Abstract
Decoration proteins are viral accessory gene products that adorn the surfaces of some phages and viral capsids, particularly tailed dsDNA phages. These proteins often play a “cementing” role, reinforcing capsids against accumulating internal pressure due to genome packaging, or environmental insults such as extremes of temperature or pH. Many decoration proteins serve alternative functions, including target cell recognition, participation in viral assembly, capsid size determination, or modulation of host gene expression. Examples that currently have structures characterized to high-resolution fall into five main folding motifs: β-tulip, β-tadpole, OB-fold, Ig-like, and a rare knotted α-helical fold. Most of these folding motifs have structure homologs in virus and target cell proteins, suggesting horizontal gene transfer was important in their evolution. Oligomerization states of decoration proteins range from monomers to trimers, with the latter most typical. Decoration proteins bind to a variety of loci on capsids that include icosahedral 2-, 3-, and 5-fold symmetry axes, as well as pseudo-symmetry sites. These binding sites often correspond to “weak points” on the capsid lattice. Because of their unique abilities to bind virus surfaces noncovalently, decoration proteins are increasingly exploited for technology, with uses including phage display, viral functionalization, vaccination, and improved nanoparticle design for imaging and drug delivery. These applications will undoubtedly benefit from further advances in our understanding of these versatile augmenters of viral functions.
1. Introduction
Viruses and bacteriophages (phages) have their genetic material enveloped by membranes or encapsulated in proteinaceous shells called capsids. The stabilities of the frameworks that harbor the nuclei acid genomes are crucial for the successful assembly and infectivity of phages and viruses [1]. Tailed phages (Caudoviralaes) together with related viruses including herpesviruses, adenoviruses, and some archaeal and giant viruses, initially assemble immature meta-stable structures called procapsids. These undergo irreversible conformational changes upon DNA packaging to become mature capsids. An increase in volume often accompanies the process, and requires stabilization of the capsid against the buildup of internal pressure that results from genome packaging [2,3,4,5]. Stabilization of some capsids is achieved through amino acid modifications that covalently cross-link coat protein subunits, others make use of interactions between genetically inserted auxiliary domains within coat proteins (I-domains), while some have their capsids stabilized by accessory proteins [2,4,6,7,8,9].
Accessory proteins are broadly defined as virally encoded proteins that do not have a role in virus replication but rather modify the properties of the virion [10]. These proteins usually bind capsids in the final stages or after completion of viral assembly, and play a variety of roles in the infection cycle, including stabilizing the expanded capsid and modulating host interactions [11]. Among the accessory proteins are decoration [11] and cementing proteins [12]. The nomenclature has become largely interchangeable; although in the original definitions decoration proteins were designated as binding to the surfaces of mature virions [11,13], whereas cementing proteins had the additional feature of stabilizing the virion upon binding [12]. The distinction is tenuous because it presupposes measuring the effects of the accessory protein on virion stability, which is often more difficult than identifying a viral surface protein. Indeed, many of the decoration proteins for which data are available such as Dec (L) [13], gpD (λ) [14], gp56 (TW1) [15], pb10 (T5) [16], gp87 (P74-26) [17], gp88 (P23-45) [18] and Soc (T4) [19] stabilize viruses, making them operationally indistinguishable from cementing proteins. We therefore use the term “decoration proteins” for this review. For brevity, we use a naming scheme in which the decoration protein is listed first, followed by the virus that encodes it in parentheses. For example, gpD (λ) refers to the decoration protein gpD from phage λ.
To date, decoration proteins are only known to occur in viruses with DNA genomes [17]. Within the DNA viruses, they are common and span many genera including tailed phages (Caudovirales) [13,15,20], herpesviruses [21,22,23], adenoviruses [12,24,25,26], as well as archaeal viruses [17,27,28], and giant viruses [29,30,31]. They appear to be particularly common in tailed phages, which “inject” their DNA genomes, and are thus under no selective pressure to break their capsids during infection [28]. The ubiquity of decoration proteins might be underestimated. For many viruses, surface protrusions may be initially imaged only at low-resolution, so that the presence of decoration proteins could be missed until the virus is better characterized. The capsid structures of three actinobacteriophages, discovered through a collaboration with the SEA-PHAGES (Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science) program, were recently resolved to approximately 6 Å by cryo-EM. Of these phages, two had novel decoration proteins with no known sequence homologs [32]. Similarly, 7 of 16 newly discovered Shigella-infecting phages had a novel decoration protein [33]. Finally, also recently described are structures of the jumbo phages G at 6 Å [34] and ΦRSL2 (16 Å) [35], and the semi-jumbo phage ΦRP13 (9.5 Å) [35], each of which have decoration proteins, though the resolution of these reconstructions precludes knowing if they are unique decoration proteins. Although this abundance of decoration proteins may be due to their prevalence in nature, it could also reflect a partial bias in collection and isolation methods, as phages with decoration proteins are often more resistant to harsh environmental conditions.
Since a comprehensive review of decoration proteins is a daunting task, we focus here on the subset that have high-resolution structures and well-established functions. Much of our knowledge about decoration proteins owes a large debt to the groundbreaking work of Michael Rossmann [15,19,36,37,38,39,40], whose contributions and accomplishments are celebrated in this special issue of Viruses. In this review, we discuss the versatile functions of decoration proteins, classify and analyze their structures and virus binding sites, and explore the opportunities these proteins present for nanotechnology applications.
2. Functions of Decoration Proteins
Although most decoration proteins contribute to capsid stability and maturation, some have additional roles such as mediating viral targeting of host cells. The functional properties of decoration proteins are reviewed in this subsection with an overview provided in Table 1.

Table 1.
Properties and functions of decoration proteins.
2.1. Stabilization of Capsids by Decoration Proteins
A common function of decoration proteins is to increase virus stability. The phage HK97 (Hong Kong 97) coat protein fold is prevalent across dsDNA viruses [41]. The commonality of this structural motif is thought to be due to its ability to form a variety of icosahedral and prolate capsids, spanning a range of sizes [31,42]. The prototypical example, from phage HK97, features covalent cross-links between capsomers across icosahedral 3-fold symmetry axes, as well as between subunits within capsomers. [2,42,43]. This “chainmail” of covalent links reinforces the capsid against internal pressure. The dsDNA genome of HK97 is packaged to liquid crystalline density, with the ensuant pressure comparable to that of a pressurized champagne bottle [14,44,45].
To withstand internal pressure from genome packaging or external environmental insults, viruses lacking the ability to form cross-links have evolved several strategies to strengthen their capsids, including encoding stabilizing decoration proteins [6,14,46]. In some cases, such as phage λ, decoration proteins are required for maturation but the majority of decoration proteins only function to stabilize capsids, suggesting they may confer a selective advantage only under conditions of viral stress [14,47].
2.1.1. Decoration Proteins are Required for the Assembly of Some Viruses
An example of a decoration protein necessary for phage assembly is gpD (λ). gpD (λ) binds to expanded capsids of λ phage during the last stages of DNA packaging, to stabilize them while the last 10–20% of the genome needed for maturation is incorporated [49,74,75]. The cementing function of gpD (λ) in λ phage has been suggested to substitute for the covalent cross-links that form the “chainmail” structure of HK97 phage [14,49]. Atomic force microscopy (AFM) experiments indicate that gpD (λ) additionally reinforces the λ virus against mechanical pressure and collisions [48]. gpD (λ) is related to decoration proteins from other λ-like phages, including SHP (21), which shares 49% sequence identity. Interestingly, SHP (21) can bind to the λ capsid as a chimeric oligomer with gpD (λ), producing particles with varying stabilities [76]. Decoration proteins gp56 (TW1), gp87 (P74-26) along with its close relative gp88 (P23-45), and gp8.5 (φ29), as well as the Herpes Simplex Virus-1 (HSV-1) VP23/VP19C and Human Cytomegalovirus (HCMV) Tri1/Tri2 triplex proteins [21,22,23], share both the β-tulip fold and capsid-stabilizing function of gpD (λ). If these related proteins are also required for the assembly of their respective phages and viruses is currently unknown [15,17,40,77].
Herpesviruses share similarities with dsDNA phages that extend to structures and assembly mechanisms [78]. The coat protein of herpesviruses is based on the HK97-fold, and triplex proteins have the same β-tulip fold found in decoration proteins such as gpD (λ), gp87 (P74-26) and gp88 (P23-45). Similar to gpD (λ), the triplex proteins are necessary for capsid assembly but because of the larger genome, additional proteins are also used to reinforce herpesvirus capsids [79,80,81].
2.1.2. Some Decoration Proteins Provide Stability but Are Not Required for Infectivity
Although some decoration proteins are necessary for virus maturation, others function primarily to boost stability. In contrast to gpD (λ), Dec (L) is not required for phage infectivity. In addition to its natural substrate phage L, Dec (L) can also noncovalently bind and stabilize expanded heads or mature capsids of phage P22 in vitro and in vivo [13,47]. This occurs because the coat proteins of phages L and P22 are highly homologous, differing in only 4 out of 430 positions (99.6% identical) [13,57]. P22 is often substituted as a model for phage L, owing to its extremely well-characterized genetics and biochemistry [47]. Dec (L) stabilizes phage P22 in the presence of EDTA, a chelator that binds Mg2+. In the absence of Mg2+, the dsDNA genome of P22 undergoes decondensation that causes the phages to burst because of the increase in internal pressure. Without Dec (L), 90% of P22 phages are destroyed in the presence of EDTA. By contrast, EDTA has no effect on P22 phages when these are bound by Dec (L) [13]. Despite not being essential for phage viability, Dec (L) clearly plays a role in stabilizing the phage L capsid against internal pressure, and may facilitate the survival of the phage in harsh environments.
Similar to Dec (L), gp17 (N4) does not affect infectivity but offers stability under harsh conditions, including exposure to DNase I and ETDA [19]. Soc (T4) found in T4 and T4-like phages, together with pb10 (T5) [16] and gp10 (ε15) [73] are expendable for maturation and infectivity but stabilize their respective capsids in their matured states. Pb10 (T5) prevents DNA leakage under low ionic strength conditions, while Soc(T4) stabilizes the capsid against temperature and pH extremes [16,39,58].
Taken together, raised stability appears to be a key function for decoration proteins. In cases where decoration proteins are expendable for phage viability, their stabilizing effects are likely beneficial when the virus is subjected to stress [16,47,58,59].
2.2. Multifunctional Decoration Proteins
Viruses are under selective pressure to economize their genomes. Addition of a stabilizing protein could require a larger capsid to accommodate the concomitant increase in genetic material encoding the new protein. Consequently, it is perhaps not surprising that decoration proteins are often multifunctional, augmenting their roles in capsid stabilization with additional moonlighting functions that include acting as viral tape-measures for capsid size or serving as mRNA transcription anti-terminators.
