef]
- Lau, N.C.; Seto, A.G.; Kim, J.; Kuramochi-Miyagawa, S.; Nakano, T.; Bartel, D.P.; Kingston, R.E. Characterization of the piRNA complex from rat testes. Science 2006, 313, 363–367. [Google Scholar] [CrossRef]
- Ruby, J.G.; Jan, C.; Player, C.; Axtell, M.J.; Lee, W.; Nusbaum, C.; Ge, H.; Bartel, D.P. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 2006, 127, 1193–1207. [Google Scholar] [CrossRef] [PubMed]
- Szewczyk, B.; Bienkowska-Szewczyk, K.; Krol, E. Introduction to molecular biology of influenza a viruses. Acta Biochim. Pol. 2014, 61, 397–401. [Google Scholar] [CrossRef]
- Bouvier, N.M.; Palese, P. The biology of influenza viruses. Vaccine 2008, 26 (Suppl. 4), D49–D53. [Google Scholar] [CrossRef]
- Basler, C.F. Influenza viruses: Basic biology and potential drug targets. Infect. Disord. Drug Targets 2007, 7, 282–293. [Google Scholar] [CrossRef]
- Decroly, E.; Ferron, F.; Lescar, J.; Canard, B. Conventional and unconventional mechanisms for capping viral mRNA. Nat. Rev. Microbiol. 2011, 10, 51–65. [Google Scholar] [CrossRef]
- Bouloy, M.; Plotch, S.J.; Krug, R.M. Globin mRNAs are primers for the transcription of influenza viral RNA in vitro. Proc. Natl. Acad. Sci. USA 1978, 75, 4886–4890. [Google Scholar] [CrossRef]
- Krug, R.M.; Broni, B.A.; Bouloy, M. Are the 5′ ends of influenza viral mRNAs synthesized in vivo donated by host mRNAs? Cell 1979, 18, 329–334. [Google Scholar] [CrossRef]
- Dhar, R.; Chanock, R.M.; Lai, C.J. Nonviral oligonucleotides at the 5′ terminus of cytoplasmic influenza viral mRNA deduced from cloned complete genomic sequences. Cell 1980, 21, 495–500. [Google Scholar] [CrossRef]
- Plotch, S.J.; Bouloy, M.; Ulmanen, I.; Krug, R.M. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell 1981, 23, 847–858. [Google Scholar] [CrossRef]
- Shi, L.; Summers, D.F.; Peng, Q.; Galarz, J.M. Influenza A virus RNA polymerase subunit PB2 is the endonuclease which cleaves host cell mRNA and functions only as the trimeric enzyme. Virology 1995, 208, 38–47. [Google Scholar] [CrossRef][Green Version]
- Rao, P.; Yuan, W.; Krug, R.M. Crucial role of CA cleavage sites in the cap-snatching mechanism for initiating viral mRNA synthesis. EMBO J. 2003, 22, 1188–1198. [Google Scholar] [CrossRef]
- Dias, A.; Bouvier, D.; Crepin, T.; McCarthy, A.A.; Hart, D.J.; Baudin, F.; Cusack, S.; Ruigrok, R.W. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 2009, 458, 914–918. [Google Scholar] [CrossRef]
- Datta, K.; Wolkerstorfer, A.; Szolar, O.H.; Cusack, S.; Klumpp, K. Characterization of PA-N terminal domain of Influenza A polymerase reveals sequence specific RNA cleavage. Nucleic Acids Res. 2013, 41, 8289–8299. [Google Scholar] [CrossRef]
- Niedzwiecka, A.; Marcotrigiano, J.; Stepinski, J.; Jankowska-Anyszka, M.; Wyslouch-Cieszynska, A.; Dadlez, M.; Gingras, A.C.; Mak, P.; Darzynkiewicz, E.; Sonenberg, N.; et al. Biophysical studies of eIF4E cap-binding protein: Recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J. Mol. Biol. 2002, 319, 615–635. [Google Scholar] [CrossRef]
- Pavelitz, T.; Bailey, A.D.; Elco, C.P.; Weiner, A.M. Human U2 snRNA genes exhibit a persistently open transcriptional state and promoter disassembly at metaphase. Mol. Cell Biol. 2008, 28, 3573–3588. [Google Scholar] [CrossRef][Green Version]
- Sauterer, R.A.; Feeney, R.J.; Zieve, G.W. Cytoplasmic assembly of snRNP particles from stored proteins and newly transcribed snRNA’s in L929 mouse fibroblasts. Exp. Cell Res. 1988, 176, 344–359. [Google Scholar] [CrossRef]
