Keeping It Together: Structures, Functions, and Applications of Viral Decoration Proteins
Abstract
1. Introduction
2. Functions of Decoration Proteins
2.1. Stabilization of Capsids by Decoration Proteins
2.1.1. Decoration Proteins are Required for the Assembly of Some Viruses
2.1.2. Some Decoration Proteins Provide Stability but Are Not Required for Infectivity
2.2. Multifunctional Decoration Proteins
2.2.1. Decoration Proteins that Act as ‘Tape-Measures’ to Determine Virus Size
2.2.2. The Psu Decoration Protein Moonlights as a Transcription Antiterminator
2.3. Participation of Decoration Proteins in Host Attachment
2.3.1. Decoration Proteins with Ig-Like Domains Can Participate in Host Adhesion
2.3.2. Head Fibers May Coordinate Cell Attachment
3. Decoration Protein Structures
3.1. Capsid-Binding Modes and Oligomerization States
3.1.1. Decoration Proteins Bind to a Variety of Symmetry and/or Pseudo-Symmetry Axes
3.1.2. Oligomerization of Some Decoration Proteins May Require Capsid Binding
3.2. Current Decoration Protein Structures Fall into Five Main Folding Motifs
3.2.1. The β-Tulip Motif Has Three Subfamilies
3.2.2. Dec (L) Has an Oligonucleotide/Oligosaccharide-Binding (OB)-Fold
3.2.3. Soc (T4) Has a Unique β-Tadpole Fold
3.2.4. Hoc (T4) Has Multiple Immunoglobulin (Ig)-Like Domains
3.2.5. Psu (P4) Has a Unique Knotted α-Helical Fold
3.2.6. Additional Decoration Protein Structures
3.3. Structural Homology Suggests Evolution through Horizontal Gene Transfer
4. Nanotechnology Applications
4.1. Decoration Protein Platforms for Design of Novel Nanomaterials
4.2. Decoration Proteins in Phage Display and Biopanning
4.3. Decoration Proteins in Vaccine Design
4.4. Decoration Proteins used as Postmarks to Target VNP Delivery
Author Contributions
Funding
Conflicts of Interest
References
- Sobhy, H.; Scola, B.L.; Pagnier, I.; Raoult, D.; Colson, P. Identification of giant Mimivirus protein functions using RNA interference. Front. Microbiol. 2015, 6, 345. [Google Scholar] [CrossRef]
- Suhanovsky, M.M.; Teschke, C.M. Nature׳s favorite building block: Deciphering folding and capsid assembly of proteins with the HK97-fold. Virology 2015, 487–497. [Google Scholar] [CrossRef]
- Aksyuk, A.A.; Rossmann, M.G. Bacteriophage Assembly. Viruses 2011, 3, 172–203. [Google Scholar] [CrossRef]
- Prevelige, P.E. Send for reinforcements! Conserved binding of capsid decoration proteins. Structure 2008, 16, 1292–1293. [Google Scholar] [CrossRef]
- Mateu, M.G. Assembly, stability and dynamics of virus capsids. Arch. Biochem. Biophys. 2013, 531, 65–79. [Google Scholar] [CrossRef]
- Parent, K.N.; Khayat, R.; Tu, L.H.; Suhanovsky, M.M.; Cortines, J.R.; Teschke, C.M.; Johnson, J.E.; Baker, T.S. P22 Coat Protein Structures Reveal a Novel Mechanism for Capsid Maturation: Stability without Auxiliary Proteins or Chemical Crosslinks. Structure 2010, 18, 390–401. [Google Scholar] [CrossRef]
- Ross, P.D.; Cheng, N.; Conway, J.F.; Firek, B.A.; Hendrix, R.W.; Duda, R.L.; Steven, A.C. Crosslinking renders bacteriophage HK97 capsid maturation irreversible and effects an essential stabilization. EMBO J. 2005, 24, 1352–1363. [Google Scholar] [CrossRef]
- Rizzo, A.A.; Suhanovsky, M.M.; Baker, M.L.; Fraser, L.C.; Jones, L.M.; Rempel, D.L.; Gross, M.L.; Chiu, W.; Alexandrescu, A.T.; Teschke, C.M. Multiple Functional Roles of the Accessory I-Domain of Bacteriophage P22 Coat Protein Revealed by NMR Structure and CryoEM Modeling. Structure 2014, 22, 830–841. [Google Scholar] [CrossRef][Green Version]
- Tripler, T.N.; Kaplan, A.R.; Alexandrescu, A.T.; Teschke, C.M. Conservation and Divergence of the I-Domain Inserted into the Ubiquitous HK97 Coat Protein Fold in P22-Like Bacteriophages. J. Virol. 2019, 93, 9. [Google Scholar] [CrossRef]
- Swanstrom, R.; Wills, J.W. Synthesis, Assembly, and Processing of Viral Proteins. In Retroviruses; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1997. [Google Scholar]
- Casjens, S.; Hendrix, R. Control Mechanisms in dsDNA Bacteriophage Assembly. In The Bacteriophages; Calendar, R., Ed.; Springer US: Boston, MA, USA, 1988; pp. 15–91. [Google Scholar]
- Furcinitti, P.S.; Van Oostrum, J.; Burnett, R.M. Adenovirus polypeptide IX revealed as capsid cement by difference images from electron microscopy and crystallography. EMBO J. 1989, 8, 3563–3570. [Google Scholar] [CrossRef]
- Gilcrease, E.B.; Winn-Stapley, D.A.; Hewitt, F.C.; Joss, L.; Casjens, S.R. Nucleotide Sequence of the Head Assembly Gene Cluster of Bacteriophage L and Decoration Protein Characterization. J. Bacteriol. 2005, 187, 2050–2057. [Google Scholar] [CrossRef]
- Lander, G.C.; Evilevitch, A.; Jeembaeva, M.; Potter, C.S.; Carragher, B.; Johnson, J.E. Bacteriophage Lambda Stabilization by Auxiliary Protein gpD: Timing, Location, and Mechanism of Attachment Determined by Cryo-EM. Structure 2008, 16, 1399–1406. [Google Scholar] [CrossRef]