2.2.1. Decoration Proteins that Act as ‘Tape-Measures’ to Determine Virus Size
Phage PRD1 is evolutionarily related to complex viruses such as adenovirus and the giant virus PCBV-1 [37,82,83]. All these feature decoration proteins that function as both cementing stabilizers and “tape-measure” regulators of capsid size.
The decoration protein P30 (PRD1) is necessary for phage maturation and additionally forms a cage-like structure that anchors the capsid to the internal membrane encapsulating the genome [70,71]. P30 (PRD1) modulates the capsid transformations required for assembly, and has been compared to both scaffolding and tape-measure proteins, as it governs both nucleation and capsid size determination [70].
Adenovirus features four cementing proteins (IIIa, VI, VIII, and IX) that are proteolytically cleaved during the virus maturation process. The primary function of all four proteins is to buttress capsid stability by connecting capsomers both internally and externally. Some of the four proteins play additional roles in the infection cycle [12,24]. IIIa (adenovirus) has been suggested to act as a tape-measure protein, similar to P30 (PRD1) [25]. The mature form of VI (adenovirus) has an N-terminal amphipathic β-helix that allows the virus to escape from endosomes during the infection process [24].
The giant Paramecium bursaria chlorella virus, PCBV-1, has a staggering 13 minor capsid proteins (P2–P14) that provide structural stability. The minor capsid proteins cement capsomers in triangular and pentameric arrangements, known as symmetrons [37]. In addition, the minor capsid proteins P12, P13, and P14 anchor the external capsid to the internal membrane encapsulating the nucleocapsid [37,72]. The PCBV-1 minor protein P2, in addition to providing structural support, acts as a tape-measure that controls the size of the giant PCBV-1 virus [37].
2.2.2. The Psu Decoration Protein Moonlights as a Transcription Antiterminator
Polarity suppression protein Psu (P4) is a non-essential but multifunctional decoration protein in phage P4. On the capsid surface, Psu (P4) forms V-shaped dimers that cover structural gaps at the centers of hexons in the icosahedral lattice [64]. The dimerization interface of Psu (P4) forms a knotted and highly hydrophobic structure that likely imparts the decoration protein-capsid complex significant tensile strength [64]. In the infected cell, Psu (P4) additionally functions as a transcription antiterminator by inhibiting the host transcription termination factor, Rho [64,65].
2.3. Participation of Decoration Proteins in Host Attachment
Several decoration proteins, including Hoc (T4), pb10 (T5), gp12 (SPP1), and gp8.5 (φ29), contribute little to capsid structural integrity but rather enhance binding to both host and non-host cell surfaces through interactions with carbohydrates [40,60,61,63,66,67]. Mediation of these interactions by decoration proteins can serve two purposes. First, the decoration proteins can concentrate phages to the bacterial target cells, aiding their infectivity [84], or conversely disperse phages through electrostatic surface repulsion preventing their aggregation [38]. Second, binding of decoration proteins to glycans emanating from the mucus layers of metazoan cells may facilitate a symbiotic form of non-host innate immunity [85,86]. In animal cells, the mucus layer, which is rich in complex glycoproteins and antimicrobial compounds, is part of the innate immune system that forms the first line of defense against infection [85,87]. Phages can aggregate in the mucus layer via decoration protein mediated glycan binding, where they can protect animal cells by lysing invading bacteria [38,84,85,86,88].
2.3.1. Decoration Proteins with Ig-Like Domains Can Participate in Host Adhesion
A bioinformatics survey showed that 25% of tailed dsDNA phages encoded proteins with predicted immunoglobulin-like (Ig-like) β-sandwich folding motif [89]. These domains were found in five functional classes: tail fibers, baseplate wedge initiators, major tail components, major head components (such as coat protein insertion domains), and decoration proteins [89]. Highly immunogenic outer capsid proteins, such as Hoc (T4), pb10 (T5), and gp17 (N4) share an Ig-like fold, and are thought not to contribute significantly to capsid stability but instead to mediate host cell adhesion [19,38,61,63]. T4 particles missing Hoc (T4) tend to aggregate at low cation concentrations, indicating that the decoration protein could also be important for dispersal of viral particles when the host cell density is low [38].
2.3.2. Head Fibers May Coordinate Cell Attachment
Like the Hoc (T4) decoration protein, head fibers do not affect capsid stability or infectivity of the φ29 phage [40]. Phage φ29 particles decorated with head fibers, however, interact in an ordered fashion with host cell bacterial walls compared to fiber-less phages. This suggests head fibers may aid infectivity under conditions of low viral presence [40]. Although most decoration proteins are rich in β-sheet structure, the φ29 head fiber is comprised of an elongated α-helical coil-coiled, similar to that found in the T4 fibritin and P22 tail needle structures [40]. As these proteins are important for attachment to target cells, it is possible that the φ29 head fiber is evolutionarily related to them. Likewise, some proteins predicted to be collagen-like, such as those in the elongated trimeric spike gp12 (SPP1), may be important for cell surface recognition [66,67,68].
3. Decoration Protein Structures
Decoration proteins show considerable structural versatility both in terms of the folds they adopt as well as their capsid-binding mechanisms. In this subsection we review the main structural motifs of decoration proteins, their capsid-binding modes, and possible evolutionary relationships suggested by structural homology.
3.1. Capsid-Binding Modes and Oligomerization States
Decoration proteins bind on the surfaces of viruses as exemplified by the cryo-EM image of Dec (L) in Figure 1. As such, decoration proteins can have significant effects on the ruggedness and patterning of the viral surface [9], which in turn can affect virus recognition, including interactions with target cells or host defenses.
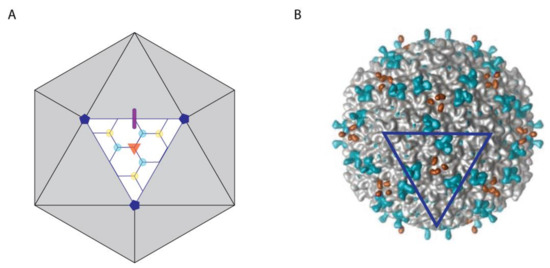
Figure 1.
Symmetry of capsid-binding sites for decoration proteins. (A) Model of a T = 7 icosahedral capsid with symmetry sites highlighted on one facet. The 2-, 3-, and 5-fold symmetry axes are shown by a thick purple line, an orange triangle, and dark blue pentagons, respectively. Additionally, two types of quasi-3-fold axes are indicated. The first connects only hexons and is shown by cyan dots, the second connects two hexons and a penton and is shown using yellow dots. (B) Surface map of the phage L capsid bound by the decoration protein Dec (L) at 3-fold (orange) and type I quasi-3-fold (cyan) sites, corresponding to those illustrated in panel A. The dark blue triangle outlines a facet in the same orientation as in panel A. The figure is adapted from Tang et al. [47].
3.1.1. Decoration Proteins Bind to a Variety of Symmetry and/or Pseudo-Symmetry Axes
The icosahedral frameworks of spherical and prolate capsids have a basis set of 2-, 3-, and 5-fold symmetry axes, as summarized in the schematic of Figure 1A. In addition to these true symmetry axes, there exist quasi-three-fold sites [57]. The first type occurs between hexamers on icosahedral facets as indicated by the cyan dots in Figure 1A. A second type of quasi-three-fold site lies between pentons and hexons surrounding each vertex, as indicated by yellow dots Figure 1B. The differences between three-fold (orange) and quasi-three-fold (cyan, yellow) sites is illustrated with the cryo-EM [47] surface map of Dec (L) bound to phage L in Figure 1B. A summary of capsid-binding-site symmetries for different types of decoration proteins is given in Table 1.
Most decoration proteins bind to three-fold and quasi-three-fold symmetry axes [14,15,28]. These are the sites reinforced by covalent cross-links in the HK97 capsid [41], and thought to correspond to weak points in the icosahedral lattice [28,90]. Consistently the three-folds sites, which occur between the icosahedral hexamers, have been shown to be mechanical weak points susceptible to bursting at increased internal DNA pressure in modeling studies [91]. There are, however, decoration proteins with alternative preferential binding sites. Thus, P30 (PRD1), for example, is a proline-rich, mostly disordered, extended protein that when capsid-bound dimerizes through an N-terminal hook at the icosahedral 2-fold axis of symmetry [70]. The network of dimers forms a chainmail-like structure surrounding the capsid ~12 Å above the viral membrane [70].
Several decoration proteins bind at both true and quasi-symmetry sites, or prefer one type of site over others. For example, Dec (L) binds type I quasi-three-fold sites between hexons 1000 times more strongly than true three-folds [92]. Structural data from cryo-EM suggests that Dec (L) discriminates binding-site topologies by forming a larger number of contacts with the higher avidity quasi-three-fold site [57]. By contrast, trimers of YSD1_16 (YSD1) create a non-covalent chainmail-like structure that includes binding sites at both three-fold and quasi-three-fold symmetry axes [54]. Both Soc (T4) and Soc (RB69), from phage RB59 a close relative of T4, bind their respective capsid as trimers. The tadpole-like heads of the Soc decoration proteins point to quasi-two-fold axes relating adjacent hexamers, and their tails are located near quasi-three-fold axes [39]. Trimers of gp8.5 (φ29) bind the mature capsid at quasi-3-fold axes of symmetry where they interact with the Ig-like coat protein insertion domain, BIG2 [77]. The elongated decoration protein gp17 (N4) comprised of three Ig-like domains [19], binds as a monomer to both types of quasi-three-fold axes (cyan and yellow in Fig 1B). By contrast, the decoration protein Hoc (T4), which also has an elongated shape consisting of three Ig-like domains and an N-terminal capsid-binding domain, binds as a monomer preferentially to the quasi-6-fold axis at the center of hexons [61]. Adding to the wide diversity of decoration protein structures are those that are α-helical, including Psu (P4) [64] and gp12 (SPP1) [66,67]. Both proteins feature coiled-coil structures that bind to the center of hexons, corresponding to a quasi-6-fold symmetry axis.
Rather than showing a unifying capsid-binding theme, the interactions between decoration proteins and capsids seem to be structurally opportunistic, employing a wide variety of binding modes that depend on the distinct structures of the decoration proteins and of the coat proteins that make up the cognate capsids.
3.1.2. Oligomerization of Some Decoration Proteins May Require Capsid Binding
The oligomerization states of capsid-bound decoration proteins are summarized in Table 1. Known examples include monomers, dimers, and trimers, with the latter being the most common. Most recent structural models of decoration proteins have come from cryo-EM studies, where only the capsid-bound state is determined. To have information on both the capsid-bound and -unbound structures is much rarer; however, these data exist for gpD (λ), SHP (21), Dec (L), and Soc (RB69). In three of four cases the decoration protein is a trimer when capsid-bound but can exist as a monomer in solution. Soc is a monomer in solution by analytical ultracentrifugation [62] and only becomes a trimer when capsid-bound [39]. Similarly, gpD (λ) is a monomer and only trimerizes on capsids, or in crystals [50,51]. Dec (L) forms a monomer in solution upon acidification that consists of a folded N-terminal domain and a disordered C-terminal tail [56]. The C-terminal tail is thought to fold into a three-stranded α-helix structure in the capsid-bound state [57], since if the tail is deleted Dec(L) can no longer bind capsids (ATA and CMT, unpublished observation). These observations raise the question of whether some decoration proteins only oligomerize in their capsid-bound states. By contrast SHP(21), which is homologous to gpD (λ), is a trimer in both solution and capsid-bound states [52].
