Impact of Microbiota: A Paradigm for Evolving Herd Immunity against Viral Diseases
Abstract
1. Introduction
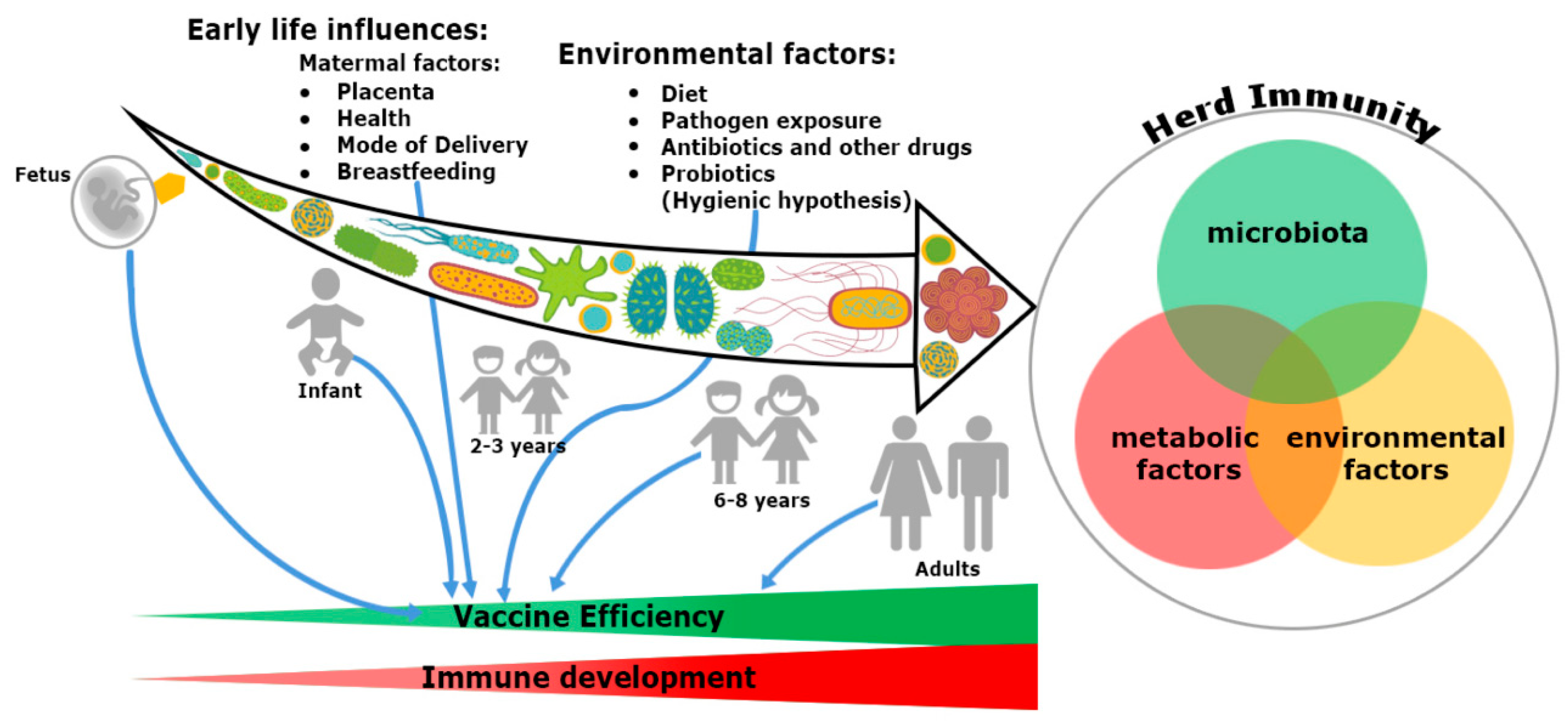
2. Trinity of the Immune System Development: Microbiome, Nutrients and Environmental Factors
2.1. Putting the Microbiome, the Second Brain to Develop the Immune System
2.2. Impact of Nutrients
2.3. Impact of Environmental Factors
3. Route of Immunization Determines the Efficacy of Herd Immunity
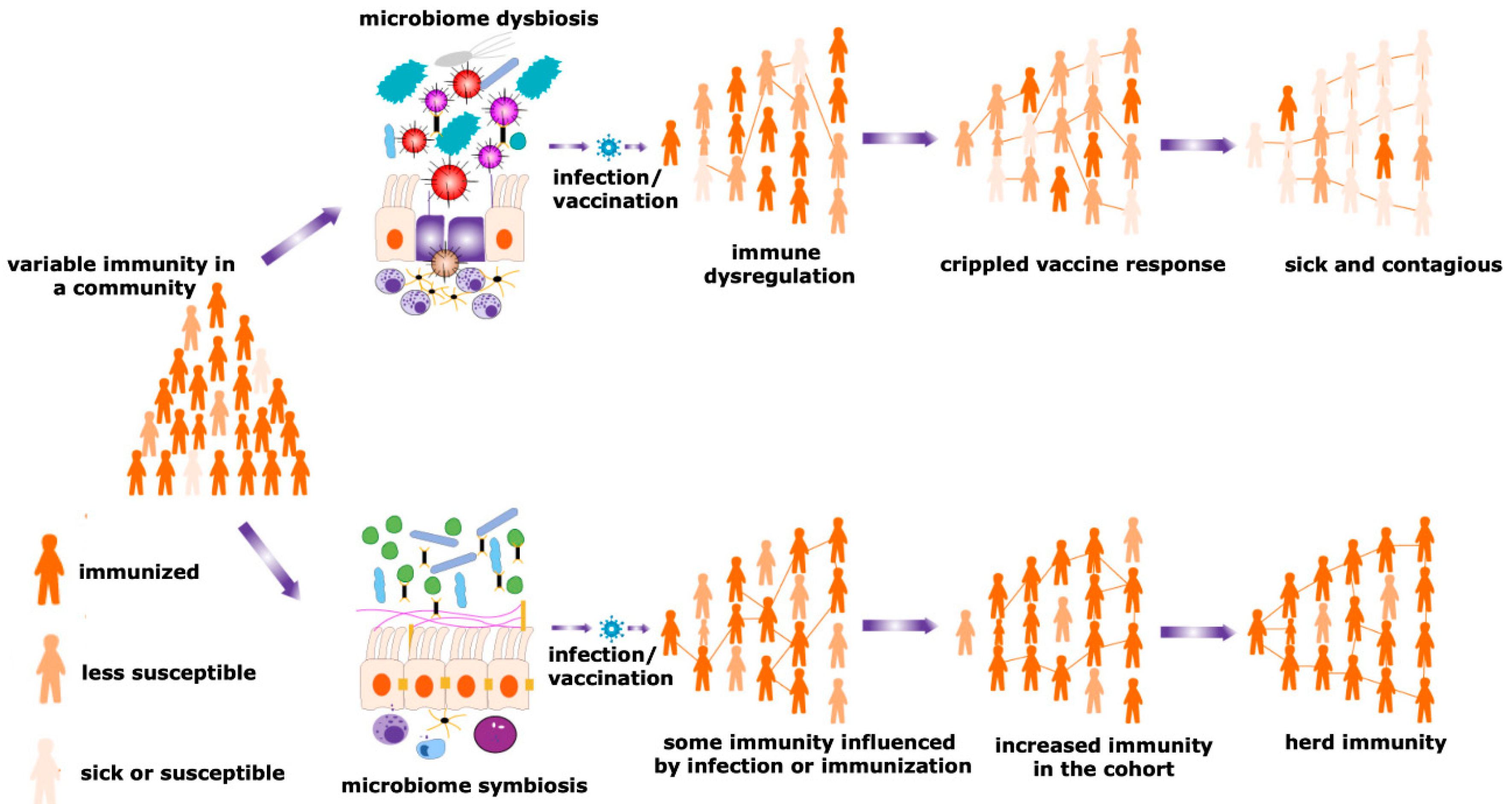
4. Potential Role of the Microbiome in the Individual Vaccination to Community Immunity
4.1. Differential Expression of Microbiota Decides the Fate of Vaccination
4.2. Effect of EE on Vaccination
4.3. Cardinal Effect of Probiotics and Antibiotics on Avidity of Vaccination
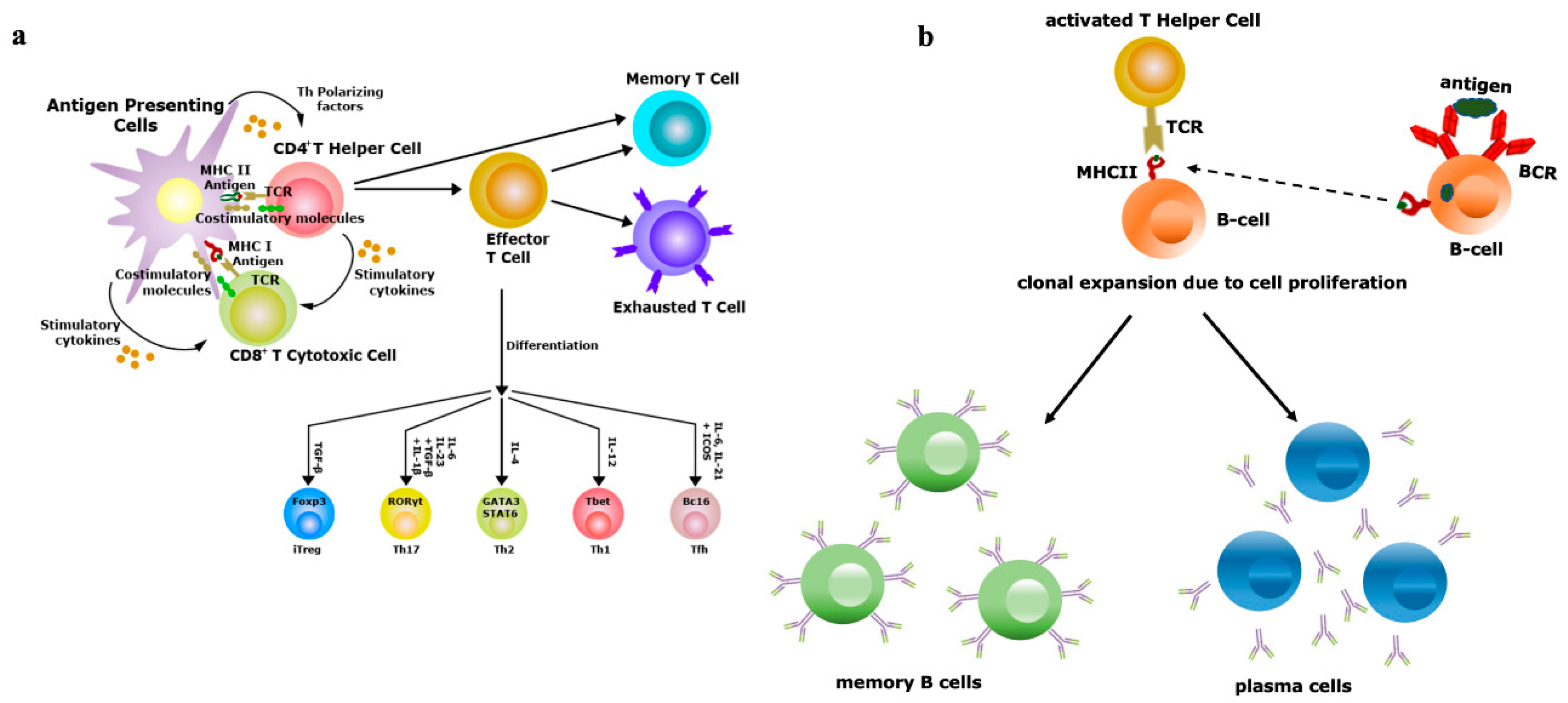
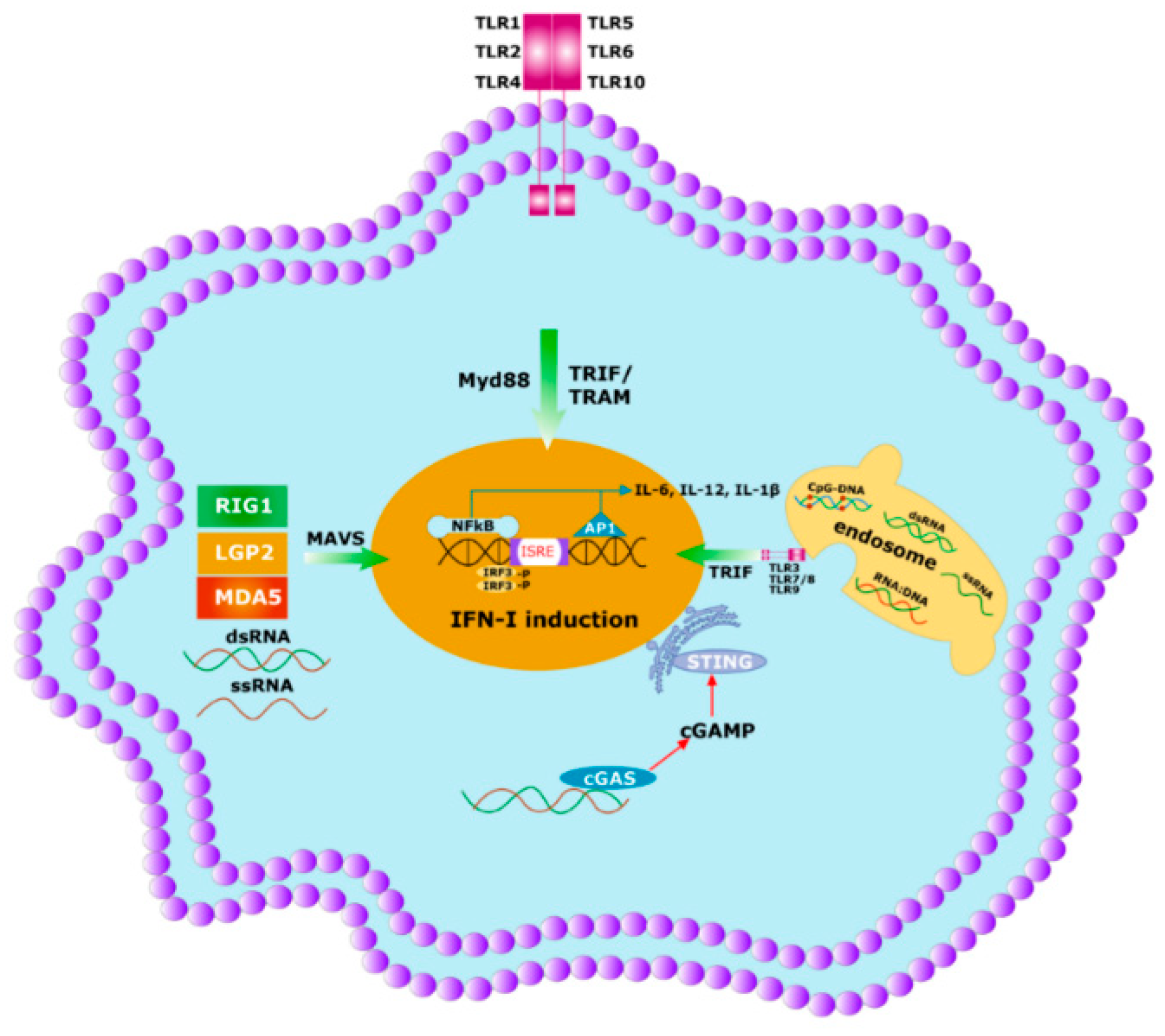
5. Innate Immunity Controls Antibody Titter
6. The Specificity of Herd Immunity Relies on Innate Immunity
7. Realm of Herd Immunity Can Be Established by Heterologous Immunity
8. Heterologous Immunity Provides Significant Protection against SARS-CoV-2 Infection in India
8.1. Herd Immunity against Coronavirus and Impact of Microbiota
8.2. Microbiome: New Songs in Old Music
8.3. Herd Immunity: Friends or Fes for Covid-19
8.4. Heterologous Immunity: Poliovirus Vaccine May Provide Protection against SARS-CoV-2 Infection
8.5. In Silico Comparison of SARS-CoV-2 with OPV and BCG
9. Perspective and Future Opportunity
10. Conclusions
- Herd immunity is the most critical prophylactic intervention, delivering the protective immunity against several infectious diseases such as smallpox, poliovirus, measles, etc. in the past. The new paradigm of evolving herd immunity during a pandemic situation cannot be disregarded.