- Wang, Z.; Hardies, S.C.; Fokine, A.; Klose, T.; Jiang, W.; Cho, B.C.; Rossmann, M.G. Structure of the Marine Siphovirus TW1: Evolution of Capsid-Stabilizing Proteins and Tail Spikes. Structure 2018, 26, 238–248. [Google Scholar] [CrossRef]
- Vernhes, E.; Renouard, M.; Gilquin, B.; Cuniasse, P.; Durand, D.; England, P.; Hoos, S.; Huet, A.; Conway, J.F.; Glukhov, A.; et al. High affinity anchoring of the decoration protein pb10 onto the bacteriophage T5 capsid. Sci. Rep. 2017, 7, 41662. [Google Scholar] [CrossRef]
- Stone, N.P.; Hilbert, B.J.; Hidalgo, D.; Halloran, K.T.; Lee, J.; Sontheimer, E.J.; Kelch, B.A. A Hyperthermophilic Phage Decoration Protein Suggests Common Evolutionary Origin with Herpesvirus Triplex Proteins and an Anti-CRISPR Protein. Structure 2018, 26, 936–947. [Google Scholar] [CrossRef]
- Bayfield, O.W.; Klimuk, E.; Winkler, D.C.; Hesketh, E.L.; Chechik, M.; Cheng, N.; Dykeman, E.C.; Minakhin, L.; Ranson, N.A.; Severinov, K.; et al. Cryo-EM structure and in vitro DNA packaging of a thermophilic virus with supersized T=7 capsids. Proc. Natl. Acad. Sci. USA 2019, 116, 3556–3561. [Google Scholar] [CrossRef]
- Choi, K.H.; McPartland, J.; Kaganman, I.; Bowman, V.D.; Rothman-Denes, L.B.; Rossmann, M.G. Insight into DNA and Protein Transport in Double-Stranded DNA Viruses: The Structure of Bacteriophage N4. J. Mol. Biol. 2008, 378, 726–736. [Google Scholar] [CrossRef]
- Dokland, T.; Murialdo, H. Structural Transitions during Maturation of Bacteriophage Lambda Capsids. J. Mol. Biol. 1993, 233, 682–694. [Google Scholar] [CrossRef]
- Adamson, W.E.; McNab, D.; Preston, V.G.; Rixon, F.J. Mutational Analysis of the Herpes Simplex Virus Triplex Protein VP19C. J. Virol. 2006, 80, 1537–1548. [Google Scholar] [CrossRef]
- Kirkitadze, M.D.; Barlow, P.N.; Price, N.C.; Kelly, S.M.; Boutell, C.J.; Rixon, F.J.; McClelland, D.A. The Herpes Simplex Virus Triplex Protein, VP23, Exists as a Molten Globule. J. Virol. 1998, 72, 10066–10072. [Google Scholar] [CrossRef]
- Yu, X.; Jih, J.; Jiang, J.; Zhou, Z.H. Atomic structure of the human cytomegalovirus capsid with its securing tegument layer of pp150. Science 2017, 356, eaam6892. [Google Scholar] [CrossRef]
- Dai, X.; Wu, L.; Sun, R.; Zhou, Z.H. Atomic Structures of Minor Proteins VI and VII in Human Adenovirus. J. Virol. 2017, 91, e00850-17. [Google Scholar] [CrossRef]
- Reddy, V.S.; Nemerow, G.R. Structures and organization of adenovirus cement proteins provide insights into the role of capsid maturation in virus entry and infection. Proc. Natl. Acad. Sci. USA 2014, 111, 11715. [Google Scholar] [CrossRef]
- Yu, X.; Veesler, D.; Campbell, M.G.; Barry, M.E.; Asturias, F.J.; Barry, M.A.; Reddy, V.S. Cryo-EM structure of human adenovirus D26 reveals the conservation of structural organization among human adenoviruses. Sci. Adv. 2017, 3, e1602670. [Google Scholar] [CrossRef]
- Pietilä, M.K.; Laurinmäki, P.; Russell, D.A.; Ko, C.-C.; Jacobs-Sera, D.; Butcher, S.J.; Bamford, D.H.; Hendrix, R.W. Insights into Head-Tailed Viruses Infecting Extremely Halophilic Archaea. J. Virol. 2013, 87, 3248–3260. [Google Scholar] [CrossRef]
- Stone, N.P.; Demo, G.; Agnello, E.; Kelch, B.A. Principles for enhancing virus capsid capacity and stability from a thermophilic virus capsid structure. Nat. Commun. 2019, 10, 4471–13. [Google Scholar] [CrossRef]
- Wagemans, J.; Tsonos, J.; Holtappels, D.; Fortuna, K.; Hernalsteens, J.-P.; De Greve, H.; Estrozi, L.F.; Bacia-Verloop, M.; Moriscot, C.; Noben, J.-P.; et al. Structural Analysis of Jumbo Coliphage phAPEC6. Int. J. Mol. Sci. 2020, 21, 3119. [Google Scholar] [CrossRef]
- Klose, T.; Fau-Rossmann, M.G.; Rossmann, M.G. Structure of large dsDNA viruses. Biol. Chem. 2014, 395, 711–719. [Google Scholar] [CrossRef]
- Hua, J.; Huet, A.; Lopez, C.A.; Toropova, K.; Pope, W.H.; Duda, R.L.; Hendrix, R.W.; Conway, J.F. Capsids and Genomes of Jumbo-Sized Bacteriophages Reveal the Evolutionary Reach of the HK97 Fold. mBio 2017, 8, e01579-17. [Google Scholar] [CrossRef]
- Podgorski, J.; Calabrese, J.; Alexandrescu, L.; Jacobs-Sera, D.; Pope, W.H.; Hatfull, G.F.; White, S.J. Structures of Three Actinobacteriophage Capsids: Roles of Symmetry and Accessory Proteins. Viruses 2020, 12, 294. [Google Scholar] [CrossRef]
- Doore, S.M.; Schrad, J.R.; Dean, W.F.; Dover, J.A.; Parent, K.N. Shigella Phages Isolated during a Dysentery Outbreak Reveal Uncommon Structures and Broad Species Diversity. J. Virol. 2018, 92, e02117–17. [Google Scholar] [CrossRef]
- González, B.; Monroe, L.; Li, K.; Yan, R.; Wright, E.; Walter, T.; Kihara, D.; Weintraub, S.T.; Thomas, J.A.; Serwer, P.; et al. Phage G Structure at 6.1 Å Resolution, Condensed DNA, and Host Identity Revision to a Lysinibacillus. J. Mol. Biol. 2020, 432, 4139–4153. [Google Scholar] [CrossRef]