- Burgui, I.; Yanguez, E.; Sonenberg, N.; Nieto, A. Influenza virus mRNA translation revisited: Is the eIF4E cap-binding factor required for viral mRNA translation? J. Virol. 2007, 81, 12427–12438. [Google Scholar] [CrossRef]
- Yedavalli, V.S.; Jeang, K.T. Trimethylguanosine capping selectively promotes expression of Rev-dependent HIV-1 RNAs. Proc. Natl. Acad. Sci. USA 2010, 107, 14787–14792. [Google Scholar] [CrossRef]
- Core, L.J.; Waterfall, J.J.; Lis, J.T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 2008, 322, 1845–1848. [Google Scholar] [CrossRef]
- Tani, H.; Mizutani, R.; Salam, K.A.; Tano, K.; Ijiri, K.; Wakamatsu, A.; Isogai, T.; Suzuki, Y.; Akimitsu, N. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 2012, 22, 947–956. [Google Scholar] [CrossRef]
- McGrath, P.T.; Lee, H.; Zhang, L.; Iniesta, A.A.; Hottes, A.K.; Tan, M.H.; Hillson, N.J.; Hu, P.; Shapiro, L.; McAdams, H.H. High-throughput identification of transcription start sites, conserved promoter motifs and predicted regulons. Nat. Biotechnol. 2007, 25, 584–592. [Google Scholar] [CrossRef]
- Sandelin, A.; Carninci, P.; Lenhard, B.; Ponjavic, J.; Hayashizaki, Y.; Hume, D.A. Mammalian RNA polymerase II core promoters: Insights from genome-wide studies. Nat. Rev. Genet. 2007, 8, 424–436. [Google Scholar] [CrossRef]
- Juven-Gershon, T.; Hsu, J.Y.; Theisen, J.W.; Kadonaga, J.T. The RNA polymerase II core promoter—The gateway to transcription. Curr. Opin. Cell Biol. 2008, 20, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Seila, A.C.; Calabrese, J.M.; Levine, S.S.; Yeo, G.W.; Rahl, P.B.; Flynn, R.A.; Young, R.A.; Sharp, P.A. Divergent transcription from active promoters. Science 2008, 322, 1849–1851. [Google Scholar] [CrossRef]
- Affymetrix/ENCODE_Transcriptome_Project. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature 2009, 457, 1028–1032. [Google Scholar] [CrossRef]
- Engelhardt, O.G.; Smith, M.; Fodor, E. Association of the influenza A virus RNA-dependent RNA polymerase with cellular RNA polymerase II. J. Virol. 2005, 79, 5812–5818. [Google Scholar] [CrossRef]
- Ho, J.S.Y.; Angel, M.; Ma, Y.; Sloan, E.; Wang, G.; Martinez-Romero, C.; Alenquer, M.; Roudko, V.; Chung, L.; Zheng, S.; et al. Hybrid Gene Origination Creates Human-Virus Chimeric Proteins during Infection. Cell 2020, 181, 1502–1517.e23. [Google Scholar] [CrossRef]
- Mogensen, T.H. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 2009, 22, 240–273. [Google Scholar] [CrossRef]
- Lester, S.N.; Li, K. Toll-like receptors in antiviral innate immunity. J. Mol. Biol. 2014, 426, 1246–1264. [Google Scholar] [CrossRef]
- Wu, M.H.; Zhang, P.; Huang, X. Toll-like receptors in innate immunity and infectious diseases. Front. Med. China 2010, 4, 385–393. [Google Scholar] [CrossRef]
- Kato, H.; Takeuchi, O.; Sato, S.; Yoneyama, M.; Yamamoto, M.; Matsui, K.; Uematsu, S.; Jung, A.; Kawai, T.; Ishii, K.J.; et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 2006, 441, 101–105. [Google Scholar] [CrossRef]
- Hornung, V.; Ellegast, J.; Kim, S.; Brzozka, K.; Jung, A.; Kato, H.; Poeck, H.; Akira, S.; Conzelmann, K.K.; Schlee, M.; et al. 5′-triphosphate RNA is the ligand for RIG-I. Science 2006, 314, 994–997. [Google Scholar] [CrossRef]