- Crosstalk among microbiota, metabolism and environmental factors is critical for developing a competent immune system, which is a prerequisite for evolving herd immunity against any contagious infections;
- The importance of herd immunity has been documented with the current context of a pandemic scenario due to transmissible infection of SARS-CoV-2 virus;
- In India, the infection rate of SARS-CoV-2 is unexpectedly very low, i.e., only 0.37% of total population. The prevalence of heterologous immunity due to rigorous vaccination programs at the grass-root level may provide protection against the SARS-CoV-2 pandemic;
- Comparisons between SARS-CoV-2 Orf7a protein epitope (KHVYQLRARS) and the human poliovirus type 3 Sabin strain epitope (RHVVQRRSRS) from VP1 protein offer a great insights into the concept of heterologous immunity, which can be an alternative providing prophylactic intervention against both the poliovirus and COVID-19.
Author Contributions
Funding
Conflicts of Interest
References
- Nokes, D.J.; Anderson, R.M. The use of mathematical models in the epidemiological study of infectious diseases and in the design of mass immunization programmes. Epidemiol. Infect. 1988, 101, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.M.; May, R.M. Vaccination and herd immunity to infectious diseases. Nature 1985, 318, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Fine, P.; Eames, K.; Heymann, D.L. “Herd immunity”: A rough guide. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2011, 52, 911–916. [Google Scholar] [CrossRef] [PubMed]
- John, T.J.; Samuel, R. Herd immunity and herd effect: New insights and definitions. Eur. J. Epidemiol. 2000, 16, 601–606. [Google Scholar] [CrossRef]
- Lane, J.M. Mass vaccination and surveillance/containment in the eradication of smallpox. Curr. Top. Microbiol. Immunol. 2006, 304, 17–29. [Google Scholar] [CrossRef]
- Ali, M.; Emch, M.; von Seidlein, L.; Yunus, M.; Sack, D.A.; Rao, M.; Holmgren, J.; Clemens, J.D. Herd immunity conferred by killed oral cholera vaccines in Bangladesh: A reanalysis. Lancet 2005, 366, 44–49. [Google Scholar] [CrossRef]
- Mast, T.C.; Wang, F.T.; Su, S.; Seeger, J.D. Evidence of herd immunity and sustained impact of rotavirus vaccination on the reduction of rotavirus-related medical encounters among infants from 2006 through 2011 in the United States. Pediatr. Infect. Dis. J. 2015, 34, 615–620. [Google Scholar] [CrossRef]
- Kim, T.H.; Johnstone, J.; Loeb, M. Vaccine herd effect. Scand. J. Infect. Dis. 2011, 43, 683–689. [Google Scholar] [CrossRef]
- Metcalf, C.J.E.; Ferrari, M.; Graham, A.L.; Grenfell, B.T. Understanding herd immunity. Trends Immunol. 2015, 36, 753–755. [Google Scholar] [CrossRef]
- Liu, Y.; Gayle, A.A.; Wilder-Smith, A.; Rocklöv, J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J. Travel Med. 2020, 27. [Google Scholar] [CrossRef]
- Kwok, K.O.; Lai, F.; Wei, W.I.; Wong, S.Y.S.; Tang, J.W.T. Herd immunity—Estimating the level required to halt the COVID-19 epidemics in affected countries. J. Infect. 2020, 80, e32–e33. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.B.; Antunes, L.C.; Finlay, B.B. Should the human microbiome be considered when developing vaccines? PLoS Pathog. 2010, 6, e1001190. [Google Scholar] [CrossRef] [PubMed]
- Lynn, D.J.; Pulendran, B. The potential of the microbiota to influence vaccine responses. J. Leukoc. Biol. 2018, 103, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Guttinger, S. The anti-vaccination debate and the microbiome: How paradigm shifts in the life sciences create new challenges for the vaccination debate. EMBO Rep. 2019, 20. [Google Scholar] [CrossRef] [PubMed]
- Lederberg, J.; McCray, A.T. Ome SweetOmics—A genealogical treasury of words. Scientist 2001, 15, 8. [Google Scholar]
- Ley, R.E.; Peterson, D.A.; Gordon, J.I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 2006, 124, 837–848. [Google Scholar] [CrossRef]
- Cerf-Bensussan, N.; Gaboriau-Routhiau, V. The immune system and the gut microbiota: Friends or foes? Nat. Rev. Immunol. 2010, 10, 735–744. [Google Scholar] [CrossRef]
- Wu, H.J.; Wu, E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes 2012, 3, 4–14. [Google Scholar] [CrossRef]
- Grice, E.A.; Segre, J.A. The human microbiome: Our second genome. Annu. Rev. Genom. Hum. Genet. 2012, 13, 151–170. [Google Scholar] [CrossRef]
- Gensollen, T.; Iyer, S.S.; Kasper, D.L.; Blumberg, R.S. How colonization by microbiota in early life shapes the immune system. Science 2016, 352, 539–544. [Google Scholar] [CrossRef]
- Magwira, C.A.; Taylor, M.B. Composition of gut microbiota and its influence on the immunogenicity of oral rotavirus vaccines. Vaccine 2018, 36, 3427–3433. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, I.I.; Honda, K. Intestinal commensal microbes as immune modulators. Cell Host Microbe 2012, 12, 496–508. [Google Scholar] [CrossRef] [PubMed]
- Mazmanian, S.K.; Liu, C.H.; Tzianabos, A.O.; Kasper, D.L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 2005, 122, 107–118. [Google Scholar] [CrossRef]
- Round, J.L.; Mazmanian, S.K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. USA 2010, 107, 12204–12209. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef]
- Gaboriau-Routhiau, V.; Rakotobe, S.; Lécuyer, E.; Mulder, I.; Lan, A.; Bridonneau, C.; Rochet, V.; Pisi, A.; De Paepe, M.; Brandi, G.; et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 2009, 31, 677–689. [Google Scholar] [CrossRef] [PubMed]
- Mueller, N.T.; Bakacs, E.; Combellick, J.; Grigoryan, Z.; Dominguez-Bello, M.G. The infant microbiome development: Mom matters. Trends Mol. Med. 2015, 21, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Penders, J.; Thijs, C.; Vink, C.; Stelma, F.F.; Snijders, B.; Kummeling, I.; van den Brandt, P.A.; Stobberingh, E.E. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 2006, 118, 511–521. [Google Scholar] [CrossRef]