- Neumann, E.; Kawasaki, T.; Effantin, G.; Estrozi, L.F.; Chatchawankanphanich, O.; Eyamada, T.; Schoehn, G. 3D structure of three jumbo phage heads. J. Gen. Virol. 2020, jgv001487. [Google Scholar] [CrossRef]
- Chen, Z.; Sun, L.; Zhang, Z.; Fokine, A.; Padilla-Sanchez, V.; Hanein, D.; Jiang, W.; Rossmann, M.G.; Rao, V.B. Cryo-EM structure of the bacteriophage T4 isometric head at 3.3-Å resolution and its relevance to the assembly of icosahedral viruses. Proc. Natl. Acad. Sci. USA 2017, 14, E81–E84. [Google Scholar] [CrossRef]
- Fang, Q.; Zhu, D.; Agarkova, I.; Adhikari, J.; Klose, T.; Liu, Y.; Chen, Z.; Sun, Y.; Gross, M.L.; Van Etten, J.L.; et al. Near-atomic structure of a giant virus. Nat. Commun. 2019, 10, 388. [Google Scholar] [CrossRef]
- Fokine, A.; Islam, M.Z.; Zhang, Z.; Bowman, V.D.; Rao, V.B.; Rossmann, M.G. Structure of the Three N-Terminal Immunoglobulin Domains of the Highly Immunogenic Outer Capsid Protein from a T4-Like Bacteriophage. J. Virol. 2011, 85, 8141–8148. [Google Scholar] [CrossRef]
- Qin, L.; Fokine, A.; O’Donnell, E.; Rao, V.B.; Rossmann, M.G. Structure of the Small Outer Capsid Protein, Soc: A Clamp for Stabilizing Capsids of T4-like Phages. J. Mol. Biol. 2010, 395, 728–741. [Google Scholar] [CrossRef]
- Xiang, Y.; Rossmann, M.G. Structure of bacteriophage phi29 head fibers has a supercoiled triple repeating helix-turn-helix motif. Proc. Natl. Acad. Sci. USA 2011, 108, 4806. [Google Scholar] [CrossRef]
- Wikoff, W.R.; Liljas, L.; Duda, R.L.; Tsuruta, H.; Hendrix, R.W.; Johnson, J.E. Topologically Linked Protein Rings in the Bacteriophage HK97 Capsid. Science 2000, 289, 2129–2133. [Google Scholar] [CrossRef]
- Duda, R.L.; Teschke, C.M. The amazing HK97 fold: Versatile results of modest differences. Curr. Opin. Virol. 2019, 36, 9–16. [Google Scholar] [CrossRef]
- Helgstrand, C.; Wikoff, W.R.; Duda, R.L.; Hendrix, R.W.; Johnson, J.E.; Liljas, L. The Refined Structure of a Protein Catenane: The HK97 Bacteriophage Capsid at 3.44 Å Resolution. J. Mol. Biol. 2003, 334, 885–899. [Google Scholar] [CrossRef]
- Conway, J.F.; Wikoff, W.R.; Cheng, N.; Duda, R.L.; Hendrix, R.W.; Johnson, J.E.; Steven, A.C. Virus Maturation Involving Large Subunit Rotations and Local Refolding. Science 2001, 292, 744–748. [Google Scholar] [CrossRef]
- Gertsman, I.; Gan, L.; Guttman, M.; Lee, K.; Speir, J.A.; Duda, R.L.; Hendrix, R.W.; Komives, E.A.; Johnson, J.E. An unexpected twist in viral capsid maturation. Nature 2009, 458, 646–650. [Google Scholar] [CrossRef]
- Sae-Ueng, U.; Liu, T.; Catalano, C.E.; Huffman, J.B.; Homa, F.L.; Evilevitch, A. Major capsid reinforcement by a minor protein in herpesviruses and phage. Nucleic Acids Res. 2014, 42, 9096–9107. [Google Scholar] [CrossRef]
- Tang, L.; Gilcrease, E.B.; Casjens, S.R.; Johnson, J.E. Highly Discriminatory Binding of Capsid-Cementing Proteins in Bacteriophage L. Structure 2006, 14, 837–845. [Google Scholar] [CrossRef]
- Hernando-Pérez, M.; Lambert, S.; Nakatani-Webster, E.; Catalano, C.E.; De Pablo, P.J. Cementing proteins provide extra mechanical stabilization to viral cages. Nat. Commun. 2014, 5, 4520. [Google Scholar] [CrossRef]
- Yang, Q.; Maluf, N.K.; Catalano, C.E. Packaging of a Unit-Length Viral Genome: The Role of Nucleotides and the gpD Decoration Protein in Stable Nucleocapsid Assembly in Bacteriophage λ. J. Mol. Biol. 2008, 383, 1037–1048. [Google Scholar] [CrossRef]
- Yang, F.; Forrer, P.; Dauter, Z.; Conway, J.F.; Cheng, N.; Cerritelli, M.E.; Steven, A.C.; Plückthun, A.; Wlodawer, A. Novel fold and capsid-binding properties of the λ-phage display platform protein gpD. Nat. Struct. Biol. 2000, 7, 230–237. [Google Scholar]
- Iwai, H.; Forrer, P.; Pluckthun, A.; Guntert, P. NMR solution structure of the monomeric form of the bacteriophage lambda capsid stabilizing protein gpD. J. Biomol. NMR 2005, 31, 351–356. [Google Scholar] [CrossRef]
- Forrer, P.; Chang, C.; Ott, D.; Wlodawer, A.; Plückthun, A. Kinetic Stability and Crystal Structure of the Viral Capsid Protein SHP. J. Mol. Biol. 2004, 344, 179–193. [Google Scholar] [CrossRef]
- Shin, H.; Lee, J.-H.; Ahn, C.S.; Ryu, S.; Cho, B.C. Complete genome sequence of marine bacterium Pseudoalteromonas phenolica bacteriophage TW1. Arch. Virol. 2014, 159, 159–162. [Google Scholar] [CrossRef]
- Hardy, J.M.; Dunstan, R.A.; Grinter, R.; Belousoff, M.J.; Wang, J.; Pickard, D.; Venugopal, H.; Dougan, G.; Lithgow, T.; Coulibaly, F. The architecture and stabilisation of flagellotropic tailed bacteriophages. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef]