- Yoneyama, M.; Kikuchi, M.; Matsumoto, K.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Foy, E.; Loo, Y.M.; Gale, M., Jr.; Akira, S.; et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 2005, 175, 2851–2858. [Google Scholar] [CrossRef]
- Tabara, H.; Yigit, E.; Siomi, H.; Mello, C.C. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell 2002, 109, 861–871. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, R.; Wang, J.; Ding, S.W.; Lu, R. Homologous RIG-I-like helicase proteins direct RNAi-mediated antiviral immunity in C. elegans by distinct mechanisms. Proc. Natl. Acad. Sci. USA 2013, 110, 16085–16090. [Google Scholar] [CrossRef] [PubMed]
- Ashe, A.; Belicard, T.; Le Pen, J.; Sarkies, P.; Frezal, L.; Lehrbach, N.J.; Felix, M.A.; Miska, E.A. A deletion polymorphism in the Caenorhabditis elegans RIG-I homolog disables viral RNA dicing and antiviral immunity. Elife 2013, 2, e00994. [Google Scholar] [CrossRef] [PubMed]
- Felix, M.A.; Ashe, A.; Piffaretti, J.; Wu, G.; Nuez, I.; Belicard, T.; Jiang, Y.; Zhao, G.; Franz, C.J.; Goldstein, L.D.; et al. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 2011, 9, e1000586. [Google Scholar] [CrossRef] [PubMed]
- Coffman, S.R.; Lu, J.; Guo, X.; Zhong, J.; Jiang, H.; Broitman-Maduro, G.; Li, W.X.; Lu, R.; Maduro, M.; Ding, S.W. Caenorhabditis elegans RIG-I Homolog Mediates Antiviral RNA Interference Downstream of Dicer-Dependent Biogenesis of Viral Small Interfering RNAs. mBio 2017, 8. [Google Scholar] [CrossRef]
- Deshpande, T.; Takagi, T.; Hao, L.; Buratowski, S.; Charbonneau, H. Human PIR1 of the protein-tyrosine phosphatase superfamily has RNA 5′-triphosphatase and diphosphatase activities. J. Biol. Chem. 1999, 274, 16590–16594. [Google Scholar] [CrossRef]
- Sankhala, R.S.; Lokareddy, R.K.; Cingolani, G. Structure of human PIR1, an atypical dual-specificity phosphatase. Biochemistry 2014, 53, 862–871. [Google Scholar] [CrossRef]
- Takagi, T.; Taylor, G.S.; Kusakabe, T.; Charbonneau, H.; Buratowski, S. A protein tyrosine phosphatase-like protein from baculovirus has RNA 5′-triphosphatase and diphosphatase activities. Proc. Natl. Acad. Sci. USA 1998, 95, 9808–9812. [Google Scholar] [CrossRef]
- Yuan, Y.; Li, D.M.; Sun, H. PIR1, a novel phosphatase that exhibits high affinity to RNA ribonucleoprotein complexes. J. Biol. Chem. 1998, 273, 20347–20353. [Google Scholar] [CrossRef]
- Li, L.; Dai, H.; Nguyen, A.P.; Gu, W. A convenient strategy to clone modified/unmodified small RNA and mRNA for high throughput sequencing. RNA 2020, 26, 218–227. [Google Scholar] [CrossRef]
- Chaves, D.A.; Dai, H.; Li, L.; Moresco, J.J.; Eun Oh, M.; Conte, D.J.; Yates, J.R.I.; Mello, C.C.; Gu, W. The RNA phosphatase PIR-1 regulates endogenous small RNA pathways in C. elegans. bioRxiv 2020. [Google Scholar] [CrossRef]
- Dai, H.; Gu, W. Strategies and Best Practice in Cloning Small RNAs. Gene Technol. 2020, 9, 151. [Google Scholar]
- Burke, J.M.; Kincaid, R.P.; Nottingham, R.M.; Lambowitz, A.M.; Sullivan, C.S. DUSP11 activity on triphosphorylated transcripts promotes Argonaute association with noncanonical viral microRNAs and regulates steady-state levels of cellular noncoding RNAs. Genes Dev. 2016, 30, 2076–2092. [Google Scholar] [CrossRef] [PubMed]
- Burke, J.M.; Sullivan, C.S. DUSP11-An RNA phosphatase that regulates host and viral non-coding RNAs in mammalian cells. RNA Biol. 2017, 14, 1457–1465. [Google Scholar] [CrossRef]