- Aagaard, K.; Ma, J.; Antony, K.M.; Ganu, R.; Petrosino, J.; Versalovic, J. The placenta harbors a unique microbiome. Sci. Transl. Med. 2014, 6, 237ra265. [Google Scholar] [CrossRef]
- Gritz, E.C.; Bhandari, V. The human neonatal gut microbiome: A brief review. Front. Pediatr. 2015, 3, 17. [Google Scholar] [CrossRef]
- Sood, R.; Zehnder, J.L.; Druzin, M.L.; Brown, P.O. Gene expression patterns in human placenta. Proc. Natl. Acad. Sci. USA 2006, 103, 5478–5483. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef] [PubMed]
- Pantoja-Feliciano, I.G.; Clemente, J.C.; Costello, E.K.; Perez, M.E.; Blaser, M.J.; Knight, R.; Dominguez-Bello, M.G. Biphasic assembly of the murine intestinal microbiota during early development. ISME J. 2013, 7, 1112–1115. [Google Scholar] [CrossRef] [PubMed]
- Biasucci, G.; Rubini, M.; Riboni, S.; Morelli, L.; Bessi, E.; Retetangos, C. Mode of delivery affects the bacterial community in the newborn gut. Early Hum. Dev. 2010, 86 (Suppl. S1), 13–15. [Google Scholar] [CrossRef]
- Madan, J.C.; Farzan, S.F.; Hibberd, P.L.; Karagas, M.R. Normal neonatal microbiome variation in relation to environmental factors, infection and allergy. Curr. Opin. Pediatr. 2012, 24, 753–759. [Google Scholar] [CrossRef]
- Hunt, K.M.; Foster, J.A.; Forney, L.J.; Schütte, U.M.; Beck, D.L.; Abdo, Z.; Fox, L.K.; Williams, J.E.; McGuire, M.K.; McGuire, M.A. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS ONE 2011, 6, e21313. [Google Scholar] [CrossRef]
- Balmer, S.E.; Wharton, B.A. Diet and faecal flora in the newborn: Breast milk and infant formula. Arch. Dis. Child. 1989, 64, 1672–1677. [Google Scholar] [CrossRef]
- Harmsen, H.J.; Wildeboer-Veloo, A.C.; Raangs, G.C.; Wagendorp, A.A.; Klijn, N.; Bindels, J.G.; Welling, G.W. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 2000, 30, 61–67. [Google Scholar] [CrossRef]
- Yoshioka, H.; Iseki, K.; Fujita, K. Development and differences of intestinal flora in the neonatal period in breast-fed and bottle-fed infants. Pediatrics 1983, 72, 317–321. [Google Scholar]
- Pérez-Pérez, A.; Vilariño-García, T.; Fernández-Riejos, P.; Martín-González, J.; Segura-Egea, J.J.; Sánchez-Margalet, V. Role of leptin as a link between metabolism and the immune system. Cytokine Growth Factor Rev. 2017, 35, 71–84. [Google Scholar] [CrossRef]
- La Cava, A.; Matarese, G. The weight of leptin in immunity. Nat. Rev. Immunol. 2004, 4, 371–379. [Google Scholar] [CrossRef] [PubMed]
- De Rosa, V.; Procaccini, C.; Calì, G.; Pirozzi, G.; Fontana, S.; Zappacosta, S.; La Cava, A.; Matarese, G. A key role of leptin in the control of regulatory T cell proliferation. Immunity 2007, 26, 241–255. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.H.; Park, J.; Kim, M. Gut microbiota-derived short-chain Fatty acids, T cells, and inflammation. Immune Netw. 2014, 14, 277–288. [Google Scholar] [CrossRef]
- Fukuda, S.; Toh, H.; Hase, K.; Oshima, K.; Nakanishi, Y.; Yoshimura, K.; Tobe, T.; Clarke, J.M.; Topping, D.L.; Suzuki, T.; et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011, 469, 543–547. [Google Scholar] [CrossRef]
- Peng, L.; He, Z.; Chen, W.; Holzman, I.R.; Lin, J. Effects of butyrate on intestinal barrier function in a Caco-2 cell monolayer model of intestinal barrier. Pediatr. Res. 2007, 61, 37–41. [Google Scholar] [CrossRef]
- Sommer, A.; Tarwotjo, I.; Katz, J. Increased risk of xerophthalmia following diarrhea and respiratory disease. Am. J. Clin. Nutr. 1987, 45, 977–980. [Google Scholar] [CrossRef]
- Cha, H.R.; Chang, S.Y.; Chang, J.H.; Kim, J.O.; Yang, J.Y.; Kim, C.H.; Kweon, M.N. Downregulation of Th17 cells in the small intestine by disruption of gut flora in the absence of retinoic acid. J. Immunol. 2010, 184, 6799–6806. [Google Scholar] [CrossRef]
- Shapiro, H.; Thaiss, C.A.; Levy, M.; Elinav, E. The cross talk between microbiota and the immune system: Metabolites take center stage. Curr. Opin. Immunol. 2014, 30, 54–62. [Google Scholar] [CrossRef]
- Pawankar, R.; Canonica, G.W.; Holgate, S.T.; Lockey, R.F.; Blaiss, M.S. WAO White Book on Allergy; World Allergy Organization: Milwaukee, WI, USA, 2011; Volume 3. [Google Scholar]
- Platts-Mills, T.A. The allergy epidemics: 1870–2010. J. Allergy Clin. Immunol. 2015, 136, 3–13. [Google Scholar] [CrossRef]
- Ritchel, M. Your environment is cleaner. Your immune system has never been so unprepared. The New York Times, 12 March 2019; D1. [Google Scholar]
- Strachan, D.P. Hay fever, hygiene, and household size. BMJ Clin. Res. Ed. 1989, 299, 1259–1260. [Google Scholar] [CrossRef] [PubMed]
- Apostol, A.C.; Jensen, K.D.C.; Beaudin, A.E. Training the fetal immune system through maternal inflammation-A layered hygiene hypothesis. Front. Immunol. 2020, 11, 123. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, M.; Wang, S.; Han, R.; Cao, Y.; Hua, W.; Mao, Y.; Zhang, X.; Pang, X.; Wei, C.; et al. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 2010, 4, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Deehan, E.C.; Walter, J. The fiber gap and the disappearing gut microbiome: Implications for human nutrition. Trends Endocrinol. Metab. 2016, 27, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Sonnenburg, E.D.; Smits, S.A.; Tikhonov, M.; Higginbottom, S.K.; Wingreen, N.S.; Sonnenburg, J.L. Diet-induced extinctions in the gut microbiota compound over generations. Nature 2016, 529, 212–215. [Google Scholar] [CrossRef]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef]
- Andre, F.E.; Booy, R.; Bock, H.L.; Clemens, J.; Datta, S.K.; John, T.J.; Lee, B.W.; Lolekha, S.; Peltola, H.; Ruff, T.A.; et al. Vaccination greatly reduces disease, disability, death and inequity worldwide. Bull. World Health Organ. 2008, 86, 140–146. [Google Scholar] [CrossRef]
- Levine, M.M. Immunogenicity and efficacy of oral vaccines in developing countries: Lessons from a live cholera vaccine. BMC Biol. 2010, 8, 129. [Google Scholar] [CrossRef]