- Schwarz, B.; Madden, P.; Avera, J.; Gordon, B.; Larson, K.; Miettinen, H.M.; Uchida, M.; Lafrance, B.; Basu, G.; Rynda-Apple, A.; et al. Symmetry Controlled, Genetic Presentation of Bioactive Proteins on the P22 Virus-like Particle Using an External Decoration Protein. ACS Nano 2015, 9, 9134–9147. [Google Scholar] [CrossRef]
- Newcomer, R.L.; Belato, H.B.; Teschke, C.M.; Alexandrescu, A.T. NMR assignments for monomeric phage L decoration protein. Biomol. NMR Assign. 2018, 12, 339–343. [Google Scholar] [CrossRef]
- Newcomer, R.L.; Schrad, J.R.; Gilcrease, I.B.; Casjens, S.R.; Feig, M.; Teschke, C.M.; Alexandrescu, A.T.; Parent, K.N. The phage L capsid decoration protein has a novel OB-fold and an unusual capsid binding strategy. eLife 2019, 8. [Google Scholar] [CrossRef]
- Steven, A.C.; Greenstone, H.L.; Booy, F.P.; Black, L.W.; Ross, P.D. Conformational changes of a viral capsid protein: Thermodynamic rationale for proteolytic regulation of bacteriophage T4 capsid expansion, co-operativity, and super-stabilization by soc binding. J. Mol. Biol. 1992, 228, 870–884. [Google Scholar] [CrossRef]
- Ishii, T.; Yanagida, M. The two dispensable structural proteins (soc and hoc) of the T4 phage capsid; their purification and properties, isolation and characterization of the defective mutants, and their binding with the defective heads in vitro. J. Mol. Biol. 1977, 109, 487–514. [Google Scholar] [CrossRef]
- Olson, N.H.; Gingery, M.; Eiserling, F.A.; Baker, T.S. The Structure of Isometric Capsids of Bacteriophage T4. Virology 2001, 279, 385–391. [Google Scholar] [CrossRef]
- Sathaliyawala, T.; Islam, M.Z.; Li, Q.; Fokine, A.; Rossmann, M.G.; Rao, V.B. Functional analysis of the highly antigenic outer capsid protein, Hoc, a virus decoration protein from T4-like bacteriophages. Mol. Microbiol. 2010, 77, 444–455. [Google Scholar] [CrossRef]
- Iwasaki, K.; Trus, B.L.; Wingfield, P.T.; Cheng, N.; Campusano, G.; Rao, V.B.; Steven, A.C. Molecular Architecture of Bacteriophage T4 Capsid: Vertex Structure and Bimodal Binding of the Stabilizing Accessory Protein, Soc. Virology 2000, 271, 321–333. [Google Scholar] [CrossRef][Green Version]
- Effantin, G.; Boulanger, P.; Neumann, E.; Letellier, L.; Conway, J.F. Bacteriophage T5 Structure Reveals Similarities with HK97 and T4 Suggesting Evolutionary Relationships. J. Mol. Biol. 2006, 361, 993–1002. [Google Scholar] [CrossRef]
- Banerjee, R.; Nath, S.; Ranjan, A.; Khamrui, S.; Pani, B.; Sen, R.; Sen, U. The First Structure of Polarity Suppression Protein, Psu from Enterobacteria Phage P4, Reveals a Novel Fold and a Knotted Dimer. J. Biol. Chem. 2012, 287, 44667–44675. [Google Scholar] [CrossRef]
- Linderoth, N.A.; Calendar, R.L. The Psu protein of bacteriophage P4 is an antitermination factor for rho-dependent transcription termination. J. Bacteriol. 1991, 173, 6722–6731. [Google Scholar] [CrossRef][Green Version]
- White, H.E.; Sherman, M.B.; Brasilès, S.; Jacquet, E.; Seavers, P.; Tavares, P.; Orlova, E.V. Capsid Structure and Its Stability at the Late Stages of Bacteriophage SPP1 Assembly. J. Virol. 2012, 86, 6768. [Google Scholar] [CrossRef]
- Zairi, M.; Stiege, A.C.; Nhiri, N.; Jacquet, E.; Tavares, P. The Collagen-like Protein gp12 is a Temperature-dependent Reversible Binder of SPP1 Viral Capsids. J. Biol. Chem. 2014, 289, 27169–27181. [Google Scholar] [CrossRef]
- Ignatiou, A.; Brasilès, S.; Fadel, M.E.S.; Bürger, J.; Mielke, T.; Topf, M.; Tavares, P.; Orlova, E.V. Structural transitions during the scaffolding-driven assembly of a viral capsid. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Rosa-Calatrava, M.; Grave, L.; Puvion-Dutilleul, F.; Chatton, B.; Kedinger, C. Functional Analysis of Adenovirus Protein IX Identifies Domains Involved in Capsid Stability, Transcriptional Activity, and Nuclear Reorganization. J. Virol. 2001, 75, 7131–7141. [Google Scholar] [CrossRef]
- Abrescia, N.G.A.; Cockburn, J.J.B.; Grimes, J.M.; Sutton, G.C.; Diprose, J.M.; Butcher, S.J.; Fuller, S.D.; Martín, C.S.; Burnett, R.M.; Stuart, D.I.; et al. Insights into assembly from structural analysis of bacteriophage PRD1. Nature 2004, 432, 68–74. [Google Scholar] [CrossRef]
- Rydman, P.S.; Bamford, J.K.; Bamford, D.H. A minor capsid protein P30 is essential for bacteriophage PRD1 capsid assembly. J. Mol. Biol. 2001, 313, 785–795. [Google Scholar] [CrossRef]
- Dunigan, D.D.; Cerny, R.L.; Bauman, A.T.; Roach, J.C.; Lane, L.C.; Agarkova, I.V.; Wulser, K.; Yanai-Balser, G.M.; Gurnon, J.R.; Vitek, J.C.; et al. Paramecium bursaria Chlorella Virus 1 Proteome Reveals Novel Architectural and Regulatory Features of a Giant Virus. J. Virol. 2012, 86, 8821–8834. [Google Scholar] [CrossRef]
- Chang, J.T.; Schmid, M.F.; Haase-Pettingell, C.; Weigele, P.R.; King, J.A.; Chiu, W. Visualizing the Structural Changes of Bacteriophage Epsilon15 and Its Salmonella Host during Infection. J. Mol. Biol. 2010, 402, 731–740. [Google Scholar] [CrossRef]