3.2. Current Decoration Protein Structures Fall into Five Main Folding Motifs
Protein structure is arranged hierarchically. Segments of hydrogen-bonded secondary structure such as α-helices and β-strands coalesce into higher-order “super-secondary structure”—a concept originated by Michael Rossmann [93]. These super-secondary structure modules can govern tertiary folding topology, which often provides clues about the functions and evolutionary relationships of proteins. For example, the Rossmann-fold—one of the most ubiquitous protein folds in nature (named after Michael Rossmann)—is an α/β structure that is typically found in proteins and enzymes with nucleotide-binding functions [94,95].
The most common structural motifs of decoration proteins are summarized in Figure 2 and Table 2. Here we consider only those structures for which high-resolution models are available in the Protein Data Bank (PDB). Many of the decoration proteins structures were novel folds when they were first determined. Novel folds appear to be more commonly represented in viruses, possibly because the proteomes of viruses are less well studied and because viral genomes are subject to higher mutation rates, affording more structural innovation [96,97].
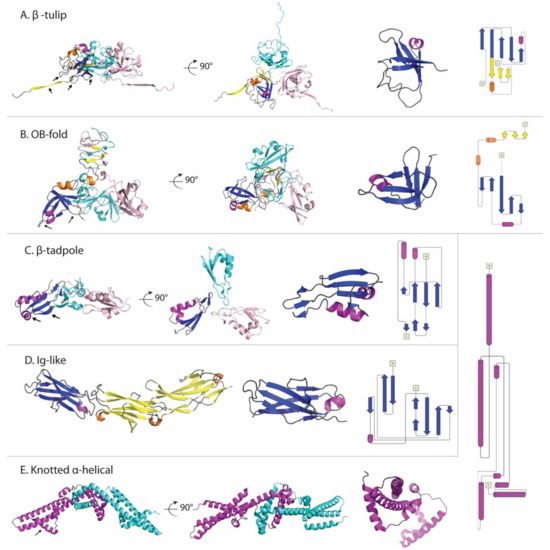
Figure 2.
Comparison of decoration protein structures. (A) β-Tulip fold illustrated by gp87 (P74-26), PDB 6O3H. (B) OB-fold in Dec (L), PDB 6E3C. (C) β−Tadpole fold in Soc (T4), PDB 3IG9. (D) Ig-like fold exemplified by Hoc (T4), PDB 3SHS. (E) Knotted α-helix fold shown by Psu (P4), PDB 3RX6. The first two views in each panel are related by a 90° x-axis rotation. The first is parallel to the capsid surface, with the bottom of each structure corresponding to the parts of the protomer (denoted by arrows) involved in contacting the capsid surface. The second view is looking down towards the surface of the capsid. The last two panels show the protein fold, and a corresponding topology diagram. For each structure except the last, one protomer is colored with blue and purple indicating β-strands and α-helices within the conserved fold, while yellow and orange highlight β-strands and α-helices in non-conserved structure.

Table 2.
Decoration Protein Structural Homology a.
3.2.1. The β-Tulip Motif Has Three Subfamilies
The first high-resolution structure of a decoration protein was that of gpD (λ) [50]. At the time the gpD (λ) structure was a previously unobserved novel fold. The motif was named the “β-tulip” fold some 18 years later, in the context of the structurally related decoration protein gp87 (P74-26) [17]. Besides gpD (λ), gp87 (P74-26), gp88 (P23-45) and their structural homologs (Table 2), a third branch of the β-tulip fold family occurs in a domain of the head-fiber decoration protein gp8.5 (φ29) [98]. gp8.5 (φ29) has a complex elongated multi-domain structure, consisting of an N-terminal β-tulip “base” domain that contacts the capsid, and a C-terminal extended three-stranded helix-turn-helix supercoil that forms the “spike” domain emanating from the virus surface [98].
A β-tulip domain consists of a 5-stranded anti-parallel β-barrel with an α-helix intervening between strands 3 and 4. This fold is illustrated by the representative structure of gp87 (P74-26) in Figure 2A. To date most occurrences of the β-tulip folding motif have been found in virus proteins, with the exception of MoeA a molybdenum-binding protein from E. coli [17]. The β-tulip motif has a “bloom” side corresponding to the end of the barrel that is flared open, and a “stem” side at the opposite end that is capped by loops [17]. The β-tulip motif (blue and magenta in Figure 2A) is conjoined within a mixed α+β subdomain (yellow and orange in Figure 2A) to form a larger structure in the three subfamilies represented by gpD (λ), gp8.5 (φ29), gp87 (P74-26), and gp88 (P23-45) [17,18,51,98].
Each of the three β-tulip decoration protein subfamilies forms trimers in their capsid-bound states. The bloom side of the β-tulip interacts with the mixed α + β subdomain of the neighboring protomer to buttress the trimer [17]. Capsid-binding (illustrated by the arrows in Figure 2A) primarily involves the N-terminus of the decoration protein (‘Dec-arm’), which in the case of gp87 (P74-26) is disordered in the crystal structure but visible in the cryo-EM structure of the capsid-bound protein [28]. The N-arm of gp88 (P23-45) is also ordered when bound to the capsid [18]. This suggests the stabilization of the N-terminal segment is coupled to capsid binding. In addition to forming interactions with the capsid, the N-terminal Dec-arm also links neighboring trimers across the icosahedral three-fold and quasi-three-fold axes, forming an interlocked decoration protein chainmail surrounding the capsid [28].
3.2.2. Dec (L) Has an Oligonucleotide/Oligosaccharide-Binding (OB)-Fold
The OB-fold was initially identified in proteins with oligonucleotide or oligosaccharide-binding functions but now includes proteins with functions as varied as proteinase inhibitors, chemotaxis, and molybdenum-binding proteins [99,100,101]. The fold consists of a five-stranded Greek Key β-barrel (Figure 2B) that is closed by an anti-parallel connection between strands 1 and 4 and a short parallel connection between strands 3 and 5. Typically, an α-helix between strands 3 and 4, provides a hydrophobic plug residue for one side of the β-barrel. In the Dec (L) structure [56] the OB-fold is distorted so that strands 3 and 5 are too far apart to hydrogen bond, and the α-helix is displaced relative to the axis of the barrel. The capsid-binding site in Dec (L) is comprised of the α-helix between strands β3 and β4 and two clasp-like prongs formed by the hairpin loops linking strands β1-β2 and β4-β5 (arrows in Figure 2B).
It is interesting to note that the OB-fold and β-tulip motifs are similar. Both are anti-parallel five-stranded β-barrels with an α-helix between strands β3 and β4 (Figure 2A,B). The principal difference is in the hydrogen-bonded pairing of the β-strands. It is thus conceivable that the β-tulip and OB-fold could be evolutionarily related.
As in the previously discussed examples of β-tulip proteins, the OB-fold in Dec (L) is also part of a more elaborate structure. In the Dec (L) protomers, the OB-fold is flanked by a short N-terminal strand, a short C-terminal α-helix, and a long 40 residue disordered tail [56]. The folded parts of the protomers act as the legs of a tripod that sits on the capsid [56]. Trimerization exclusively involves the 40-residue C-terminal tail, which forms a three-stranded β-helix spike in the capsid-bound structure but is unfolded in the monomers. The globular OB-fold domains are too far from each other to account for any stabilizing contacts [56]. Both the OB-fold and α-helix parts of the structure could be potential binding sites for polysaccharides [57]. This is interesting because Dec (L) has been suggested to interact with bacterial cell surfaces [13], which could be mediated through carbohydrate binding.
3.2.3. Soc (T4) Has a Unique β-Tadpole Fold
The proteins Soc (T4) and Hoc (T4) simultaneously decorate the capsids of mature T4 and T4-like phages, with 870 Soc and 155 Hoc proteins per capsid. Both the Soc (T4) and Hoc (T4) structures were determined in Michael Rossmann’s lab [36,38,39,61]. The Soc (T4) structure was a novel fold [39]. The elongated fold called a ‘β-tadpole’ consists of a head subdomain, formed by an anti-parallel three-stranded β-sheet packed against two α-helices. A β-hairpin that extends out from strands 1 and 2 in the head subdomain, forms the tail of the tadpole. The tail subdomain is primarily involved in trimerization, while the head forms the capsid-binding site (arrows in Figure 2C). The Soc (T4) trimers act as clamps, linking neighboring capsomers in a chainmail structure that surrounds the capsid and stabilizes it against temperature and pH fluctuations [39]. To illustrate the complexity of decoration protein-capsid complexes we have chosen the Rossmann lab’s cryo-EM structure of phage T4 isometric heads complexed with Soc and Hoc, shown in Figure 3 [36].
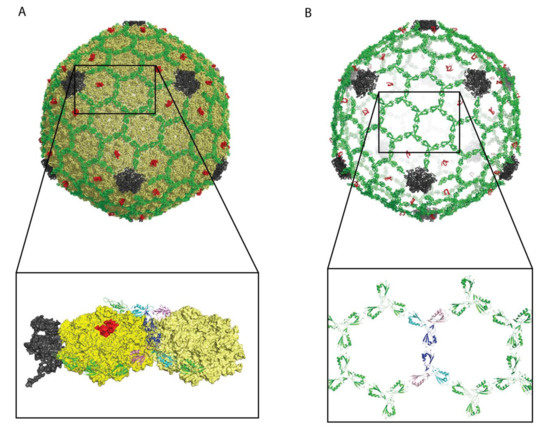
Figure 3.
Structure of isometric T4 heads bound by Soc (T4) and Hoc (T4). The data are from PDB file 5VF3 [28]. (A) View of the complex: coat protein—yellow, vertex protein—grey, Soc (T4)—green, Hoc (T4)—red. Please note that only a small part of the HocT4 protein was visible in the 3.3 Å cryo-EM data. (B) View of the structure showing only Soc (T4) and Hoc (T4). The Soc (T4) decoration protein forms a chainmail-like structure surrounding the capsid. This is not the case for every decoration protein, for example the Dec (L) trimers in Figure 1B are isolated from each other. The expansion in (B) shows the Soc (T4) molecules surrounding two of the T4 hexons. For clarity two of the Soc (T4) trimers at the center of the hexons have their three protomers colored in blue, cyan, and purple.