- Huda, M.N.; Lewis, Z.; Kalanetra, K.M.; Rashid, M.; Ahmad, S.M.; Raqib, R.; Qadri, F.; Underwood, M.A.; Mills, D.A.; Stephensen, C.B. Stool microbiota and vaccine responses of infants. Pediatrics 2014, 134, e362–e372. [Google Scholar] [CrossRef]
- John, T.J.; Jayabal, P. Oral polio vaccination of children in the tropics: I. The poor seroconversion rates and the absence of viral interference. Am. J. Epidemiol. 1972, 96, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Patriarca, P.A.; Wright, P.F.; John, T.J. Factors affecting the immunogenicity of oral poliovirus vaccine in developing countries: Review. Rev. Infect. Dis. 1991, 13, 926–939. [Google Scholar] [CrossRef] [PubMed]
- Hallander, H.O.; Paniagua, M.; Espinoza, F.; Askelöf, P.; Corrales, E.; Ringman, M.; Storsaeter, J. Calibrated serological techniques demonstrate significant different serum response rates to an oral killed cholera vaccine between Swedish and Nicaraguan children. Vaccine 2002, 21, 138–145. [Google Scholar] [CrossRef]
- Levine, M.M.; Kotloff, K.L.; Barry, E.M.; Pasetti, M.F.; Sztein, M.B. Clinical trials of Shigella vaccines: Two steps forward and one step back on a long, hard road. Nat. Rev. Microbiol. 2007, 5, 540–553. [Google Scholar] [CrossRef] [PubMed]
- Lanata, C.F.; Midthun, K.; Black, R.E.; Butron, B.; Huapaya, A.; Penny, M.E.; Ventura, G.; Gil, A.; Jett-Goheen, M.; Davidson, B.L. Safety, immunogenicity, and protective efficacy of one and three doses of the tetravalent rhesus rotavirus vaccine in infants in Lima, Peru. J. Infect. Dis. 1996, 174, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Gotuzzo, E.; Butron, B.; Seas, C.; Penny, M.; Ruiz, R.; Losonsky, G.; Lanata, C.F.; Wasserman, S.S.; Salazar, E.; Kaper, J.B.; et al. Safety, immunogenicity, and excretion pattern of single-dose live oral cholera vaccine CVD 103-HgR in Peruvian adults of high and low socioeconomic levels. Infect. Immun. 1993, 61, 3994–3997. [Google Scholar] [CrossRef] [PubMed]
- Simanjuntak, C.; Witham, N.; Punjabi, N.; Heppner, D.G.; Losonsky, G.; Totosudirjo, H.; Rifai, A.R.; Clemens, J.; Lim, Y.L.; Sorenson, K.; et al. Safety and immunogenicity of single-dose live oral cholera vaccine CVD 103-HgR in 5–9-year-old Indonesian children. Lancet 1992, 340, 689–694. [Google Scholar] [CrossRef]
- McDermott, A.J.; Huffnagle, G.B. The microbiome and regulation of mucosal immunity. Immunology 2014, 142, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhao, S.; Liu, M.; Zhao, Z.; Xu, Y.; Wang, P.; Lin, M.; Xu, Y.; Huang, B.; Zuo, X.; et al. ACE2 expression by colonic epithelial cells is associated with viral infection, immunity and energy metabolism. medRxiv 2020. [Google Scholar] [CrossRef]
- Zimmermann, P.; Curtis, N. The influence of the intestinal microbiome on vaccine responses. Vaccine 2018, 36, 4433–4439. [Google Scholar] [CrossRef] [PubMed]
- Eloe-Fadrosh, E.A.; McArthur, M.A.; Seekatz, A.M.; Drabek, E.F.; Rasko, D.A.; Sztein, M.B.; Fraser, C.M. Impact of oral typhoid vaccination on the human gut microbiota and correlations with s. Typhi-specific immunological responses. PLoS ONE 2013, 8, e62026. [Google Scholar] [CrossRef] [PubMed]
- Harris, V.; Ali, A.; Fuentes, S.; Korpela, K.; Kazi, M.; Tate, J.; Parashar, U.; Wiersinga, W.J.; Giaquinto, C.; de Weerth, C.; et al. Rotavirus vaccine response correlates with the infant gut microbiota composition in Pakistan. Gut Microbes 2018, 9, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Harris, V.C.; Armah, G.; Fuentes, S.; Korpela, K.E.; Parashar, U.; Victor, J.C.; Tate, J.; de Weerth, C.; Giaquinto, C.; Wiersinga, W.J.; et al. Significant correlation between the infant gut microbiome and rotavirus vaccine response in rural ghana. J. Infect. Dis. 2017, 215, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Moreau, M.C.; Routhiau, V.; Dubuquoy, C.; Bisetti, N.; Bouley, C.; Prévoteau, H. Modulating properties of intestinal bacterial strains, Escherichia coli and bifidobacterium, on two specific immune responses generated by the gut, i.e. oral tolerance to ovalbumin and intestinal IgA anti-rotavirus response, in gnotobiotic mice. In Proceedings of the 10th International Congress of Immunology, New Delhi, India, 1–6 November 1998. [Google Scholar]
- Mullié, C.; Yazourh, A.; Thibault, H.; Odou, M.F.; Singer, E.; Kalach, N.; Kremp, O.; Romond, M.B. Increased poliovirus-specific intestinal antibody response coincides with promotion of Bifidobacterium longum-infantis and Bifidobacterium breve in infants: A randomized, double-blind, placebo-controlled trial. Pediatr. Res. 2004, 56, 791–795. [Google Scholar] [CrossRef] [PubMed]
- Pabst, O.; Hornef, M. Gut microbiota: A natural adjuvant for vaccination. Immunity 2014, 41, 349–351. [Google Scholar] [CrossRef] [PubMed]
- Gilmartin, A.A.; Petri, W.A., Jr. Exploring the role of environmental enteropathy in malnutrition, infant development and oral vaccine response. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2015, 370. [Google Scholar] [CrossRef] [PubMed]
- Harper, K.M.; Mutasa, M.; Prendergast, A.J.; Humphrey, J.; Manges, A.R. Environmental enteric dysfunction pathways and child stunting: A systematic review. PLoS Negl. Trop. Dis. 2018, 12, e0006205. [Google Scholar] [CrossRef] [PubMed]
- McKay, S.; Gaudier, E.; Campbell, D.I.; Prentice, A.M.; Albers, R. Environmental enteropathy: New targets for nutritional interventions. Int. Health 2010, 2, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Xavier, R.J. Growth and the Microbiome—Integrating global health with basic science. N. Engl. J. Med. 2020, 383, 391–393. [Google Scholar] [CrossRef] [PubMed]
- Dahan, S.; Roth-Walter, F.; Arnaboldi, P.; Agarwal, S.; Mayer, L. Epithelia: Lymphocyte interactions in the gut. Immunol. Rev. 2007, 215, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.M.; Garrett, W.S. The gut microbiota and mucosal T cells. Front. Microbiol. 2011, 2, 111. [Google Scholar] [CrossRef] [PubMed]