- Fuller, D.N.; Raymer, D.M.; Rickgauer, J.P.; Robertson, R.M.; Catalano, C.E.; Anderson, D.L.; Grimes, S.; Smith, D.E. Measurements of Single DNA Molecule Packaging Dynamics in Bacteriophage λ Reveal High Forces, High Motor Processivity, and Capsid Transformations. J. Mol. Biol. 2007, 373, 1113–1122. [Google Scholar] [CrossRef]
- Imber, R.; Tsugita, A.; Würtz, M.; Hohn, T. Outer surface protein of bacteriophage lambda. J. Mol. Biol. 1980, 139, 277–295. [Google Scholar] [CrossRef]
- Wendt, J.L.; Feiss, M. A fragile lattice: Replacing bacteriophage λ’s head stability gene D with the shp gene of phage 21 generates the Mg2+-dependent virus, λ shp. Virology 2004, 326, 41–46. [Google Scholar] [CrossRef][Green Version]
- Morais, M.C.; Choi, K.H.; Koti, J.S.; Chipman, P.R.; Anderson, D.L.; Rossmann, M.G. Conservation of the Capsid Structure in Tailed dsDNA Bacteriophages: The Pseudoatomic Structure of ϕ29. Mol. Cell 2005, 18, 149–159. [Google Scholar] [CrossRef]
- Baker, M.L.; Jiang, W.; Rixon, F.J.; Chiu, W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J. Virol. 2005, 79, 14967. [Google Scholar] [CrossRef]
- Brown, J.C.; Newcomb, W.W. Herpesvirus capsid assembly: Insights from structural analysis. Curr. Opin. Virol. 2011, 1, 142–149. [Google Scholar] [CrossRef]
- Dai, X.; Zhou, Z.H. Structure of the herpes simplex virus 1 capsid with associated tegument protein complexes. Science 2018, 360, eaao7298. [Google Scholar] [CrossRef]
- Yuan, S.; Wang, J.; Zhu, D.; Wang, N.; Gao, Q.; Chen, W.; Tang, H.; Wang, J.; Zhang, X.; Liu, H.; et al. Cryo-EM structure of a herpesvirus capsid at 3.1 Å. Science 2018, 360, eaao7283. [Google Scholar] [CrossRef]
- Belnap, D.M.; Steven, A.C. ‘Déjà vu all over again’: The similar structures of bacteriophage PRD1 and adenovirus. Trends Microbiol. 2000, 8, 91–93. [Google Scholar] [CrossRef]
- Benson, S.D.; Bamford, J.K.; Bamford, D.H.; Burnett, R.M. Does Common Architecture Reveal a Viral Lineage Spanning All Three Domains of Life? Mol. Cell 2004, 16, 673–685. [Google Scholar] [CrossRef]
- Barr, J.J.; Auro, R.; Sam-Soon, N.; Kassegne, S.; Peters, G.; Bonilla, N.; Hatay, M.; Mourtada, S.; Bailey, B.; Youle, M.; et al. Subdiffusive motion of bacteriophage in mucosal surfaces increases the frequency of bacterial encounters. Proc. Natl. Acad. Sci. USA 2015, 112, 13675. [Google Scholar] [CrossRef]
- Barr, J.J.; Youle, M.; Rohwer, F. Innate and acquired bacteriophage-mediated immunity. Bacteriophage 2013, 3, e25857. [Google Scholar] [CrossRef]
- Barr, J.J.; Auro, R.; Furlan, M.; Whiteson, K.L.; Erb, M.L.; Pogliano, J.; Stotland, A.; Wolkowicz, R.; Cutting, A.S.; Doran, K.S.; et al. Bacteriophage adhering to mucus provide a non–host-derived immunity. Proc. Natl. Acad. Sci. USA 2013, 110, 10771. [Google Scholar] [CrossRef]
- Bansil, R.; Turner, B.S. The biology of mucus: Composition, synthesis and organization. Adv. Drug Deliv. Rev. 2018, 124, 3–15. [Google Scholar] [CrossRef]
- Silveira, C.B.; Rohwer, F.L. Piggyback-the-Winner in host-associated microbial communities. NPJ Biofilms Microbiomes 2016, 2, 16010. [Google Scholar] [CrossRef]
- Fraser, J.S.; Maxwell, K.L.; Davidson, A.R. Immunoglobulin-like domains on bacteriophage: Weapons of modest damage? Curr. Opin. Microbiol. 2007, 10, 382–387. [Google Scholar] [CrossRef]
- Wilson, D.P. Protruding Features of Viral Capsids Are Clustered on Icosahedral Great Circles. PLoS ONE 2016, 11, e0152319. [Google Scholar] [CrossRef][Green Version]
- Zandi, R.; Reguera, D. Mechanical properties of viral capsids. Phys. Rev. E 2005, 72, 021917. [Google Scholar] [CrossRef]
- Parent, K.N.; Deedas, C.T.; Egelman, E.H.; Casjens, S.R.; Baker, T.S.; Teschke, C.M. Stepwise molecular display utilizing icosahedral and helical complexes of phage coat and decoration proteins in the development of robust nanoscale display vehicles. Biomaterials 2012, 33, 5628–5637. [Google Scholar] [CrossRef]
- Rao, S.; Rossmann, M.G. Comparison of super-secondary structures in proteins. J. Mol. Biol. 1973, 76, 241–256. [Google Scholar] [CrossRef]
- Rossmann, M.G. Super-secondary Structure: A Historical Perspective. Methods Mol. Biol. 2013, 932, 1–4. [Google Scholar]
- Shin, W.-H.; Kihara, D. 55 Years of the Rossmann Fold. Methods Mol. Biol. 2019, 1958, 1–13. [Google Scholar]
- Neuman, B.W.; Chamberlain, P.; Bowden, F.; Joseph, J. Atlas of coronavirus replicase structure. Virus Res. 2014, 194, 49–66. [Google Scholar] [CrossRef]
- Abroi, A.; Gough, J. Are viruses a source of new protein folds for organisms? - Virosphere structure space and evolution. BioEssays 2011, 33, 626–635. [Google Scholar] [CrossRef]