- Duchaine, T.F.; Wohlschlegel, J.A.; Kennedy, S.; Bei, Y.X.; Conte, D.; Pang, K.M.; Brownell, D.R.; Harding, S.; Mitani, S.; Ruvkun, G.; et al. Functional proteomics reveals the biochemical niche of C-elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 2006, 124, 343–354. [Google Scholar] [CrossRef]
- Han, T.; Manoharan, A.P.; Harkins, T.T.; Bouffard, P.; Fitzpatrick, C.; Chu, D.S.; Thierry-Mieg, D.; Thierry-Mieg, J.; Kim, J.K. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2009, 106, 18674–18679. [Google Scholar] [CrossRef]
- Conine, C.C.; Batista, P.J.; Gu, W.; Claycomb, J.M.; Chaves, D.A.; Shirayama, M.; Mello, C.C. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2010, 107, 3588–3593. [Google Scholar] [CrossRef]
- Gent, J.I.; Lamm, A.T.; Pavelec, D.M.; Maniar, J.M.; Parameswaran, P.; Tao, L.; Kennedy, S.; Fire, A.Z. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol. Cell 2010, 37, 679–689. [Google Scholar] [CrossRef]
- Vasale, J.J.; Gu, W.; Thivierge, C.; Batista, P.J.; Claycomb, J.M.; Youngman, E.M.; Duchaine, T.F.; Mello, C.C.; Conte, D. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc. Natl. Acad. Sci. USA 2010, 107, 3582–3587. [Google Scholar] [CrossRef]
- Blumenfeld, A.L.; Jose, A.M. Reproducible features of small RNAs in C. elegans reveal NU RNAs and provide insights into 22G RNAs and 26G RNAs. RNA 2016, 22, 184–192. [Google Scholar] [CrossRef]
- Lu, R.; Maduro, M.; Li, F.; Li, H.W.; Broitman-Maduro, G.; Li, W.X.; Ding, S.W. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature 2005, 436, 1040–1043. [Google Scholar] [CrossRef] [PubMed]
- Welker, N.C.; Pavelec, D.M.; Nix, D.A.; Duchaine, T.F.; Kennedy, S.; Bass, B.L. Dicer’s helicase domain is required for accumulation of some, but not all, C. elegans endogenous siRNAs. RNA 2010, 16, 893–903. [Google Scholar] [CrossRef] [PubMed]
- Welker, N.C.; Maity, T.S.; Ye, X.; Aruscavage, P.J.; Krauchuk, A.A.; Liu, Q.; Bass, B.L. Dicer’s helicase domain discriminates dsRNA termini to promote an altered reaction mode. Mol. Cell 2011, 41, 589–599. [Google Scholar] [CrossRef] [PubMed]
- Claycomb, J.M.; Batista, P.J.; Pang, K.M.; Gu, W.; Vasale, J.J.; van Wolfswinkel, J.C.; Chaves, D.A.; Shirayama, M.; Mitani, S.; Ketting, R.F.; et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 2009, 139, 123–134. [Google Scholar] [CrossRef]
- Gu, W.; Shirayama, M.; Conte, D.; Vasale, J.; Batista, P.J.; Claycomb, J.M.; Moresco, J.J.; Youngman, E.M.; Keys, J.; Stoltz, M.J.; et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell 2009, 36, 231–244. [Google Scholar] [CrossRef]
- Ashe, A.; Sapetschnig, A.; Weick, E.M.; Mitchell, J.; Bagijn, M.P.; Cording, A.C.; Doebley, A.L.; Goldstein, L.D.; Lehrbach, N.J.; Le Pen, J.; et al. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 2012, 150, 88–99. [Google Scholar] [CrossRef]
- Grentzinger, T.; Armenise, C.; Brun, C.; Mugat, B.; Serrano, V.; Pelisson, A.; Chambeyron, S. piRNA-mediated transgenerational inheritance of an acquired trait. Genome Res. 2012, 22, 1877–1888. [Google Scholar] [CrossRef]
- Shen, E.Z.; Chen, H.; Ozturk, A.R.; Tu, S.; Shirayama, M.; Tang, W.; Ding, Y.H.; Dai, S.Y.; Weng, Z.; Mello, C.C. Identification of piRNA Binding Sites Reveals the Argonaute Regulatory Landscape of the C. elegans Germline. Cell 2018, 172, 937–951.e18. [Google Scholar] [CrossRef]
- Zhang, D.; Tu, S.; Stubna, M.; Wu, W.S.; Huang, W.C.; Weng, Z.; Lee, H.C. The piRNA targeting rules and the resistance to piRNA silencing in endogenous genes. Science 2018, 359, 587–592. [Google Scholar] [CrossRef] [PubMed]