- Becattini, S.; Taur, Y.; Pamer, E.G. Antibiotic-induced changes in the intestinal microbiota and disease. Trends Mol. Med. 2016, 22, 458–478. [Google Scholar] [CrossRef] [PubMed]
- Bach, J.F. The effect of infections on susceptibility to autoimmune and allergic diseases. N. Engl. J. Med. 2002, 347, 911–920. [Google Scholar] [CrossRef] [PubMed]
- West, C.E. Gut microbiota and allergic disease: New findings. Curr. Opin. Clin. Nutr. Metab. Care 2014, 17, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Fagundes-Neto, U.; Viaro, T.; Wehba, J.; Patrício, F.R.; Machado, N.L. Tropical enteropathy (environmental enteropathy) in early childhood: A syndrome caused by contaminated environment. J. Trop. Pediatr. 1984, 30, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Taniuchi, M.; Sobuz, S.U.; Begum, S.; Platts-Mills, J.A.; Liu, J.; Yang, Z.; Wang, X.Q.; Petri, W.A., Jr.; Haque, R.; Houpt, E.R. Etiology of diarrhea in Bangladeshi infants in the first year of life analyzed using molecular methods. J. Infect. Dis. 2013, 208, 1794–1802. [Google Scholar] [CrossRef] [PubMed]
- Lagos, R.; Fasano, A.; Wasserman, S.S.; Prado, V.; San Martin, O.; Abrego, P.; Losonsky, G.A.; Alegria, S.; Levine, M.M. Effect of small bowel bacterial overgrowth on the immunogenicity of single-dose live oral cholera vaccine CVD 103-HgR. J. Infect. Dis. 1999, 180, 1709–1712. [Google Scholar] [CrossRef][Green Version]
- Clem, A.S. Fundamentals of vaccine immunology. J. Glob. Infect. Dis. 2011, 3, 73–78. [Google Scholar] [CrossRef]
- Hand, T.W. The role of the microbiota in shaping infectious immunity. Trends Immunol. 2016, 37, 647–658. [Google Scholar] [CrossRef]
- Netea, M.G.; Joosten, L.A.; Latz, E.; Mills, K.H.; Natoli, G.; Stunnenberg, H.G.; O’Neill, L.A.; Xavier, R.J. Trained immunity: A program of innate immune memory in health and disease. Science 2016, 352, aaf1098. [Google Scholar] [CrossRef]
- Netea, M.G.; van Crevel, R. BCG-induced protection: Effects on innate immune memory. Semin. Immunol. 2014, 26, 512–517. [Google Scholar] [CrossRef] [PubMed]
- Coster, T.S.; Hoge, C.W.; VanDeVerg, L.L.; Hartman, A.B.; Oaks, E.V.; Venkatesan, M.M.; Cohen, D.; Robin, G.; Fontaine-Thompson, A.; Sansonetti, P.J.; et al. Vaccination against shigellosis with attenuated Shigella flexneri 2a strain SC602. Infect. Immun. 1999, 67, 3437–3443. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Future needs and directions for Shigella vaccines. Relev. Epidemiol. Hebd. 2006, 81, 51–58. [Google Scholar]
- Kirkpatrick, B.D.; Colgate, E.R.; Mychaleckyj, J.C.; Haque, R.; Dickson, D.M.; Carmolli, M.P.; Nayak, U.; Taniuchi, M.; Naylor, C.; Qadri, F.; et al. The “Performance of Rotavirus and Oral Polio Vaccines in Developing Countries” (PROVIDE) study: Description of methods of an interventional study designed to explore complex biologic problems. Am. J. Trop. Med. Hyg. 2015, 92, 744–751. [Google Scholar] [CrossRef] [PubMed]
- Naylor, C.; Lu, M.; Haque, R.; Mondal, D.; Buonomo, E.; Nayak, U.; Mychaleckyj, J.C.; Kirkpatrick, B.; Colgate, R.; Carmolli, M.; et al. Environmental enteropathy, oral vaccine failure and growth faltering in infants in Bangladesh. EBioMedicine 2015, 2, 1759–1766. [Google Scholar] [CrossRef] [PubMed]
- Taniuchi, M.; Platts-Mills, J.A.; Begum, S.; Uddin, M.J.; Sobuz, S.U.; Liu, J.; Kirkpatrick, B.D.; Colgate, E.R.; Carmolli, M.P.; Dickson, D.M.; et al. Impact of enterovirus and other enteric pathogens on oral polio and rotavirus vaccine performance in Bangladeshi infants. Vaccine 2016, 34, 3068–3075. [Google Scholar] [CrossRef]
- Nakaya, H.I.; Wrammert, J.; Lee, E.K.; Racioppi, L.; Marie-Kunze, S.; Haining, W.N.; Means, A.R.; Kasturi, S.P.; Khan, N.; Li, G.M.; et al. Systems biology of vaccination for seasonal influenza in humans. Nat. Immunol. 2011, 12, 786–795. [Google Scholar] [CrossRef]
- Oh, J.Z.; Ravindran, R.; Chassaing, B.; Carvalho, F.A.; Maddur, M.S.; Bower, M.; Hakimpour, P.; Gill, K.P.; Nakaya, H.I.; Yarovinsky, F.; et al. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity 2014, 41, 478–492. [Google Scholar] [CrossRef]
- Benyacoub, J.; Rochat, F.; Saudan, K.Y.; Rochat, I.; Antille, N.; Cherbut, C.; von der Weid, T.; Schiffrin, E.J.; Blum, S. Feeding a diet containing a fructooligosaccharide mix can enhance salmonella vaccine efficacy in mice. J. Nutr. 2008, 138, 123–129. [Google Scholar] [CrossRef][Green Version]
- Ciabattini, A.; Olivieri, R.; Lazzeri, E.; Medaglini, D. Role of the microbiota in the modulation of vaccine immune responses. Front. Microbiol. 2019, 10, 1305. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Russell, S.L.; Gold, M.J.; Hartmann, M.; Willing, B.P.; Thorson, L.; Wlodarska, M.; Gill, N.; Blanchet, M.R.; Mohn, W.W.; McNagny, K.M.; et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012, 13, 440–447. [Google Scholar] [CrossRef] [PubMed]
- Vijay-Kumar, M.; Aitken, J.D.; Carvalho, F.A.; Cullender, T.C.; Mwangi, S.; Srinivasan, S.; Sitaraman, S.V.; Knight, R.; Ley, R.E.; Gewirtz, A.T. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 2010, 328, 228–231. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, H.; Barber, G.N. The STING pathway and regulation of innate immune signaling in response to DNA pathogens. Cell. Mol. Life Sci. 2011, 68, 1157–1165. [Google Scholar] [CrossRef]
- Thompson, M.R.; Kaminski, J.J.; Kurt-Jones, E.A.; Fitzgerald, K.A. Pattern recognition receptors and the innate immune response to viral infection. Viruses 2011, 3, 920–940. [Google Scholar] [CrossRef]
- Kato, H.; Takeuchi, O.; Sato, S.; Yoneyama, M.; Yamamoto, M.; Matsui, K.; Uematsu, S.; Jung, A.; Kawai, T.; Ishii, K.J.; et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 2006, 441, 101–105. [Google Scholar] [CrossRef]
- NIH. Radiation-Killed Bacteria Vaccine Induces Broad Immune Response in Mice; News, N., Ed.; National Institute of Allergy and Infectious Diseases: Bethesda, MD, USA, 2006.