- Xu, J.; Wang, D.; Gui, M.; Xiang, Y. Structural assembly of the tailed bacteriophage varphi29. Nat. Commun. 2019, 10, 2366. [Google Scholar] [CrossRef]
- Alexandrescu, A.T.; Gittis, A.G.; Abeygunawardana, C.; Shortle, D. NMR Structure of a Stable “OB-fold” Sub-domain Isolated from Staphylococcal Nuclease. J. Mol. Biol. 1995, 250, 134–143. [Google Scholar] [CrossRef]
- Guardino, K.M.; Sheftic, S.R.; Slattery, R.E.; Alexandrescu, A.T. Relative Stabilities of Conserved and Non-Conserved Structures in the OB-Fold Superfamily. Int. J. Mol. Sci. 2009, 10, 2412–2430. [Google Scholar] [CrossRef]
- Murzin, A.G. OB (oligonucleotide/oligosaccharide binding)-fold: Common structural and functional solution for non-homologous sequences. EMBO J. 1993, 12, 861–867. [Google Scholar] [CrossRef]
- Holm, L.; Rosenstr�MP. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010, 38, W545–W549. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Hendrix, R.W. Bacteriophage genomics. Curr. Opin. Microbiol. 2003, 6, 506–511. [Google Scholar] [CrossRef]
- Krupovic, M.; Koonin, E.V. Multiple origins of viral capsid proteins from cellular ancestors. Proc. Natl. Acad. Sci. USA 2017, 114, E2401–E2410. [Google Scholar] [CrossRef]
- Xiang, Y.; Leiman, P.G.; Li, L.; Grimes, S.; Anderson, D.L.; Rossmann, M.G. Crystallographic Insights into the Autocatalytic Assembly Mechanism of a Bacteriophage Tail Spike. Mol. Cell 2009, 34, 375–386. [Google Scholar] [CrossRef]
- Asija, K.; Teschke, C.M. Lessons from bacteriophages part 2: A saga of scientific breakthroughs and prospects for their use in human health. PLoS Pathog. 2018, 14, e1006970. [Google Scholar] [CrossRef]
- Asija, K.; Teschke, C.M. Lessons from bacteriophages part 1: Deriving utility from protein structure, function, and evolution. PLoS Pathog. 2018, 14, e1006971. [Google Scholar] [CrossRef]
- Sarikaya, M.; Tamerler, C.; Jen, A.K.-Y.; Schulten, K.; Baneyx, F. Molecular biomimetics: Nanotechnology through biology. Nat. Mater. 2003, 2, 577–585. [Google Scholar] [CrossRef]
- Vera-Robles, L.I.; González-Gracida, J.; Hernández-Gordillo, A.; Campero, A. Using the M13 Phage as a Biotemplate to Create Mesoporous Structures Decorated with Gold and Platinum Nanoparticles. Langmuir 2015, 31, 9188–9197. [Google Scholar] [CrossRef]
- Chatterji, A.; Ochoa, W.F.; Ueno, T.; Lin, T.; Johnson, J.E. A Virus-Based Nanoblock with Tunable Electrostatic Properties. Nano Lett. 2005, 5, 597–602. [Google Scholar] [CrossRef]
- Klem, M.T.; Young, M.; Douglas, T. Biomimetic magnetic nanoparticles. Mater. Today 2005, 8, 28–37. [Google Scholar] [CrossRef]
- Cho, C.F.; Shukla, S.; Simpson, E.J.; Steinmetz, N.F.; Luyt, L.G.; Lewis, J.D. Molecular targeted viral nanoparticles as tools for imaging cancer. Methods Mol. Biol. 2014, 1108, 211–230. [Google Scholar]
- Li, Q.; Shivachandra, S.B.; Zhang, Z.; Rao, V.B. Assembly of the Small Outer Capsid Protein, Soc, on Bacteriophage T4: A Novel System for High Density Display of Multiple Large Anthrax Toxins and Foreign Proteins on Phage Capsid. J. Mol. Biol. 2007, 370, 1006–1019. [Google Scholar] [CrossRef]
- Van Rosmalen, M.G.M.; Nemerow, G.R.; Wuite, G.J.L.; Roos, W.H. A single point mutation in precursor protein VI doubles the mechanical strength of human adenovirus. J. Biol. Phys. 2017, 44, 119–132. [Google Scholar] [CrossRef]
- Srivastava, S.; Nykypanchuk, D.; Fukuto, M.; Halverson, J.D.; Tkachenko, A.V.; Yager, K.G.; Gang, O. Two-Dimensional DNA-Programmable Assembly of Nanoparticles at Liquid Interfaces. J. Am. Chem. Soc. 2014, 136, 8323–8332. [Google Scholar] [CrossRef]
- Zhang, S.; Li, Z.; Samarajeewa, S.; Sun, G.; Yang, C.; Wooley, K.L. Orthogonally Dual-Clickable Janus Nanoparticles via a Cyclic Templating Strategy. J. Am. Chem. Soc. 2011, 133, 11046–11049. [Google Scholar] [CrossRef][Green Version]
- Braunschweig, A.B.; Huo, F.; Mirkin, C.A. Molecular printing. Nat. Chem. 2009, 1, 353–358. [Google Scholar] [CrossRef]
- Alberti, S.; Soler-Illia, G.J.; Azzaroni, O. Gated supramolecular chemistry in hybrid mesoporous silica nanoarchitectures: Controlled delivery and molecular transport in response to chemical, physical and biological stimuli. Chem. Commun. (Camb.) 2015, 51, 6050–6075. [Google Scholar] [CrossRef]
- The Nobel Prize is Chemistry 2018. Available online: https://www.nobelprize.org/prizes/chemistry/2018/summary/ (accessed on 1 August 2020).
- Smith, G.P. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, 1315–1317. [Google Scholar] [CrossRef]
- Jiang, J.; Abu-Shilbayeh, L.; Rao, V.B. Display of a PorA peptide from Neisseria meningitidis on the bacteriophage T4 capsid surface. Infect. Immun. 1997, 65, 4770–4777. [Google Scholar] [CrossRef]
- Li, Q.; Shivachandra, S.B.; Leppla, S.H.; Rao, V.B. Bacteriophage T4 Capsid: A Unique Platform for Efficient Surface Assembly of Macromolecular Complexes. J. Mol. Biol. 2006, 363, 577–588. [Google Scholar] [CrossRef]