- Seth, M.; Shirayama, M.; Gu, W.; Ishidate, T.; Conte, D.; Mello, C.C. The C. elegans CSR-1 argonaute pathway counteracts epigenetic silencing to promote germline gene expression. Dev. Cell 2013, 27, 656–663. [Google Scholar] [CrossRef]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef]
- van der Veen, A.G.; Maillard, P.V.; Schmidt, J.M.; Lee, S.A.; Deddouche-Grass, S.; Borg, A.; Kjaer, S.; Snijders, A.P.; Reis e Sousa, C. The RIG-I-like receptor LGP2 inhibits Dicer-dependent processing of long double-stranded RNA and blocks RNA interference in mammalian cells. EMBO J. 2018, 37, e97479. [Google Scholar]
- Li, Y.; Shi, X. MicroRNAs in the regulation of TLR and RIG-I pathways. Cell Mol. Immunol. 2013, 10, 65–71. [Google Scholar] [CrossRef]
- Takahashi, T.; Nakano, Y.; Onomoto, K.; Yoneyama, M.; Ui-Tei, K. Virus Sensor RIG-I Represses RNA Interference by Interacting with TRBP through LGP2 in Mammalian Cells. Genes 2018, 9, 511. [Google Scholar] [CrossRef]
- Li, Y.; Lu, J.F.; Han, Y.H.; Fan, X.X.; Ding, S.W. RNA Interference Functions as an Antiviral Immunity Mechanism in Mammals. Science 2013, 342, 231–234. [Google Scholar] [CrossRef] [PubMed]
- Maillard, P.V.; Ciaudo, C.; Marchais, A.; Li, Y.; Jay, F.; Ding, S.W.; Voinnet, O. Antiviral RNA interference in mammalian cells. Science 2013, 342, 235–238. [Google Scholar] [CrossRef]
- Xu, Y.P.; Qiu, Y.; Zhang, B.; Chen, G.; Chen, Q.; Wang, M.; Mo, F.; Xu, J.; Wu, J.; Zhang, R.R.; et al. Zika virus infection induces RNAi-mediated antiviral immunity in human neural progenitors and brain organoids. Cell Res. 2019, 29, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Basavappa, M.; Lu, J.; Dong, S.; Cronkite, D.A.; Prior, J.T.; Reinecker, H.C.; Hertzog, P.; Han, Y.; Li, W.X.; et al. Induction and suppression of antiviral RNA interference by influenza A virus in mammalian cells. Nat. Microbiol. 2016, 2, 16250. [Google Scholar] [CrossRef]
- Qiu, Y.; Xu, Y.P.; Zhang, Y.; Zhou, H.; Deng, Y.Q.; Li, X.F.; Miao, M.; Zhang, Q.; Zhong, B.; Hu, Y.Y.; et al. Human Virus-Derived Small RNAs Can Confer Antiviral Immunity in Mammals. Immunity 2017, 46, 780–781. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Xu, Y.P.; Wang, M.; Miao, M.; Zhou, H.; Xu, J.Y.; Kong, J.; Zheng, D.; Li, R.T.; Zhang, R.R.; et al. Flavivirus induces and antagonizes antiviral RNA interference in both mammals and mosquitoes. Sci. Adv. 2020, 6, eaax7989. [Google Scholar] [CrossRef]
- Jia, D.; Rahbar, R.; Chan, R.W.; Lee, S.M.; Chan, M.C.; Wang, B.X.; Baker, D.P.; Sun, B.; Peiris, J.S.; Nicholls, J.M.; et al. Influenza virus non-structural protein 1 (NS1) disrupts interferon signaling. PLoS ONE 2010, 5, e13927. [Google Scholar] [CrossRef]
- Marc, D. Influenza virus non-structural protein NS1: Interferon antagonism and beyond. J. Gen. Virol. 2014, 95 Pt 12, 2594–2611. [Google Scholar] [CrossRef]
- Schuster, S.; Overheul, G.J.; Bauer, L.; van Kuppeveld, F.J.M.; van Rij, R.P. No evidence for viral small RNA production and antiviral function of Argonaute 2 in human cells. Sci. Rep. UK 2019, 9, 13752. [Google Scholar] [CrossRef]
- Seo, G.J.; Kincaid, R.P.; Phanaksri, T.; Burke, J.M.; Pare, J.M.; Cox, J.E.; Hsiang, T.Y.; Krug, R.M.; Sullivan, C.S. Reciprocal Inhibition between Intracellular Antiviral Signaling and the RNAi Machinery in Mammalian Cells. Cell Host Microbe 2013, 14, 435–445. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).