3.2.4. Hoc (T4) Has Multiple Immunoglobulin (Ig)-Like Domains
Although Soc (T4) stabilizes the capsid, Hoc (T4) has little or no effect on capsid stability but is used for cell attachment including to the T4 phage target E. coli [38]. Free Hoc (T4) has an elongated 4-domain structure (Figure 2D). The first three N-terminal domains have immunoglobulin (Ig)-like structures [38]. The Ig-fold consists of 7–9 anti-parallel β-strands arranged into a two-stack β-sandwich. Ig-folds are common cell-attachment modules that mediate interactions either through binding proteins or carbohydrates. A cryo-EM reconstruction showed that Hoc (T4) binds to a central depression in the T4 hexameric capsomere, as a dumbbell-shaped monomer with both Ig domain 1 and the non-Ig domain 4 contacting the capsid [61]. Unfortunately, domain 4 could not be fully seen in either the X-ray structure of the free protein nor the cryo-EM structure of capsid-bound Hoc (T4) [38,61].
3.2.5. Psu (P4) Has a Unique Knotted α-Helical Fold
The α-helical decoration protein Psu (P4), which doubles as a transcription antiterminator, has a novel V-shaped knotted dimer structure (Figure 2E) [64]. Knotted protein structures are extremely rare. When they occur, they are associated with extremely high stability. This may be why this unusual motif was selected as a viral decoration protein. The C-terminal α-helix 7 (arrows in Figure 2E) is thought to be responsible for capsid-binding at the center of P4 hexameric capsomers [64].
3.2.6. Additional Decoration Protein Structures
Besides the decoration proteins described above, there are examples that either are not well structurally characterized or do not fit the definition of a globular structure. These includes cases where only low-resolution structure data are currently available [66,73], proteins with substantial intrinsic disorder [25,70], and proteins that are parts of large hetero-oligomeric complexes [25,37,70].
3.3. Structural Homology Suggests Evolution through Horizontal Gene Transfer
Viruses and their target cells are constantly swapping genetic material through horizontal gene transfer processes leading to virus-host coevolution. Moreover, genome data suggests that typical phages are mosaics of genes generated by nonhomologous recombination of ancestral sequences [104]. Thus, horizontal transfer is likely to have occurred both between viruses and cells, and between viruses. An analysis of sequences and structures of viral capsid proteins found evidence that these probably evolved from cellular organisms on multiple occasions [105].
We performed a simple analysis to try to investigate the evolutionary origins of decoration proteins. Starting with the basis set of five well-characterized decoration protein folds (Table 2), we submitted the representative structure for each fold to a PDB-BLAST search [103] that looks for amino acid sequence homologs in the PDB database of known structures. All the hits in this search are decoration proteins with known structures that are sequence homologs of the representative set of decoration proteins (Table 2). For example, gpD (λ) was found to be a sequence homolog of SHP (P21), as previously described in the literature [52].
We next submitted the representative proteins to a DALI structural homology search [102]. This algorithm identifies structural homologs that have no sequence homology to the query structures [102]. We restricted this search to proteins that belong to phages or viruses. For example, gpD (λ) had a structural similarity hit to the PDB entry 3SUC, which is a preneck appendage protein of the phage φ29 tailspike [106]. Structural similarity between decoration and tailspike proteins has been reported previously [63,89]. In fact, the decoration protein pb10 (T5) was initially classified as a tail protein due to its predicted Ig-like domains [16]. Moreover, decoration proteins suggested to play roles in cell attachment may have evolved from tailspike proteins to facilitate host recognition [63,89]. As shown in Table 2, we find that structural homology to decoration proteins is not restricted to tailspike proteins, but occurs for a range of viral proteins including capsid and envelope proteins.
Finally, we did a DALI structural homology search restricting results to proteins that are found in the hosts of the respective phages (Table 2). For each of the decoration proteins that represent the five main folds, we found a structural homolog in the host. For example, gpD (λ) has homology to the E. coli molybdenum-binding protein MoeA, which as described earlier is the only example of the β-tulip fold not found in a virus. In cases where we could not find a structural homolog, probably since not all organisms are equally well-represented in the structure database, we did a BLAST search to look for sequence homologs in the host organism to one of the phage structure homologs. Thus, while gp87 (P74-26) does not have any known structural homologs in T. thermophilus, the structurally homologous φ29 protein 3SUC has sequence homology to a hypothetical protein in T. thermophilus. Taken together, the homology relationships in Table 2 suggest decoration proteins, and perhaps the cellular homologs, likely evolved through horizontal gene transfer either within phages/viruses during co-infection events, or between phages/viruses and their host organisms through recombination.
4. Nanotechnology Applications
Viruses and phages are exploited for a wide variety of uses in the fields of medicine, materials science, and nanotechnology. Among other applications, phages are being employed to treat antibiotic-resistant bacterial infections, to screen for potential drugs with phage display technology, and to deliver drugs via viral nanoparticle (VNP) vehicles [107,108]. Conjugating VNPs with moieties such as metals, polymers, or diagnostic imaging dyes is opening avenues to produce novel materials, including catalysts, biomimetics, and “smart” imaging agents (Figure 4A) [109,110,111,112,113].
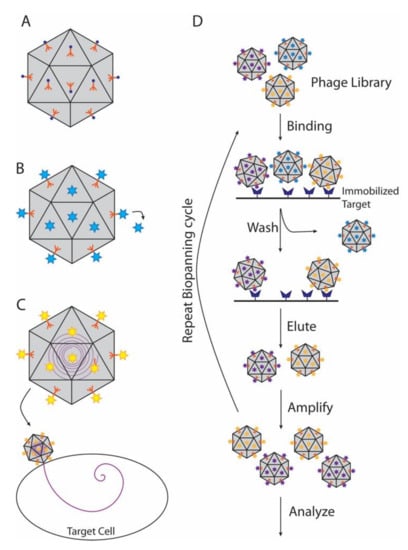
Figure 4.
Nanotechnology applications of decoration proteins. Viral nanoparticles (VNPs) are in grey, decoration proteins in orange, and cargo molecules are shown as multi-colored circles and stars. (A) Functionalization of VNPs. In this example metal ions are attached to decoration proteins on VNPs, to create magnetic or conductive nanoparticles or nanowires. (B) A wide range of molecules can be attached to decoration proteins for phage display or cargo delivery. (C) Decoration proteins can be used to target VNPs carrying internal cargos to specific cells. (D) Biopanning can be used to find novel therapeutics.
4.1. Decoration Protein Platforms for Design of Novel Nanomaterials
Although most efforts to develop novel VNPs have focused on viral coat proteins, decoration proteins offer unique advantages. First, since decoration proteins are accessory surface molecules that do not interfere with the assembly of VNPs, they can tolerate much larger cargo molecules than coat proteins [114]. Because different decoration proteins bind at different types of symmetry sites on the icosahedral surface, it should be possible to control the patterning of cargo displayed on VNPs [92]. Stability over a range of external environments is desirable for VNPs in medical applications, especially if a drug cargo needs to be delivered orally. In this regard, the S28C mutant of the adenovirus cementing precursor protein VI, has been shown to modulate the stability of the viral capsid without impacting the infection process, thus potentially allowing for VNPs with controllable stabilities [115].
Most importantly, decoration proteins bind viruses non-covalently, making it possible to tune their binding affinities through mutagenesis or by changes in solution conditions. The non-covalent binding of decoration carrier proteins affords the opportunity to control their functionalized cargo molecule activity. For example, activity could be “turned off” by substitution of a functionalized decoration protein for the wild type, or the activity could be “swapped” by substituting a decoration protein derivatized with one type of cargo for another. Possible applications for nanomaterials with dissociable decoration protein subunits include the rational design of switchable nanomaterials such as pores that assume different diameters depending on the cargo displayed [116], multi-functionalized nanomaterials [117], nanolithography [118], and nanomaterials with temporally controlled properties [119]. Moreover, nanomaterial design need not be limited to icosahedral VNPs. Subtle changes in interactions between coat protein subunits, for example altered through site-directed mutagenesis, can divert assembly from icosahedra to other types of lattice structures such as nanotubes or nanosheets that can be similarly functionalized through decoration protein binding [92].
4.2. Decoration Proteins in Phage Display and Biopanning
A major application of decoration and cementing proteins is phage display (Figure 4B), a technique developed over the last four decades and highlighted by the chemistry Nobel prize in 2018 [120]. In this technique, the gene for a protein or peptide is inserted into a phage coat or decoration protein gene, producing a fusion protein that is displayed on the outside of the capsid [121,122,123]. Decoration proteins, such as Hoc (T4), Soc (T4), and IX (adenovirus), are particularly useful for display of large protein molecules or complexes in high copy numbers, which if fused to the alternative coat proteins could disrupt capsid assembly [114,123,124,125]. Thus, T4 Hoc and Soc displaying short, random peptide sequences have been used to discover additional phage proteins that bind the terminase protein, gp17, via selection method called biopanning that uses multiple rounds of screening to find molecules that bind with high affinity to chosen target (Figure 4D) [126]. Other applications include screening vaccine candidates, drug discovery, or as biosensors to detect specific antigens [127,128,129,130,131,132].
4.3. Decoration Proteins in Vaccine Design
Over millions of years of evolution animal immune systems have adapted to recognize viruses and virus-like particles as dangerous. Consequently, multivalent display of antigens on the surfaces of virus-like particles elicits much stronger immune responses than the corresponding free antigens [133,134]. Display of immunogenic antigens on the surfaces of VNPs through decoration protein carriers has the potential to advance rational vaccine design efforts.
Several phage display systems have been shown to stimulate an immune response in animals, including M13, λ, T7, and T4 [127]. In some systems the displayed antigen is conjugated directly to the coat protein, while in others decoration proteins were used [121,123,125]. Decoration proteins offer advantages for vaccines: (i) they can potentially accommodate larger epitopes [114]; (ii) the epitope density on the VNP surface can potentially be modulated; (iii) different molecule types can be simultaneously displayed together, such as an antigens alongside adjuvants [114,135]. Examples of decoration proteins used for vaccine development include gpD (λ), which when fused to fragments of the Circovirus 2 capsid protein forms the basis of a λ phage livestock vaccine for pigs [136]. In a second example, gpD (λ) fused to a prion protein from deer-stimulated production of IgA antibodies in a mouse model without the use of adjuvant [137]. VNPs displaying gpD (λ) fused to GP2, a peptide derivative from the overexpressed tumor protein HER2/nue, generated a strong cytotoxic T lymphocyte response that had anti-tumor activity when given prophylactically or therapeutically in a mouse model [138].
Hoc (T4) and Soc (T4) have the advantage of being able to carry large cargos [125,126] such as anthrax toxin oligomers with a 93 KDa molecular mass [114,139]. Additionally, Hoc (T4) and Soc (T4) were used to display fragments of the type I porin, PorA, from Neisseria meningitides [122], as well as the 83KDa Bacillus anthracis protective antigen (PA) [139,140], and the HIV protein, p24 [125] on T4 VNPs. In each case, mice immunized with these VNPs showed strong immunogenic responses to the displayed protein [122,125,139]. The use of decoration proteins to display antigens on VNPs presents new advantages for potential vaccine development.