- Meulenbroek, R.A.; Sargent, K.L.; Lunde, J.; Jasmin, B.J.; Parks, R.J.; Meulenbroek, R. Use of adenovirus protein IX (pIX) to display large polypeptides on the virion—Generation of fluorescent virus through the incorporation of pIX-GFP. Mol. Ther. 2004, 9, 617–624. [Google Scholar] [CrossRef]
- Sathaliyawala, T.; Rao, M.; MacLean, D.M.; Birx, D.L.; Alving, C.R.; Rao, V.B. Assembly of Human Immunodeficiency Virus (HIV) Antigens on Bacteriophage T4: A Novel In Vitro Approach To Construct Multicomponent HIV Vaccines. J. Virol. 2006, 80, 7688–7698. [Google Scholar] [CrossRef]
- Malys, N.; Chang, D.-Y.; Baumann, R.G.; Xie, D.; Black, L.W. A Bipartite Bacteriophage T4 SOC and HOC Randomized Peptide Display Library: Detection and Analysis of Phage T4 Terminase (gp17) and Late σ Factor (gp55) Interaction. J. Mol. Biol. 2002, 319, 289–304. [Google Scholar] [CrossRef]
- Aghebati-Maleki, L.; Bakhshinejad, B.; Baradaran, B.; Motallebnezhad, M.; Aghebati-Maleki, A.; Nickho, H.; Yousefi, M.; Majidi, J. Phage display as a promising approach for vaccine development. J. Biomed. Sci. 2016, 23, 66. [Google Scholar] [CrossRef]
- Lee, J.-W.; Song, J.; Hwang, M.P.; Lee, K.H. Nanoscale bacteriophage biosensors beyond phage display. Int. J. Nanomed. 2013, 8, 3917–3925. [Google Scholar] [CrossRef]
- Chen, I.-C.; Chiu, Y.-K.; Yu, C.-M.; Lee, C.-C.; Tung, C.-P.; Tsou, Y.-L.; Huang, Y.-J.; Lin, C.-L.; Chen, H.-S.; Wang, A.H.-J.; et al. High throughput discovery of influenza virus neutralizing antibodies from phage-displayed synthetic antibody libraries. Sci. Rep. 2017, 7, 14455. [Google Scholar] [CrossRef]
- Yuan, A.Q.; Zhao, L.; Bai, L.; Meng, Q.; Wen, Z.; Li, Y.; Guo, D.; Zhen, S.; Chen, X.; Yang, J.; et al. Isolation of and Characterization of Neutralizing Antibodies to Covid-19 from a Large Human Naïve scFv Phage Display Library. bioRxiv 2020. [Google Scholar] [CrossRef]
- Kang, X.; Yang, B.-A.; Hu, Y.; Zhao, H.; Xiong, W.; Yang, Y.; Si, B.; Zhu, Q. Human Neutralizing Fab Molecules against Severe Acute Respiratory Syndrome Coronavirus Generated by Phage Display. Clin. Vaccine Immunol. 2006, 13, 953–957. [Google Scholar] [CrossRef]
- Yu, H.; Jiang, L.-F.; Fang, D.-Y.; Yan, H.-J.; Zhou, J.-J.; Zhou, J.-M.; Liang, Y.; Gao, Y.; Zhao, W.; Long, B.-G. Selection of SARS-Coronavirus-specific B cell epitopes by phage peptide library screening and evaluation of the immunological effect of epitope-based peptides on mice. Virology 2007, 359, 264–274. [Google Scholar] [CrossRef]
- Karch, C.P.; Burkhard, P. Vaccine technologies: From whole organisms to rationally designed protein assemblies. Biochem. Pharmacol. 2016, 120, 1–14. [Google Scholar] [CrossRef]
- Slifka, M.K.; Amanna, I.J. Role of Multivalency and Antigenic Threshold in Generating Protective Antibody Responses. Front. Immunol. 2019, 10, 956. [Google Scholar] [CrossRef]
- Tao, P.; Mahalingam, M.; Marasa, B.S.; Zhang, Z.; Chopra, A.K.; Rao, V.B. In Vitro and In Vivo delivery of genes and proteins using the bacteriophage T4 DNA packaging machine. Proc. Natl. Acad. Sci. USA 2013, 110, 5846–5851. [Google Scholar] [CrossRef]
- Hayes, S.; Gamage, L.N.A.; Hayes, C. Dual expression system for assembling phage lambda display particle (LDP) vaccine to porcine Circovirus 2 (PCV2). Vaccine 2010, 28, 6789–6799. [Google Scholar] [CrossRef]
- Cano, P.G.; Gamage, L.N.A.; Marciniuk, K.; Hayes, C.; Napper, S.; Hayes, S.; Griebel, P.J. Lambda display phage as a mucosal vaccine delivery vehicle for peptide antigens. Vaccine 2017, 35, 7256–7263. [Google Scholar] [CrossRef]
- Razazan, A.; Nicastro, J.; Slavcev, R.; Barati, N.; Arab, A.; Mosaffa, F.; Jaafari, M.R.; Behravan, J. Lambda bacteriophage nanoparticles displaying GP2, a HER2/neu derived peptide, induce prophylactic and therapeutic activities against TUBO tumor model in mice. Sci. Rep. 2019, 9, 2221. [Google Scholar] [CrossRef]
- Shivachandra, S.B.; Li, Q.; Peachman, K.K.; Matyas, G.R.; Leppla, S.H.; Alving, C.R.; Rao, M.; Rao, V.B. Multicomponent anthrax toxin display and delivery using bacteriophage T4. Vaccine 2007, 25, 1225–1235. [Google Scholar] [CrossRef]
- Shivachandra, S.B.; Rao, M.; Janosi, L.; Sathaliyawala, T.; Matyas, G.R.; Alving, C.R.; Leppla, S.H.; Rao, V.B. In vitro binding of anthrax protective antigen on bacteriophage T4 capsid surface through Hoc–capsid interactions: A strategy for efficient display of large full-length proteins. Virology 2006, 345, 190–198. [Google Scholar] [CrossRef]
- Teschke, C.M.; Parent, K.N. ‘Let the phage do the work’: Using the phage P22 coat protein structures as a framework to understand its folding and assembly mutants. Virology 2010, 401, 119–130. [Google Scholar] [CrossRef]