- NIAID. Types of Vaccines; National Institute of Allergy and Infectious Diseases: Bethesda, MD, USA, 2009.
- Palm, N.W.; Medzhitov, R. Pattern recognition receptors and control of adaptive immunity. Immunol. Rev. 2009, 227, 221–233. [Google Scholar] [CrossRef]
- Hayashi, F.; Smith, K.D.; Ozinsky, A.; Hawn, T.R.; Yi, E.C.; Goodlett, D.R.; Eng, J.K.; Akira, S.; Underhill, D.M.; Aderem, A. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 2001, 410, 1099–1103. [Google Scholar] [CrossRef]
- Ichinohe, T.; Pang, I.K.; Kumamoto, Y.; Peaper, D.R.; Ho, J.H.; Murray, T.S.; Iwasaki, A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. USA 2011, 108, 5354–5359. [Google Scholar] [CrossRef]
- Welsh, R.M.; Che, J.W.; Brehm, M.A.; Selin, L.K. Heterologous immunity between viruses. Immunol. Rev. 2010, 235, 244–266. [Google Scholar] [CrossRef]
- Spaulding, A.C.; Kurane, I.; Ennis, F.A.; Rothman, A.L. Analysis of murine CD8(+) T-cell clones specific for the dengue virus NS3 protein: Flavivirus cross-reactivity and influence of infecting serotype. J. Virol. 1999, 73, 398. [Google Scholar] [CrossRef] [PubMed]
- Nilges, K.; Höhn, H.; Pilch, H.; Neukirch, C.; Freitag, K.; Talbot, P.J.; Maeurer, M.J. Human papillomavirus type 16 E7 peptide-directed CD8+ T cells from patients with cervical cancer are cross-reactive with the coronavirus NS2 protein. J. Virol. 2003, 77, 5464–5474. [Google Scholar] [CrossRef] [PubMed]
- Wedemeyer, H.; Mizukoshi, E.; Davis, A.R.; Bennink, J.R.; Rehermann, B. Cross-reactivity between hepatitis C virus and influenza A virus determinant-specific cytotoxic T cells. J. Virol. 2001, 75, 11392. [Google Scholar] [CrossRef] [PubMed]
- Acierno, P.M.; Newton, D.A.; Brown, E.A.; Maes, L.A.; Baatz, J.E.; Gattoni-Celli, S. Cross-reactivity between HLA-A2-restricted FLU-M1:58-66 and HIV p17 GAG:77-85 epitopes in HIV-infected and uninfected individuals. J. Transl. Med. 2003, 1, 3. [Google Scholar] [CrossRef] [PubMed]
- Cornberg, M.; Sheridan, B.S.; Saccoccio, F.M.; Brehm, M.A.; Selin, L.K. Protection against vaccinia virus challenge by CD8 memory T cells resolved by molecular mimicry. J. Virol. 2007, 81, 934. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.K.; Cornberg, M.; Wang, X.Z.; Chen, H.D.; Selin, L.K.; Welsh, R.M. Private specificities of CD8 T cell responses control patterns of heterologous immunity. J. Exp. Med. 2005, 201, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Clute, S.C.; Watkin, L.B.; Cornberg, M.; Naumov, Y.N.; Sullivan, J.L.; Luzuriaga, K.; Welsh, R.M.; Selin, L.K. Cross-reactive influenza virus-specific CD8+ T cells contribute to lymphoproliferation in Epstein-Barr virus-associated infectious mononucleosis. J. Clin. Investig. 2005, 115, 3602–3612. [Google Scholar] [CrossRef] [PubMed]
- van Stipdonk, M.J.; Lemmens, E.E.; Schoenberger, S.P. Naïve CTLs require a single brief period of antigenic stimulation for clonal expansion and differentiation. Nat. Immunol. 2001, 2, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Veiga-Fernandes, H.; Walter, U.; Bourgeois, C.; McLean, A.; Rocha, B. Response of naïve and memory CD8+ T cells to antigen stimulation in vivo. Nat. Immunol. 2000, 1, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Abbas, K.M.; Kang, G.J.; Chen, D.; Werre, S.R.; Marathe, A. Demographics, perceptions, and socioeconomic factors affecting influenza vaccination among adults in the United States. PeerJ 2018, 6, e5171. [Google Scholar] [CrossRef] [PubMed]
- Randolph, H.E.; Barreiro, L.B. Herd immunity: Understanding COVID-19. Immunity 2020, 52, 737–741. [Google Scholar] [CrossRef] [PubMed]
- Dowdy, D.; D’Souza, G. Early Herd Immunity against COVID-19: A Dangerous Misconception; Johns Hopkins Coronavirus Resource Center, Ed.; Johns Hopkins University of Medicine: Baltimore, MD, USA, 2020. [Google Scholar]
- Black, F.L. The role of herd immunity in control of measles. Yale J. Biol. Med. 1982, 55, 351–360. [Google Scholar] [PubMed]
- Ye, Q.; Wang, B.; Mao, J. The pathogenesis and treatment of the “Cytokine Storm” in COVID-19. J. Infect. 2020, 80, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Zeng, M.Y.; Cisalpino, D.; Varadarajan, S.; Hellman, J.; Warren, H.S.; Cascalho, M.; Inohara, N.; Núñez, G. Gut microbiota-induced immunoglobulin G controls systemic infection by symbiotic bacteria and pathogens. Immunity 2016, 44, 647–658. [Google Scholar] [CrossRef]