4.4. Decoration Proteins used as Postmarks to Target VNP Delivery
Another potential use of surface-bound decoration proteins is to deliver cargo molecules encapsulated in VNPs to specific cell types (Figure 4C). The phage P22 system is particularly attractive as conditions for capsid assembly and cargo encapsulation are well established [6,55,92,141,142]. As mentioned earlier in the review, phage P22 can bind Dec (L) in vitro allowing cargo fused to the decoration protein to be displayed on its surface. Dec (L), which binds tightly only to expanded VNPs that mimic the mature state of phage P22, has been used to both display receptor-binding proteins and to deliver cargo molecules encapsulated in the capsid [55,92,142]. This “inside-outside” functionalization strategy demonstrates that VNPs can be simultaneously employed for both cell-targeting and payload delivery [55].
Phage T4 is likewise suitable for this application as it can hold a large volume of genetic material, and its decoration proteins are amenable for fusion with a broad range of proteins. A “progene” approach was used to simultaneously deliver both genes (encapsulated in the phage) and proteins (displayed on the surface through a Soc(T4) fusion) specifically to antigen-presenting dendritic cells via a Hoc(T4)-fused cell penetrating peptide [135]. The work shows that “inside-outside” cargo consisting of both proteins and DNA could be delivered to specific cell types through “postmarks” attached to decoration proteins. These types of approaches offer new avenues for vaccine and therapeutic strategies.
Author Contributions
Writing, review and editing, C.L.D., C.M.T. and A.T.A. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by NIH grant R01-GM076661.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sobhy, H.; Scola, B.L.; Pagnier, I.; Raoult, D.; Colson, P. Identification of giant Mimivirus protein functions using RNA interference. Front. Microbiol. 2015, 6, 345. [Google Scholar] [CrossRef]
- Suhanovsky, M.M.; Teschke, C.M. Nature׳s favorite building block: Deciphering folding and capsid assembly of proteins with the HK97-fold. Virology 2015, 487–497. [Google Scholar] [CrossRef]
- Aksyuk, A.A.; Rossmann, M.G. Bacteriophage Assembly. Viruses 2011, 3, 172–203. [Google Scholar] [CrossRef]
- Prevelige, P.E. Send for reinforcements! Conserved binding of capsid decoration proteins. Structure 2008, 16, 1292–1293. [Google Scholar] [CrossRef]
- Mateu, M.G. Assembly, stability and dynamics of virus capsids. Arch. Biochem. Biophys. 2013, 531, 65–79. [Google Scholar] [CrossRef]
- Parent, K.N.; Khayat, R.; Tu, L.H.; Suhanovsky, M.M.; Cortines, J.R.; Teschke, C.M.; Johnson, J.E.; Baker, T.S. P22 Coat Protein Structures Reveal a Novel Mechanism for Capsid Maturation: Stability without Auxiliary Proteins or Chemical Crosslinks. Structure 2010, 18, 390–401. [Google Scholar] [CrossRef]
- Ross, P.D.; Cheng, N.; Conway, J.F.; Firek, B.A.; Hendrix, R.W.; Duda, R.L.; Steven, A.C. Crosslinking renders bacteriophage HK97 capsid maturation irreversible and effects an essential stabilization. EMBO J. 2005, 24, 1352–1363. [Google Scholar] [CrossRef]
- Rizzo, A.A.; Suhanovsky, M.M.; Baker, M.L.; Fraser, L.C.; Jones, L.M.; Rempel, D.L.; Gross, M.L.; Chiu, W.; Alexandrescu, A.T.; Teschke, C.M. Multiple Functional Roles of the Accessory I-Domain of Bacteriophage P22 Coat Protein Revealed by NMR Structure and CryoEM Modeling. Structure 2014, 22, 830–841. [Google Scholar] [CrossRef][Green Version]
- Tripler, T.N.; Kaplan, A.R.; Alexandrescu, A.T.; Teschke, C.M. Conservation and Divergence of the I-Domain Inserted into the Ubiquitous HK97 Coat Protein Fold in P22-Like Bacteriophages. J. Virol. 2019, 93, 9. [Google Scholar] [CrossRef]
- Swanstrom, R.; Wills, J.W. Synthesis, Assembly, and Processing of Viral Proteins. In Retroviruses; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1997. [Google Scholar]
- Casjens, S.; Hendrix, R. Control Mechanisms in dsDNA Bacteriophage Assembly. In The Bacteriophages; Calendar, R., Ed.; Springer US: Boston, MA, USA, 1988; pp. 15–91. [Google Scholar]
- Furcinitti, P.S.; Van Oostrum, J.; Burnett, R.M. Adenovirus polypeptide IX revealed as capsid cement by difference images from electron microscopy and crystallography. EMBO J. 1989, 8, 3563–3570. [Google Scholar] [CrossRef]
- Gilcrease, E.B.; Winn-Stapley, D.A.; Hewitt, F.C.; Joss, L.; Casjens, S.R. Nucleotide Sequence of the Head Assembly Gene Cluster of Bacteriophage L and Decoration Protein Characterization. J. Bacteriol. 2005, 187, 2050–2057. [Google Scholar] [CrossRef]
- Lander, G.C.; Evilevitch, A.; Jeembaeva, M.; Potter, C.S.; Carragher, B.; Johnson, J.E. Bacteriophage Lambda Stabilization by Auxiliary Protein gpD: Timing, Location, and Mechanism of Attachment Determined by Cryo-EM. Structure 2008, 16, 1399–1406. [Google Scholar] [CrossRef]
- Wang, Z.; Hardies, S.C.; Fokine, A.; Klose, T.; Jiang, W.; Cho, B.C.; Rossmann, M.G. Structure of the Marine Siphovirus TW1: Evolution of Capsid-Stabilizing Proteins and Tail Spikes. Structure 2018, 26, 238–248. [Google Scholar] [CrossRef]
- Vernhes, E.; Renouard, M.; Gilquin, B.; Cuniasse, P.; Durand, D.; England, P.; Hoos, S.; Huet, A.; Conway, J.F.; Glukhov, A.; et al. High affinity anchoring of the decoration protein pb10 onto the bacteriophage T5 capsid. Sci. Rep. 2017, 7, 41662. [Google Scholar] [CrossRef]
- Stone, N.P.; Hilbert, B.J.; Hidalgo, D.; Halloran, K.T.; Lee, J.; Sontheimer, E.J.; Kelch, B.A. A Hyperthermophilic Phage Decoration Protein Suggests Common Evolutionary Origin with Herpesvirus Triplex Proteins and an Anti-CRISPR Protein. Structure 2018, 26, 936–947. [Google Scholar] [CrossRef]
- Bayfield, O.W.; Klimuk, E.; Winkler, D.C.; Hesketh, E.L.; Chechik, M.; Cheng, N.; Dykeman, E.C.; Minakhin, L.; Ranson, N.A.; Severinov, K.; et al. Cryo-EM structure and in vitro DNA packaging of a thermophilic virus with supersized T=7 capsids. Proc. Natl. Acad. Sci. USA 2019, 116, 3556–3561. [Google Scholar] [CrossRef]
- Choi, K.H.; McPartland, J.; Kaganman, I.; Bowman, V.D.; Rothman-Denes, L.B.; Rossmann, M.G. Insight into DNA and Protein Transport in Double-Stranded DNA Viruses: The Structure of Bacteriophage N4. J. Mol. Biol. 2008, 378, 726–736. [Google Scholar] [CrossRef]
- Dokland, T.; Murialdo, H. Structural Transitions during Maturation of Bacteriophage Lambda Capsids. J. Mol. Biol. 1993, 233, 682–694. [Google Scholar] [CrossRef]
- Adamson, W.E.; McNab, D.; Preston, V.G.; Rixon, F.J. Mutational Analysis of the Herpes Simplex Virus Triplex Protein VP19C. J. Virol. 2006, 80, 1537–1548. [Google Scholar] [CrossRef]
- Kirkitadze, M.D.; Barlow, P.N.; Price, N.C.; Kelly, S.M.; Boutell, C.J.; Rixon, F.J.; McClelland, D.A. The Herpes Simplex Virus Triplex Protein, VP23, Exists as a Molten Globule. J. Virol. 1998, 72, 10066–10072. [Google Scholar] [CrossRef]
- Yu, X.; Jih, J.; Jiang, J.; Zhou, Z.H. Atomic structure of the human cytomegalovirus capsid with its securing tegument layer of pp150. Science 2017, 356, eaam6892. [Google Scholar] [CrossRef]
- Dai, X.; Wu, L.; Sun, R.; Zhou, Z.H. Atomic Structures of Minor Proteins VI and VII in Human Adenovirus. J. Virol. 2017, 91, e00850-17. [Google Scholar] [CrossRef]
- Reddy, V.S.; Nemerow, G.R. Structures and organization of adenovirus cement proteins provide insights into the role of capsid maturation in virus entry and infection. Proc. Natl. Acad. Sci. USA 2014, 111, 11715. [Google Scholar] [CrossRef]
- Yu, X.; Veesler, D.; Campbell, M.G.; Barry, M.E.; Asturias, F.J.; Barry, M.A.; Reddy, V.S. Cryo-EM structure of human adenovirus D26 reveals the conservation of structural organization among human adenoviruses. Sci. Adv. 2017, 3, e1602670. [Google Scholar] [CrossRef]
- Pietilä, M.K.; Laurinmäki, P.; Russell, D.A.; Ko, C.-C.; Jacobs-Sera, D.; Butcher, S.J.; Bamford, D.H.; Hendrix, R.W. Insights into Head-Tailed Viruses Infecting Extremely Halophilic Archaea. J. Virol. 2013, 87, 3248–3260. [Google Scholar] [CrossRef]
- Stone, N.P.; Demo, G.; Agnello, E.; Kelch, B.A. Principles for enhancing virus capsid capacity and stability from a thermophilic virus capsid structure. Nat. Commun. 2019, 10, 4471–13. [Google Scholar] [CrossRef]
- Wagemans, J.; Tsonos, J.; Holtappels, D.; Fortuna, K.; Hernalsteens, J.-P.; De Greve, H.; Estrozi, L.F.; Bacia-Verloop, M.; Moriscot, C.; Noben, J.-P.; et al. Structural Analysis of Jumbo Coliphage phAPEC6. Int. J. Mol. Sci. 2020, 21, 3119. [Google Scholar] [CrossRef]