- Patterson, D.P.; Rynda-Apple, A.; Harmsen, A.L.; Harmsen, A.G.; Douglas, T. Biomimetic Antigenic Nanoparticles Elicit Controlled Protective Immune Response to Influenza. ACS Nano 2013, 7, 3036–3044. [Google Scholar] [CrossRef]
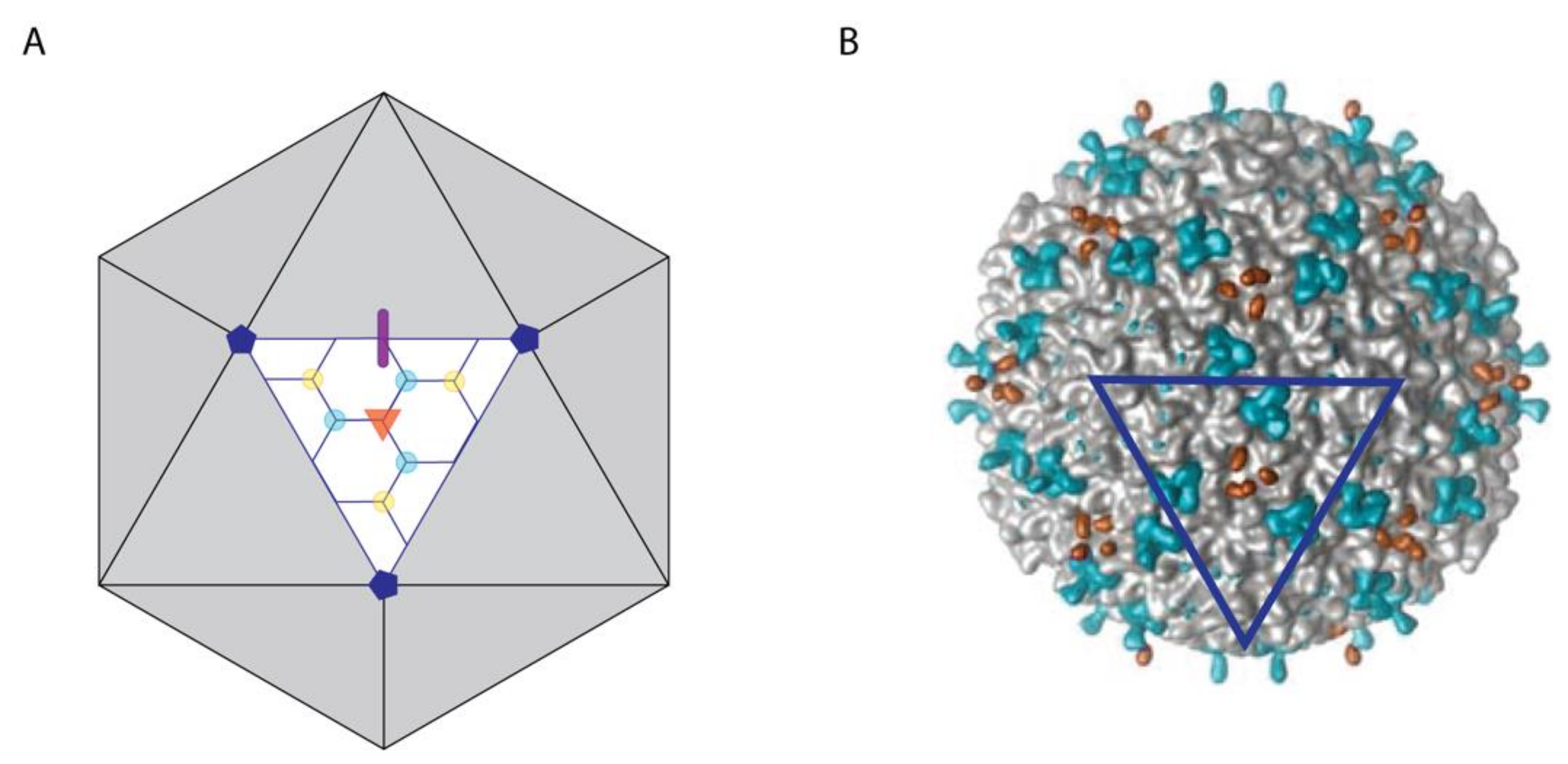
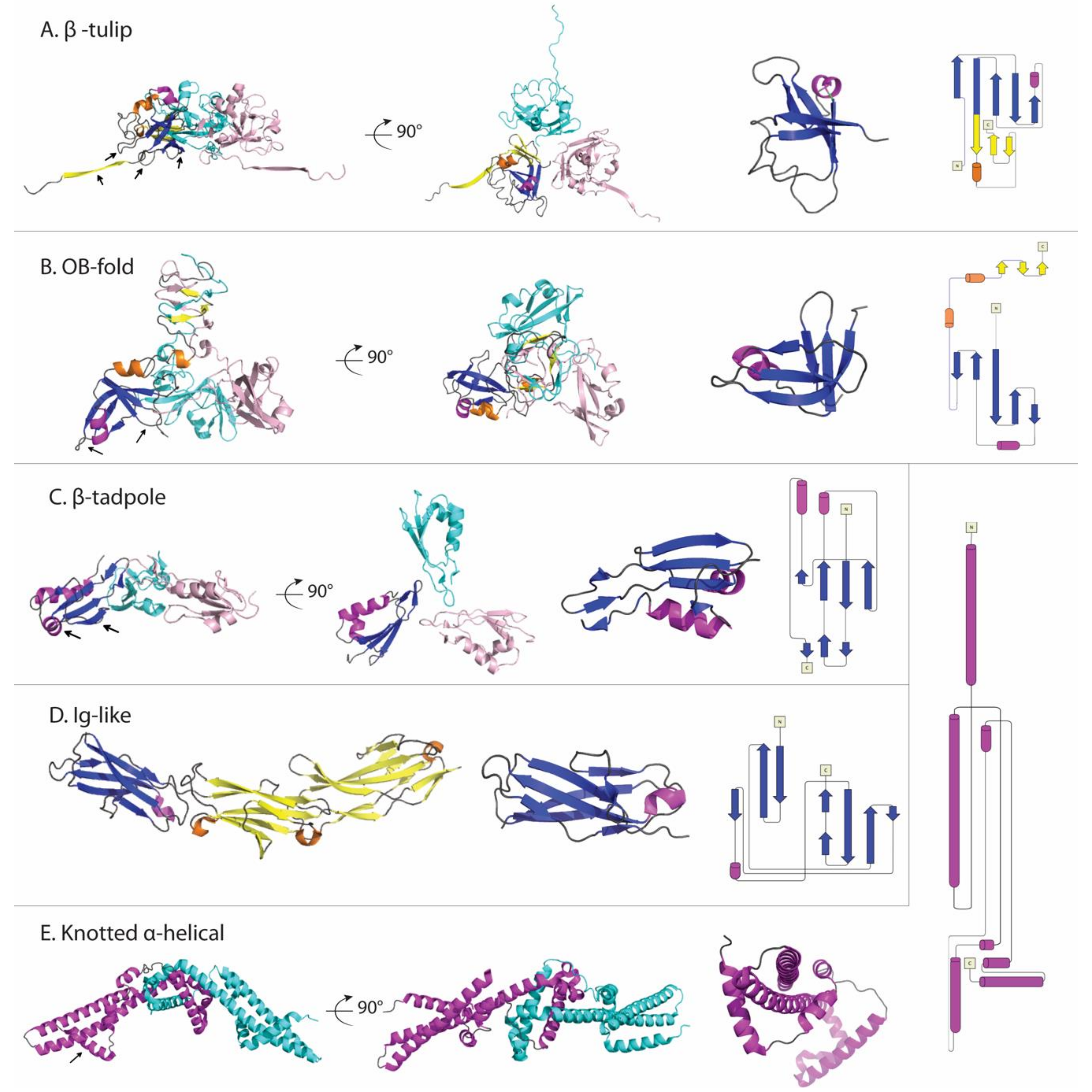
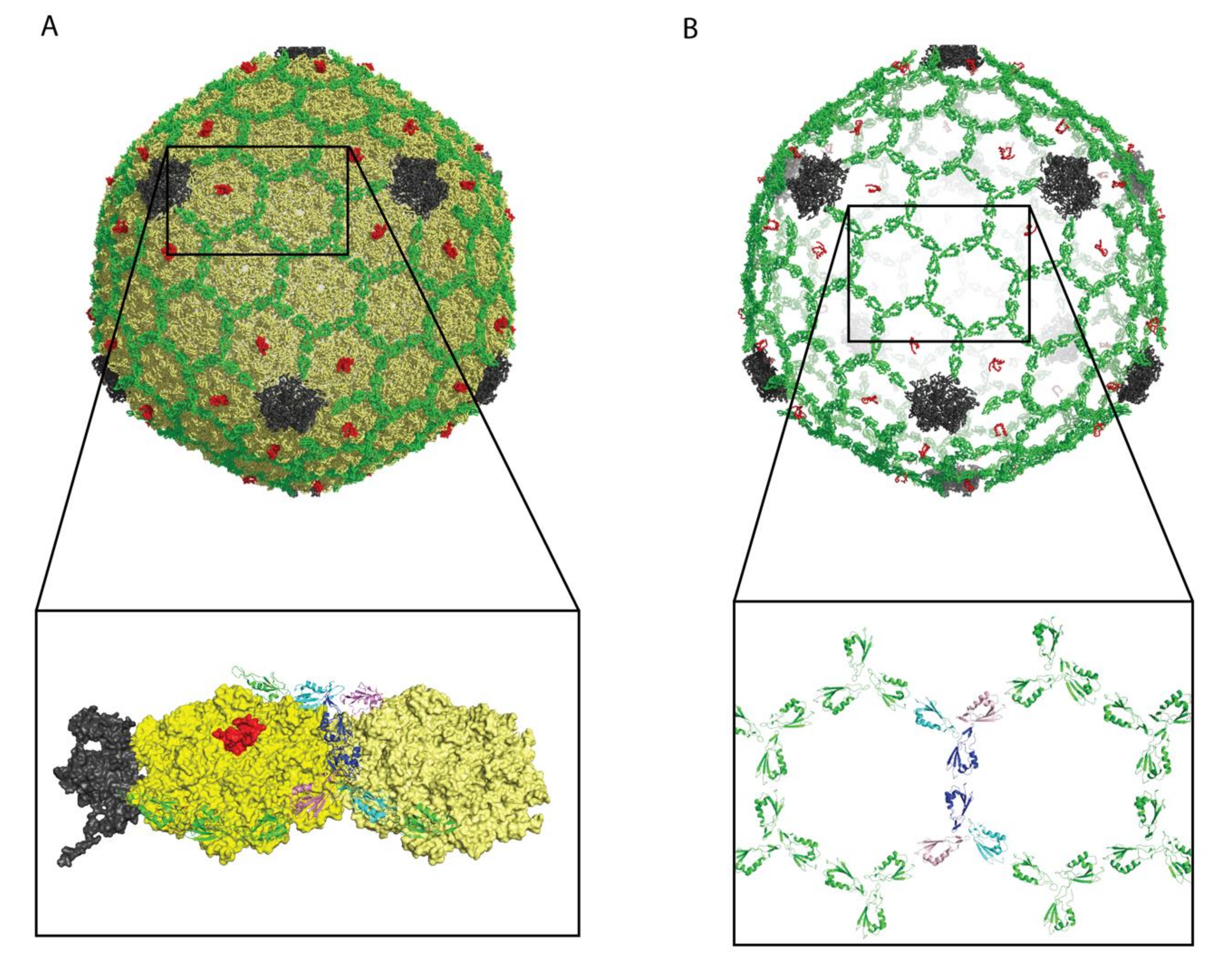
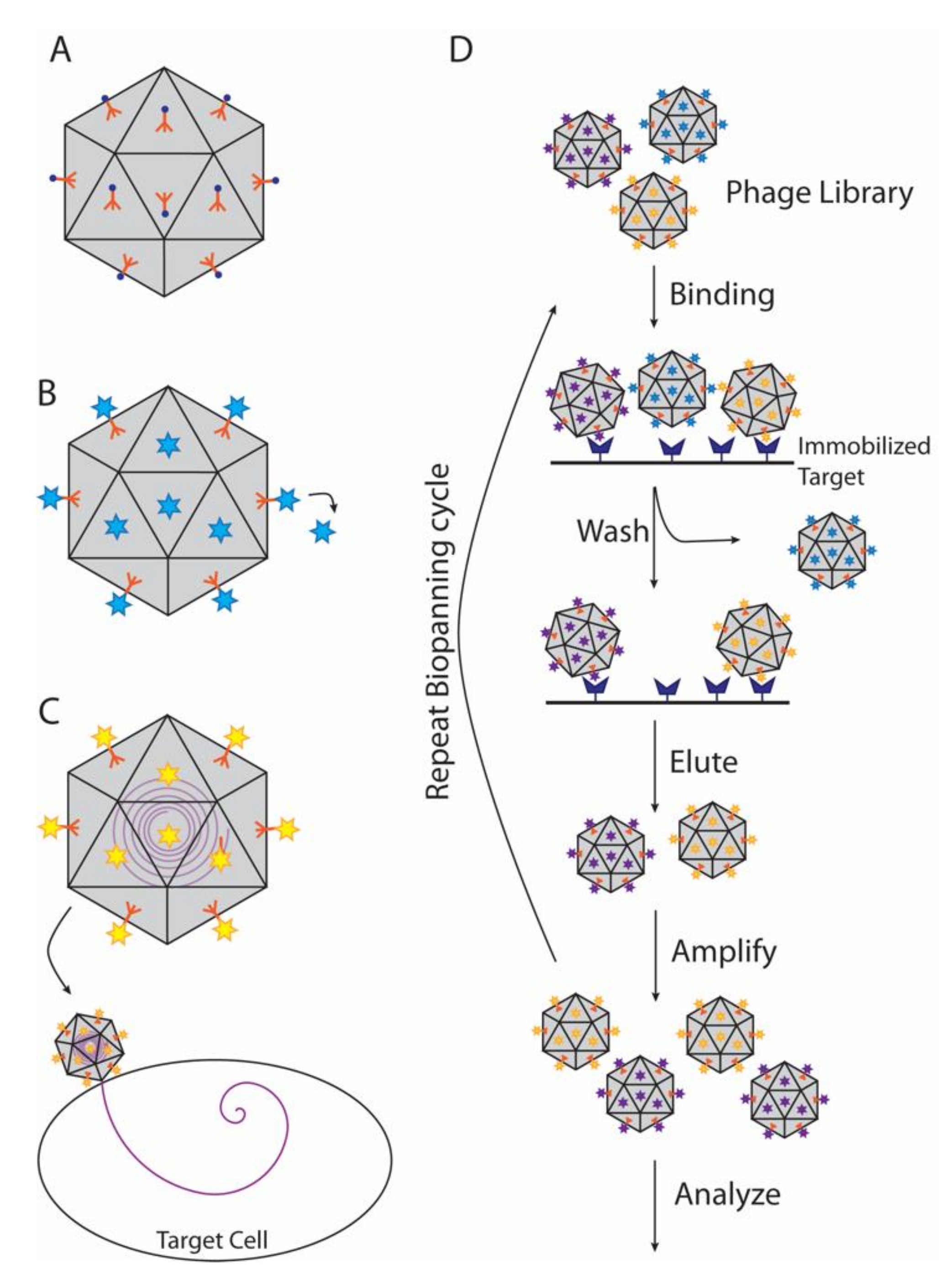
| Protein (Phage/Virus) | Host Organism | Structural Properties | Capsid Oligomer b | Binding Symmetry c | Functions | Refs |
|---|---|---|---|---|---|---|
| gpD (λ) | E. coli | β-tulip | trimer (monomer) | 3F | stability, assembly | [14,48,49,50,51] |
| SHP (21) | E. coli | trimer (trimer) | 3F | stability | [52] | |
| gp56 (TW1) | P. phenolica | trimer | q3F | stability | [15,53] | |
| gp87 (P74-26); gp88 (P23-45) | T. thermophilus | trimer | 3F | stability | [17,18] | |
| YSD1_16 (YSD1) | S. typhimurium | trimer | 3F | stability | [54] | |
| Tri1,2a,2b (HCMV) | H. sapiens | trimer | 3F | stability, assembly | [21,22,23] | |
| VP19c,23 (HSV-1) | H. sapiens | trimer | 3F and q3F | [21,22,23] | ||
| Dec (L) | S. enterica | OB-fold | trimer (monomer) | 3F & q3F | stability, host adhesion | [13,47,55,56,57] |
| Soc (T4); Soc (RB69) | E. coli | β-tadpole | trimer (monomer) | q2F and q3F | stability | [39,58] |
| Hoc (T4) | E. coli | Ig-like | monomer | q6F | host adhesion, phage dispersal | [36,38,59,60,61,62] |
| pb10 (T5) | E. coli | monomer | q6F | stability, host adhesion | [16,63] | |
| gp17 (N4) | E. coli K12 | monomer | q3F | stability, host adhesion | [19] | |
| Psu (P4) | E. coli | Knotted α-helical | dimer | q6F | stability, host transcription modulation | [64,65] |
| gp8.5 (φ29) | B. subtilis | multi-domain | trimer | q3F | host adhesion | [40] |
| gp12 (SPP1) | B. subtilis | collagen-like (predicted) | trimer | q6F | host adhesion | [66,67,68] |
| IIIa (Adenovirus) | H. sapiens | 4-helix bundle | complex | 5F | stability, capsid ‘tape-measure’ | [12,24,25,26,69] |
| IX (Adenovirus) | triskelion | complex | 3F | stability | ||
| VI (Adenovirus) | helical core, IDP a termini | complex | q6F | stability, endosome escape | ||
| VIII (Adenovirus) | IDP core | complex | 3F and 5F | stability | ||
| P30 (PRD1) | Broad host specificity | extended | dimer | 2F | stability, capsid ‘tape-measure’ | [70,71] |
| P2 through P14(PCBV-1) | C. variabilis | variable | hexagonal lattice | variable | stability, capsid ‘tape-measure’ | [37,72] |
| gp10 (ε15) | S. anatum | β-jellyroll (predicted) | dimer | 2F | stability | [73] |
| Fold | Example | PDB File | PDB-Blast Relatives b | DALI Phage/Virus Homologs c | Host: Host Homologs d |
|---|---|---|---|---|---|
| β-tulip | gpD (λ) | 1C5E | 1TD0, SHP (P21) | 6QYY, gp8.5 (φ29) 3SUC, φ29 preneck appendage | E. coli: 1C5E→ 1XI8, MoeA molybdenum biosynthesis |
| gp87 (P74-26) | 6BL5 | 6I9E-H, gp88 (P23-45) | 6XGP, YSD1_17 major capsid protein 6QYY, gp8.5(φ29) 3SUC, φ29 preneck appendage 6PPB-B, KHSV capsid vertex component | T. thermophilus: 3SUC → NHK40118.1, hypothetical protein | |
| gp8.5 (φ29) | 6QYY | None | 2JES-A, SPP1 portal protein 6BL5, gp87 (P74-26), gp88 (P23-45) 1CE5, gpD(λ) | B. subtilis: 2JES-A → WP_075218525.1, hypothetical protein | |
| OB-fold | Dec (L) | 6E3C | None | 3QR8, P2 membrane piercing | S. enterica: 6E3C→ 2OT2, chaperone (E. coli homolog) |
| β-Tadpole | Soc (T4) | 3IGE | 3IG9, Soc (RB69) | 5VF3-A, T4 capsid vertex protein gp24 | E. coli: 3IG9→ 2MCF-A, unknown function |
| Ig-like | Hoc (T4) | 3SHS | 5LXK, pb10 (T5) | 6PCI-H, ebola spike glycoprotein 6C6Q-F, norovirus VP1 capsid protein 6URH-H, hepatitis C envelope glycoprotein | E. coli: 6PCI-H → WP_168428099, hypothetical protein |
| knotted α-helix | Psu (P4) | 3RX6 | None | 1FAV-A, HIV gp41 envelope protein | E. coli: 3RX6→3AJW-A, flagellar fusion protein |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dedeo, C.L.; Teschke, C.M.; Alexandrescu, A.T. Keeping It Together: Structures, Functions, and Applications of Viral Decoration Proteins. Viruses 2020, 12, 1163. https://doi.org/10.3390/v12101163
Dedeo CL, Teschke CM, Alexandrescu AT. Keeping It Together: Structures, Functions, and Applications of Viral Decoration Proteins. Viruses. 2020; 12(10):1163. https://doi.org/10.3390/v12101163
Chicago/Turabian StyleDedeo, Corynne L., Carolyn M. Teschke, and Andrei T. Alexandrescu. 2020. "Keeping It Together: Structures, Functions, and Applications of Viral Decoration Proteins" Viruses 12, no. 10: 1163. https://doi.org/10.3390/v12101163
APA StyleDedeo, C. L., Teschke, C. M., & Alexandrescu, A. T. (2020). Keeping It Together: Structures, Functions, and Applications of Viral Decoration Proteins. Viruses, 12(10), 1163. https://doi.org/10.3390/v12101163