- Klose, T.; Fau-Rossmann, M.G.; Rossmann, M.G. Structure of large dsDNA viruses. Biol. Chem. 2014, 395, 711–719. [Google Scholar] [CrossRef]
- Hua, J.; Huet, A.; Lopez, C.A.; Toropova, K.; Pope, W.H.; Duda, R.L.; Hendrix, R.W.; Conway, J.F. Capsids and Genomes of Jumbo-Sized Bacteriophages Reveal the Evolutionary Reach of the HK97 Fold. mBio 2017, 8, e01579-17. [Google Scholar] [CrossRef]
- Podgorski, J.; Calabrese, J.; Alexandrescu, L.; Jacobs-Sera, D.; Pope, W.H.; Hatfull, G.F.; White, S.J. Structures of Three Actinobacteriophage Capsids: Roles of Symmetry and Accessory Proteins. Viruses 2020, 12, 294. [Google Scholar] [CrossRef]
- Doore, S.M.; Schrad, J.R.; Dean, W.F.; Dover, J.A.; Parent, K.N. Shigella Phages Isolated during a Dysentery Outbreak Reveal Uncommon Structures and Broad Species Diversity. J. Virol. 2018, 92, e02117–17. [Google Scholar] [CrossRef]
- González, B.; Monroe, L.; Li, K.; Yan, R.; Wright, E.; Walter, T.; Kihara, D.; Weintraub, S.T.; Thomas, J.A.; Serwer, P.; et al. Phage G Structure at 6.1 Å Resolution, Condensed DNA, and Host Identity Revision to a Lysinibacillus. J. Mol. Biol. 2020, 432, 4139–4153. [Google Scholar] [CrossRef]
- Neumann, E.; Kawasaki, T.; Effantin, G.; Estrozi, L.F.; Chatchawankanphanich, O.; Eyamada, T.; Schoehn, G. 3D structure of three jumbo phage heads. J. Gen. Virol. 2020, jgv001487. [Google Scholar] [CrossRef]
- Chen, Z.; Sun, L.; Zhang, Z.; Fokine, A.; Padilla-Sanchez, V.; Hanein, D.; Jiang, W.; Rossmann, M.G.; Rao, V.B. Cryo-EM structure of the bacteriophage T4 isometric head at 3.3-Å resolution and its relevance to the assembly of icosahedral viruses. Proc. Natl. Acad. Sci. USA 2017, 14, E81–E84. [Google Scholar] [CrossRef]
- Fang, Q.; Zhu, D.; Agarkova, I.; Adhikari, J.; Klose, T.; Liu, Y.; Chen, Z.; Sun, Y.; Gross, M.L.; Van Etten, J.L.; et al. Near-atomic structure of a giant virus. Nat. Commun. 2019, 10, 388. [Google Scholar] [CrossRef]
- Fokine, A.; Islam, M.Z.; Zhang, Z.; Bowman, V.D.; Rao, V.B.; Rossmann, M.G. Structure of the Three N-Terminal Immunoglobulin Domains of the Highly Immunogenic Outer Capsid Protein from a T4-Like Bacteriophage. J. Virol. 2011, 85, 8141–8148. [Google Scholar] [CrossRef]
- Qin, L.; Fokine, A.; O’Donnell, E.; Rao, V.B.; Rossmann, M.G. Structure of the Small Outer Capsid Protein, Soc: A Clamp for Stabilizing Capsids of T4-like Phages. J. Mol. Biol. 2010, 395, 728–741. [Google Scholar] [CrossRef]
- Xiang, Y.; Rossmann, M.G. Structure of bacteriophage phi29 head fibers has a supercoiled triple repeating helix-turn-helix motif. Proc. Natl. Acad. Sci. USA 2011, 108, 4806. [Google Scholar] [CrossRef]
- Wikoff, W.R.; Liljas, L.; Duda, R.L.; Tsuruta, H.; Hendrix, R.W.; Johnson, J.E. Topologically Linked Protein Rings in the Bacteriophage HK97 Capsid. Science 2000, 289, 2129–2133. [Google Scholar] [CrossRef]
- Duda, R.L.; Teschke, C.M. The amazing HK97 fold: Versatile results of modest differences. Curr. Opin. Virol. 2019, 36, 9–16. [Google Scholar] [CrossRef]
- Helgstrand, C.; Wikoff, W.R.; Duda, R.L.; Hendrix, R.W.; Johnson, J.E.; Liljas, L. The Refined Structure of a Protein Catenane: The HK97 Bacteriophage Capsid at 3.44 Å Resolution. J. Mol. Biol. 2003, 334, 885–899. [Google Scholar] [CrossRef]
- Conway, J.F.; Wikoff, W.R.; Cheng, N.; Duda, R.L.; Hendrix, R.W.; Johnson, J.E.; Steven, A.C. Virus Maturation Involving Large Subunit Rotations and Local Refolding. Science 2001, 292, 744–748. [Google Scholar] [CrossRef]
- Gertsman, I.; Gan, L.; Guttman, M.; Lee, K.; Speir, J.A.; Duda, R.L.; Hendrix, R.W.; Komives, E.A.; Johnson, J.E. An unexpected twist in viral capsid maturation. Nature 2009, 458, 646–650. [Google Scholar] [CrossRef]
- Sae-Ueng, U.; Liu, T.; Catalano, C.E.; Huffman, J.B.; Homa, F.L.; Evilevitch, A. Major capsid reinforcement by a minor protein in herpesviruses and phage. Nucleic Acids Res. 2014, 42, 9096–9107. [Google Scholar] [CrossRef]
- Tang, L.; Gilcrease, E.B.; Casjens, S.R.; Johnson, J.E. Highly Discriminatory Binding of Capsid-Cementing Proteins in Bacteriophage L. Structure 2006, 14, 837–845. [Google Scholar] [CrossRef]
- Hernando-Pérez, M.; Lambert, S.; Nakatani-Webster, E.; Catalano, C.E.; De Pablo, P.J. Cementing proteins provide extra mechanical stabilization to viral cages. Nat. Commun. 2014, 5, 4520. [Google Scholar] [CrossRef]
- Yang, Q.; Maluf, N.K.; Catalano, C.E. Packaging of a Unit-Length Viral Genome: The Role of Nucleotides and the gpD Decoration Protein in Stable Nucleocapsid Assembly in Bacteriophage λ. J. Mol. Biol. 2008, 383, 1037–1048. [Google Scholar] [CrossRef]
- Yang, F.; Forrer, P.; Dauter, Z.; Conway, J.F.; Cheng, N.; Cerritelli, M.E.; Steven, A.C.; Plückthun, A.; Wlodawer, A. Novel fold and capsid-binding properties of the λ-phage display platform protein gpD. Nat. Struct. Biol. 2000, 7, 230–237. [Google Scholar]
- Iwai, H.; Forrer, P.; Pluckthun, A.; Guntert, P. NMR solution structure of the monomeric form of the bacteriophage lambda capsid stabilizing protein gpD. J. Biomol. NMR 2005, 31, 351–356. [Google Scholar] [CrossRef]
- Forrer, P.; Chang, C.; Ott, D.; Wlodawer, A.; Plückthun, A. Kinetic Stability and Crystal Structure of the Viral Capsid Protein SHP. J. Mol. Biol. 2004, 344, 179–193. [Google Scholar] [CrossRef]
- Shin, H.; Lee, J.-H.; Ahn, C.S.; Ryu, S.; Cho, B.C. Complete genome sequence of marine bacterium Pseudoalteromonas phenolica bacteriophage TW1. Arch. Virol. 2014, 159, 159–162. [Google Scholar] [CrossRef]
- Hardy, J.M.; Dunstan, R.A.; Grinter, R.; Belousoff, M.J.; Wang, J.; Pickard, D.; Venugopal, H.; Dougan, G.; Lithgow, T.; Coulibaly, F. The architecture and stabilisation of flagellotropic tailed bacteriophages. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef]
- Schwarz, B.; Madden, P.; Avera, J.; Gordon, B.; Larson, K.; Miettinen, H.M.; Uchida, M.; Lafrance, B.; Basu, G.; Rynda-Apple, A.; et al. Symmetry Controlled, Genetic Presentation of Bioactive Proteins on the P22 Virus-like Particle Using an External Decoration Protein. ACS Nano 2015, 9, 9134–9147. [Google Scholar] [CrossRef]
- Newcomer, R.L.; Belato, H.B.; Teschke, C.M.; Alexandrescu, A.T. NMR assignments for monomeric phage L decoration protein. Biomol. NMR Assign. 2018, 12, 339–343. [Google Scholar] [CrossRef]
- Newcomer, R.L.; Schrad, J.R.; Gilcrease, I.B.; Casjens, S.R.; Feig, M.; Teschke, C.M.; Alexandrescu, A.T.; Parent, K.N. The phage L capsid decoration protein has a novel OB-fold and an unusual capsid binding strategy. eLife 2019, 8. [Google Scholar] [CrossRef]
- Steven, A.C.; Greenstone, H.L.; Booy, F.P.; Black, L.W.; Ross, P.D. Conformational changes of a viral capsid protein: Thermodynamic rationale for proteolytic regulation of bacteriophage T4 capsid expansion, co-operativity, and super-stabilization by soc binding. J. Mol. Biol. 1992, 228, 870–884. [Google Scholar] [CrossRef]
- Ishii, T.; Yanagida, M. The two dispensable structural proteins (soc and hoc) of the T4 phage capsid; their purification and properties, isolation and characterization of the defective mutants, and their binding with the defective heads in vitro. J. Mol. Biol. 1977, 109, 487–514. [Google Scholar] [CrossRef]
- Olson, N.H.; Gingery, M.; Eiserling, F.A.; Baker, T.S. The Structure of Isometric Capsids of Bacteriophage T4. Virology 2001, 279, 385–391. [Google Scholar] [CrossRef]
- Sathaliyawala, T.; Islam, M.Z.; Li, Q.; Fokine, A.; Rossmann, M.G.; Rao, V.B. Functional analysis of the highly antigenic outer capsid protein, Hoc, a virus decoration protein from T4-like bacteriophages. Mol. Microbiol. 2010, 77, 444–455. [Google Scholar] [CrossRef]
- Iwasaki, K.; Trus, B.L.; Wingfield, P.T.; Cheng, N.; Campusano, G.; Rao, V.B.; Steven, A.C. Molecular Architecture of Bacteriophage T4 Capsid: Vertex Structure and Bimodal Binding of the Stabilizing Accessory Protein, Soc. Virology 2000, 271, 321–333. [Google Scholar] [CrossRef][Green Version]
- Effantin, G.; Boulanger, P.; Neumann, E.; Letellier, L.; Conway, J.F. Bacteriophage T5 Structure Reveals Similarities with HK97 and T4 Suggesting Evolutionary Relationships. J. Mol. Biol. 2006, 361, 993–1002. [Google Scholar] [CrossRef]
- Banerjee, R.; Nath, S.; Ranjan, A.; Khamrui, S.; Pani, B.; Sen, R.; Sen, U. The First Structure of Polarity Suppression Protein, Psu from Enterobacteria Phage P4, Reveals a Novel Fold and a Knotted Dimer. J. Biol. Chem. 2012, 287, 44667–44675. [Google Scholar] [CrossRef]
- Linderoth, N.A.; Calendar, R.L. The Psu protein of bacteriophage P4 is an antitermination factor for rho-dependent transcription termination. J. Bacteriol. 1991, 173, 6722–6731. [Google Scholar] [CrossRef]
- White, H.E.; Sherman, M.B.; Brasilès, S.; Jacquet, E.; Seavers, P.; Tavares, P.; Orlova, E.V. Capsid Structure and Its Stability at the Late Stages of Bacteriophage SPP1 Assembly. J. Virol. 2012, 86, 6768. [Google Scholar] [CrossRef]
- Zairi, M.; Stiege, A.C.; Nhiri, N.; Jacquet, E.; Tavares, P. The Collagen-like Protein gp12 is a Temperature-dependent Reversible Binder of SPP1 Viral Capsids. J. Biol. Chem. 2014, 289, 27169–27181. [Google Scholar] [CrossRef]
- Ignatiou, A.; Brasilès, S.; Fadel, M.E.S.; Bürger, J.; Mielke, T.; Topf, M.; Tavares, P.; Orlova, E.V. Structural transitions during the scaffolding-driven assembly of a viral capsid. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Rosa-Calatrava, M.; Grave, L.; Puvion-Dutilleul, F.; Chatton, B.; Kedinger, C. Functional Analysis of Adenovirus Protein IX Identifies Domains Involved in Capsid Stability, Transcriptional Activity, and Nuclear Reorganization. J. Virol. 2001, 75, 7131–7141. [Google Scholar] [CrossRef]
- Abrescia, N.G.A.; Cockburn, J.J.B.; Grimes, J.M.; Sutton, G.C.; Diprose, J.M.; Butcher, S.J.; Fuller, S.D.; Martín, C.S.; Burnett, R.M.; Stuart, D.I.; et al. Insights into assembly from structural analysis of bacteriophage PRD1. Nature 2004, 432, 68–74. [Google Scholar] [CrossRef]
- Rydman, P.S.; Bamford, J.K.; Bamford, D.H. A minor capsid protein P30 is essential for bacteriophage PRD1 capsid assembly. J. Mol. Biol. 2001, 313, 785–795. [Google Scholar] [CrossRef]
- Dunigan, D.D.; Cerny, R.L.; Bauman, A.T.; Roach, J.C.; Lane, L.C.; Agarkova, I.V.; Wulser, K.; Yanai-Balser, G.M.; Gurnon, J.R.; Vitek, J.C.; et al. Paramecium bursaria Chlorella Virus 1 Proteome Reveals Novel Architectural and Regulatory Features of a Giant Virus. J. Virol. 2012, 86, 8821–8834. [Google Scholar] [CrossRef]
- Chang, J.T.; Schmid, M.F.; Haase-Pettingell, C.; Weigele, P.R.; King, J.A.; Chiu, W. Visualizing the Structural Changes of Bacteriophage Epsilon15 and Its Salmonella Host during Infection. J. Mol. Biol. 2010, 402, 731–740. [Google Scholar] [CrossRef]
- Fuller, D.N.; Raymer, D.M.; Rickgauer, J.P.; Robertson, R.M.; Catalano, C.E.; Anderson, D.L.; Grimes, S.; Smith, D.E. Measurements of Single DNA Molecule Packaging Dynamics in Bacteriophage λ Reveal High Forces, High Motor Processivity, and Capsid Transformations. J. Mol. Biol. 2007, 373, 1113–1122. [Google Scholar] [CrossRef]
- Imber, R.; Tsugita, A.; Würtz, M.; Hohn, T. Outer surface protein of bacteriophage lambda. J. Mol. Biol. 1980, 139, 277–295. [Google Scholar] [CrossRef]
- Wendt, J.L.; Feiss, M. A fragile lattice: Replacing bacteriophage λ’s head stability gene D with the shp gene of phage 21 generates the Mg2+-dependent virus, λ shp. Virology 2004, 326, 41–46. [Google Scholar] [CrossRef][Green Version]
- Morais, M.C.; Choi, K.H.; Koti, J.S.; Chipman, P.R.; Anderson, D.L.; Rossmann, M.G. Conservation of the Capsid Structure in Tailed dsDNA Bacteriophages: The Pseudoatomic Structure of ϕ29. Mol. Cell 2005, 18, 149–159. [Google Scholar] [CrossRef]
- Baker, M.L.; Jiang, W.; Rixon, F.J.; Chiu, W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J. Virol. 2005, 79, 14967. [Google Scholar] [CrossRef]
- Brown, J.C.; Newcomb, W.W. Herpesvirus capsid assembly: Insights from structural analysis. Curr. Opin. Virol. 2011, 1, 142–149. [Google Scholar] [CrossRef]
- Dai, X.; Zhou, Z.H. Structure of the herpes simplex virus 1 capsid with associated tegument protein complexes. Science 2018, 360, eaao7298. [Google Scholar] [CrossRef]
- Yuan, S.; Wang, J.; Zhu, D.; Wang, N.; Gao, Q.; Chen, W.; Tang, H.; Wang, J.; Zhang, X.; Liu, H.; et al. Cryo-EM structure of a herpesvirus capsid at 3.1 Å. Science 2018, 360, eaao7283. [Google Scholar] [CrossRef]
- Belnap, D.M.; Steven, A.C. ‘Déjà vu all over again’: The similar structures of bacteriophage PRD1 and adenovirus. Trends Microbiol. 2000, 8, 91–93. [Google Scholar] [CrossRef]
- Benson, S.D.; Bamford, J.K.; Bamford, D.H.; Burnett, R.M. Does Common Architecture Reveal a Viral Lineage Spanning All Three Domains of Life? Mol. Cell 2004, 16, 673–685. [Google Scholar] [CrossRef]
- Barr, J.J.; Auro, R.; Sam-Soon, N.; Kassegne, S.; Peters, G.; Bonilla, N.; Hatay, M.; Mourtada, S.; Bailey, B.; Youle, M.; et al. Subdiffusive motion of bacteriophage in mucosal surfaces increases the frequency of bacterial encounters. Proc. Natl. Acad. Sci. USA 2015, 112, 13675. [Google Scholar] [CrossRef]
- Barr, J.J.; Youle, M.; Rohwer, F. Innate and acquired bacteriophage-mediated immunity. Bacteriophage 2013, 3, e25857. [Google Scholar] [CrossRef]
- Barr, J.J.; Auro, R.; Furlan, M.; Whiteson, K.L.; Erb, M.L.; Pogliano, J.; Stotland, A.; Wolkowicz, R.; Cutting, A.S.; Doran, K.S.; et al. Bacteriophage adhering to mucus provide a non–host-derived immunity. Proc. Natl. Acad. Sci. USA 2013, 110, 10771. [Google Scholar] [CrossRef]
- Bansil, R.; Turner, B.S. The biology of mucus: Composition, synthesis and organization. Adv. Drug Deliv. Rev. 2018, 124, 3–15. [Google Scholar] [CrossRef]
- Silveira, C.B.; Rohwer, F.L. Piggyback-the-Winner in host-associated microbial communities. NPJ Biofilms Microbiomes 2016, 2, 16010. [Google Scholar] [CrossRef]
- Fraser, J.S.; Maxwell, K.L.; Davidson, A.R. Immunoglobulin-like domains on bacteriophage: Weapons of modest damage? Curr. Opin. Microbiol. 2007, 10, 382–387. [Google Scholar] [CrossRef]
- Wilson, D.P. Protruding Features of Viral Capsids Are Clustered on Icosahedral Great Circles. PLoS ONE 2016, 11, e0152319. [Google Scholar] [CrossRef]
- Zandi, R.; Reguera, D. Mechanical properties of viral capsids. Phys. Rev. E 2005, 72, 021917. [Google Scholar] [CrossRef]
- Parent, K.N.; Deedas, C.T.; Egelman, E.H.; Casjens, S.R.; Baker, T.S.; Teschke, C.M. Stepwise molecular display utilizing icosahedral and helical complexes of phage coat and decoration proteins in the development of robust nanoscale display vehicles. Biomaterials 2012, 33, 5628–5637. [Google Scholar] [CrossRef]
- Rao, S.; Rossmann, M.G. Comparison of super-secondary structures in proteins. J. Mol. Biol. 1973, 76, 241–256. [Google Scholar] [CrossRef]
- Rossmann, M.G. Super-secondary Structure: A Historical Perspective. Methods Mol. Biol. 2013, 932, 1–4. [Google Scholar]
- Shin, W.-H.; Kihara, D. 55 Years of the Rossmann Fold. Methods Mol. Biol. 2019, 1958, 1–13. [Google Scholar]
- Neuman, B.W.; Chamberlain, P.; Bowden, F.; Joseph, J. Atlas of coronavirus replicase structure. Virus Res. 2014, 194, 49–66. [Google Scholar] [CrossRef]
- Abroi, A.; Gough, J. Are viruses a source of new protein folds for organisms? - Virosphere structure space and evolution. BioEssays 2011, 33, 626–635. [Google Scholar] [CrossRef]
- Xu, J.; Wang, D.; Gui, M.; Xiang, Y. Structural assembly of the tailed bacteriophage varphi29. Nat. Commun. 2019, 10, 2366. [Google Scholar] [CrossRef]
- Alexandrescu, A.T.; Gittis, A.G.; Abeygunawardana, C.; Shortle, D. NMR Structure of a Stable “OB-fold” Sub-domain Isolated from Staphylococcal Nuclease. J. Mol. Biol. 1995, 250, 134–143. [Google Scholar] [CrossRef]
- Guardino, K.M.; Sheftic, S.R.; Slattery, R.E.; Alexandrescu, A.T. Relative Stabilities of Conserved and Non-Conserved Structures in the OB-Fold Superfamily. Int. J. Mol. Sci. 2009, 10, 2412–2430. [Google Scholar] [CrossRef]
- Murzin, A.G. OB (oligonucleotide/oligosaccharide binding)-fold: Common structural and functional solution for non-homologous sequences. EMBO J. 1993, 12, 861–867. [Google Scholar] [CrossRef]
- Holm, L.; Rosenstr�MP. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010, 38, W545–W549. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Hendrix, R.W. Bacteriophage genomics. Curr. Opin. Microbiol. 2003, 6, 506–511. [Google Scholar] [CrossRef]
- Krupovic, M.; Koonin, E.V. Multiple origins of viral capsid proteins from cellular ancestors. Proc. Natl. Acad. Sci. USA 2017, 114, E2401–E2410. [Google Scholar] [CrossRef]
- Xiang, Y.; Leiman, P.G.; Li, L.; Grimes, S.; Anderson, D.L.; Rossmann, M.G. Crystallographic Insights into the Autocatalytic Assembly Mechanism of a Bacteriophage Tail Spike. Mol. Cell 2009, 34, 375–386. [Google Scholar] [CrossRef]
- Asija, K.; Teschke, C.M. Lessons from bacteriophages part 2: A saga of scientific breakthroughs and prospects for their use in human health. PLoS Pathog. 2018, 14, e1006970. [Google Scholar] [CrossRef]
- Asija, K.; Teschke, C.M. Lessons from bacteriophages part 1: Deriving utility from protein structure, function, and evolution. PLoS Pathog. 2018, 14, e1006971. [Google Scholar] [CrossRef]
- Sarikaya, M.; Tamerler, C.; Jen, A.K.-Y.; Schulten, K.; Baneyx, F. Molecular biomimetics: Nanotechnology through biology. Nat. Mater. 2003, 2, 577–585. [Google Scholar] [CrossRef]
- Vera-Robles, L.I.; González-Gracida, J.; Hernández-Gordillo, A.; Campero, A. Using the M13 Phage as a Biotemplate to Create Mesoporous Structures Decorated with Gold and Platinum Nanoparticles. Langmuir 2015, 31, 9188–9197. [Google Scholar] [CrossRef]
- Chatterji, A.; Ochoa, W.F.; Ueno, T.; Lin, T.; Johnson, J.E. A Virus-Based Nanoblock with Tunable Electrostatic Properties. Nano Lett. 2005, 5, 597–602. [Google Scholar] [CrossRef]
- Klem, M.T.; Young, M.; Douglas, T. Biomimetic magnetic nanoparticles. Mater. Today 2005, 8, 28–37. [Google Scholar] [CrossRef]
- Cho, C.F.; Shukla, S.; Simpson, E.J.; Steinmetz, N.F.; Luyt, L.G.; Lewis, J.D. Molecular targeted viral nanoparticles as tools for imaging cancer. Methods Mol. Biol. 2014, 1108, 211–230. [Google Scholar]
- Li, Q.; Shivachandra, S.B.; Zhang, Z.; Rao, V.B. Assembly of the Small Outer Capsid Protein, Soc, on Bacteriophage T4: A Novel System for High Density Display of Multiple Large Anthrax Toxins and Foreign Proteins on Phage Capsid. J. Mol. Biol. 2007, 370, 1006–1019. [Google Scholar] [CrossRef]
- Van Rosmalen, M.G.M.; Nemerow, G.R.; Wuite, G.J.L.; Roos, W.H. A single point mutation in precursor protein VI doubles the mechanical strength of human adenovirus. J. Biol. Phys. 2017, 44, 119–132. [Google Scholar] [CrossRef]
- Srivastava, S.; Nykypanchuk, D.; Fukuto, M.; Halverson, J.D.; Tkachenko, A.V.; Yager, K.G.; Gang, O. Two-Dimensional DNA-Programmable Assembly of Nanoparticles at Liquid Interfaces. J. Am. Chem. Soc. 2014, 136, 8323–8332. [Google Scholar] [CrossRef]
- Zhang, S.; Li, Z.; Samarajeewa, S.; Sun, G.; Yang, C.; Wooley, K.L. Orthogonally Dual-Clickable Janus Nanoparticles via a Cyclic Templating Strategy. J. Am. Chem. Soc. 2011, 133, 11046–11049. [Google Scholar] [CrossRef][Green Version]
- Braunschweig, A.B.; Huo, F.; Mirkin, C.A. Molecular printing. Nat. Chem. 2009, 1, 353–358. [Google Scholar] [CrossRef]
- Alberti, S.; Soler-Illia, G.J.; Azzaroni, O. Gated supramolecular chemistry in hybrid mesoporous silica nanoarchitectures: Controlled delivery and molecular transport in response to chemical, physical and biological stimuli. Chem. Commun. (Camb.) 2015, 51, 6050–6075. [Google Scholar] [CrossRef]
- The Nobel Prize is Chemistry 2018. Available online: https://www.nobelprize.org/prizes/chemistry/2018/summary/ (accessed on 1 August 2020).
- Smith, G.P. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, 1315–1317. [Google Scholar] [CrossRef]
- Jiang, J.; Abu-Shilbayeh, L.; Rao, V.B. Display of a PorA peptide from Neisseria meningitidis on the bacteriophage T4 capsid surface. Infect. Immun. 1997, 65, 4770–4777. [Google Scholar] [CrossRef]
- Li, Q.; Shivachandra, S.B.; Leppla, S.H.; Rao, V.B. Bacteriophage T4 Capsid: A Unique Platform for Efficient Surface Assembly of Macromolecular Complexes. J. Mol. Biol. 2006, 363, 577–588. [Google Scholar] [CrossRef]
- Meulenbroek, R.A.; Sargent, K.L.; Lunde, J.; Jasmin, B.J.; Parks, R.J.; Meulenbroek, R. Use of adenovirus protein IX (pIX) to display large polypeptides on the virion—Generation of fluorescent virus through the incorporation of pIX-GFP. Mol. Ther. 2004, 9, 617–624. [Google Scholar] [CrossRef]
- Sathaliyawala, T.; Rao, M.; MacLean, D.M.; Birx, D.L.; Alving, C.R.; Rao, V.B. Assembly of Human Immunodeficiency Virus (HIV) Antigens on Bacteriophage T4: A Novel In Vitro Approach To Construct Multicomponent HIV Vaccines. J. Virol. 2006, 80, 7688–7698. [Google Scholar] [CrossRef]
- Malys, N.; Chang, D.-Y.; Baumann, R.G.; Xie, D.; Black, L.W. A Bipartite Bacteriophage T4 SOC and HOC Randomized Peptide Display Library: Detection and Analysis of Phage T4 Terminase (gp17) and Late σ Factor (gp55) Interaction. J. Mol. Biol. 2002, 319, 289–304. [Google Scholar] [CrossRef]
- Aghebati-Maleki, L.; Bakhshinejad, B.; Baradaran, B.; Motallebnezhad, M.; Aghebati-Maleki, A.; Nickho, H.; Yousefi, M.; Majidi, J. Phage display as a promising approach for vaccine development. J. Biomed. Sci. 2016, 23, 66. [Google Scholar] [CrossRef]
- Lee, J.-W.; Song, J.; Hwang, M.P.; Lee, K.H. Nanoscale bacteriophage biosensors beyond phage display. Int. J. Nanomed. 2013, 8, 3917–3925. [Google Scholar] [CrossRef]
- Chen, I.-C.; Chiu, Y.-K.; Yu, C.-M.; Lee, C.-C.; Tung, C.-P.; Tsou, Y.-L.; Huang, Y.-J.; Lin, C.-L.; Chen, H.-S.; Wang, A.H.-J.; et al. High throughput discovery of influenza virus neutralizing antibodies from phage-displayed synthetic antibody libraries. Sci. Rep. 2017, 7, 14455. [Google Scholar] [CrossRef]
- Yuan, A.Q.; Zhao, L.; Bai, L.; Meng, Q.; Wen, Z.; Li, Y.; Guo, D.; Zhen, S.; Chen, X.; Yang, J.; et al. Isolation of and Characterization of Neutralizing Antibodies to Covid-19 from a Large Human Naïve scFv Phage Display Library. bioRxiv 2020. [Google Scholar] [CrossRef]
- Kang, X.; Yang, B.-A.; Hu, Y.; Zhao, H.; Xiong, W.; Yang, Y.; Si, B.; Zhu, Q. Human Neutralizing Fab Molecules against Severe Acute Respiratory Syndrome Coronavirus Generated by Phage Display. Clin. Vaccine Immunol. 2006, 13, 953–957. [Google Scholar] [CrossRef]
- Yu, H.; Jiang, L.-F.; Fang, D.-Y.; Yan, H.-J.; Zhou, J.-J.; Zhou, J.-M.; Liang, Y.; Gao, Y.; Zhao, W.; Long, B.-G. Selection of SARS-Coronavirus-specific B cell epitopes by phage peptide library screening and evaluation of the immunological effect of epitope-based peptides on mice. Virology 2007, 359, 264–274. [Google Scholar] [CrossRef]
- Karch, C.P.; Burkhard, P. Vaccine technologies: From whole organisms to rationally designed protein assemblies. Biochem. Pharmacol. 2016, 120, 1–14. [Google Scholar] [CrossRef]
- Slifka, M.K.; Amanna, I.J. Role of Multivalency and Antigenic Threshold in Generating Protective Antibody Responses. Front. Immunol. 2019, 10, 956. [Google Scholar] [CrossRef]
- Tao, P.; Mahalingam, M.; Marasa, B.S.; Zhang, Z.; Chopra, A.K.; Rao, V.B. In Vitro and In Vivo delivery of genes and proteins using the bacteriophage T4 DNA packaging machine. Proc. Natl. Acad. Sci. USA 2013, 110, 5846–5851. [Google Scholar] [CrossRef]
- Hayes, S.; Gamage, L.N.A.; Hayes, C. Dual expression system for assembling phage lambda display particle (LDP) vaccine to porcine Circovirus 2 (PCV2). Vaccine 2010, 28, 6789–6799. [Google Scholar] [CrossRef]
- Cano, P.G.; Gamage, L.N.A.; Marciniuk, K.; Hayes, C.; Napper, S.; Hayes, S.; Griebel, P.J. Lambda display phage as a mucosal vaccine delivery vehicle for peptide antigens. Vaccine 2017, 35, 7256–7263. [Google Scholar] [CrossRef]
- Razazan, A.; Nicastro, J.; Slavcev, R.; Barati, N.; Arab, A.; Mosaffa, F.; Jaafari, M.R.; Behravan, J. Lambda bacteriophage nanoparticles displaying GP2, a HER2/neu derived peptide, induce prophylactic and therapeutic activities against TUBO tumor model in mice. Sci. Rep. 2019, 9, 2221. [Google Scholar] [CrossRef]
- Shivachandra, S.B.; Li, Q.; Peachman, K.K.; Matyas, G.R.; Leppla, S.H.; Alving, C.R.; Rao, M.; Rao, V.B. Multicomponent anthrax toxin display and delivery using bacteriophage T4. Vaccine 2007, 25, 1225–1235. [Google Scholar] [CrossRef]
- Shivachandra, S.B.; Rao, M.; Janosi, L.; Sathaliyawala, T.; Matyas, G.R.; Alving, C.R.; Leppla, S.H.; Rao, V.B. In vitro binding of anthrax protective antigen on bacteriophage T4 capsid surface through Hoc–capsid interactions: A strategy for efficient display of large full-length proteins. Virology 2006, 345, 190–198. [Google Scholar] [CrossRef]
- Teschke, C.M.; Parent, K.N. ‘Let the phage do the work’: Using the phage P22 coat protein structures as a framework to understand its folding and assembly mutants. Virology 2010, 401, 119–130. [Google Scholar] [CrossRef]
- Patterson, D.P.; Rynda-Apple, A.; Harmsen, A.L.; Harmsen, A.G.; Douglas, T. Biomimetic Antigenic Nanoparticles Elicit Controlled Protective Immune Response to Influenza. ACS Nano 2013, 7, 3036–3044. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).