- Gou, W.; Fu, Y.; Yue, L.; Chen, G.-D.; Cai, X.; Shuai, M.; Xu, F.; Yi, X.; Chen, H.; Zhu, Y.J.; et al. Gut microbiota may underlie the predisposition of healthy individuals to COVID-19. medRxiv 2020. [Google Scholar] [CrossRef]
- Raftis, E.J.; Delday, M.I.; Cowie, P.; McCluskey, S.M.; Singh, M.D.; Ettorre, A.; Mulder, I.E. Bifidobacterium breve MRx0004 protects against airway inflammation in a severe asthma model by suppressing both neutrophil and eosinophil lung infiltration. Sci. Rep. 2018, 8, 12024. [Google Scholar] [CrossRef]
- 4dpharmaplc. Clinical Update- Phase II COVID-19 Study; 4dpharmaplc: Leeds, UK, 2020. [Google Scholar]
- Xiao, F.; Tang, M.; Zheng, X.; Liu, Y.; Li, X.; Shan, H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology 2020, 158, 1831–1833.e1833. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef]
- Zuo, T.; Zhang, F.; Lui, G.C.Y.; Yeoh, Y.K.; Li, A.Y.L.; Zhan, H.; Wan, Y.; Chung, A.; Cheung, C.P.; Chen, N.; et al. Alterations in gut microbiota of patients With COVID-19 during time of hospitalization. Gastroenterology 2020, 159, 944–955.e948. [Google Scholar] [CrossRef]
- Moore, S.C.; Penrice-Randal, R.; Alruwaili, M.; Dong, X.; Pullan, S.T.; Carter, D.; Bewley, K.; Zhao, Q.; Sun, Y.; Hartley, C.; et al. Amplicon based MinION sequencing of SARS-CoV-2 and metagenomic characterisation of nasopharyngeal swabs from patients with COVID-19. medRxiv 2020. [Google Scholar] [CrossRef]
- Miquel, S.; Martín, R.; Rossi, O.; Bermúdez-Humarán, L.G.; Chatel, J.M.; Sokol, H.; Thomas, M.; Wells, J.M.; Langella, P. Faecalibacterium prausnitzii and human intestinal health. Curr. Opin. Microbiol. 2013, 16, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Verdu, E.F.; Hayes, C.L.; O’ Mahony, S.M. Chapter 9—Importance of the microbiota in early life and influence on future health. In The Gut-Brain Axis; Hyland, N., Stanton, C., Eds.; Academic Press: Cambridge, MA, USA, 2016; pp. 159–184. [Google Scholar] [CrossRef]
- Bao, L.; Zhang, C.; Dong, J.; Zhao, L.; Li, Y.; Sun, J. Oral microbiome and SARS-CoV-2: Beware of lung co-infection. Front. Microbiol. 2020, 11, 1840. [Google Scholar] [CrossRef] [PubMed]
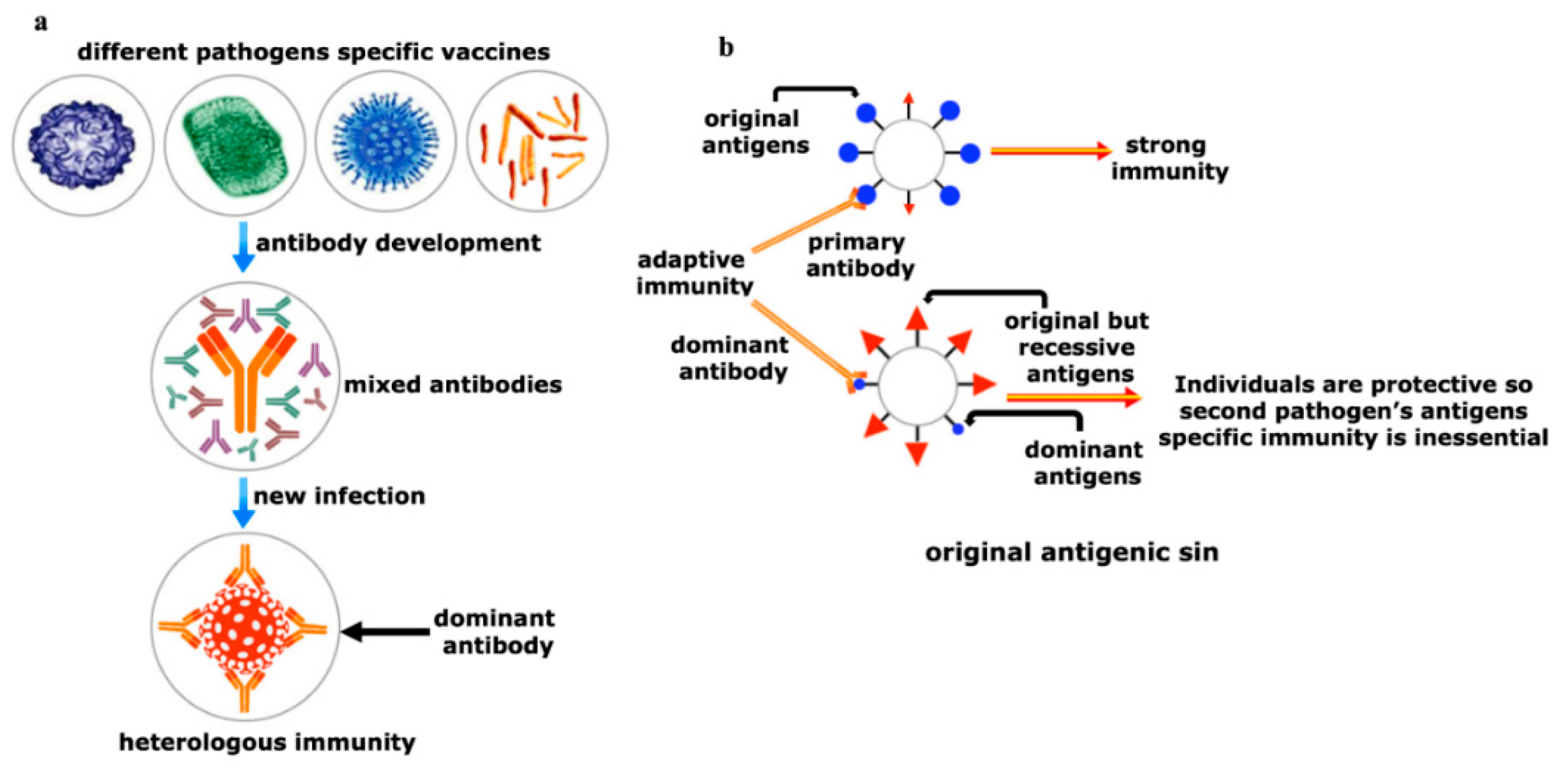
- Vatti, A.; Monsalve, D.M.; Pacheco, Y.; Chang, C.; Anaya, J.M.; Gershwin, M.E. Original antigenic sin: A comprehensive review. J. Autoimmun. 2017, 83, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Stacey, H.D.; Mullarkey, C.E.; Miller, M.S. Original antigenic sin: How first exposure shapes lifelong anti-influenza virus immune responses. J. Immunol. 2019, 202, 335–340. [Google Scholar] [CrossRef]
- Gursel, M.; Gursel, I. Is global BCG vaccination coverage relevant to the progression of SARS-CoV-2 pandemic? Med. Hypotheses 2020. [Google Scholar] [CrossRef]
- Johny, R. A review on possibility of polio vaccine to combat Covid-19. Int. J. Innov. Sci. Res. Technol. 2020, 5, 2020. [Google Scholar]
- Arts, R.J.W.; Moorlag, S.; Novakovic, B.; Li, Y.; Wang, S.Y.; Oosting, M.; Kumar, V.; Xavier, R.J.; Wijmenga, C.; Joosten, L.A.B.; et al. BCG vaccination protects against experimental viral infection in humans through the induction of cytokines associated with trained immunity. Cell Host Microbe 2018, 23, 89–100 e105. [Google Scholar] [CrossRef]
- Roth, A.; Jensen, H.; Garly, M.L.; Djana, Q.; Martins, C.L.; Sodemann, M.; Rodrigues, A.; Aaby, P. Low birth weight infants and Calmette-Guerin bacillus vaccination at birth: Community study from Guinea-Bissau. Pediatr. Infect. Dis. J. 2004, 23, 544–550. [Google Scholar] [CrossRef]
- Jensen, K.J.; Larsen, N.; Biering-Sorensen, S.; Andersen, A.; Eriksen, H.B.; Monteiro, I.; Hougaard, D.; Aaby, P.; Netea, M.G.; Flanagan, K.L.; et al. Heterologous immunological effects of early BCG vaccination in low-birth-weight infants in Guinea-Bissau: A randomized-controlled trial. J. Infect. Dis. 2015, 211, 956–967. [Google Scholar] [CrossRef]
- Chen, Y.W.; Yiu, C.B.; Wong, K.Y. Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CL (pro)) structure: Virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Research 2020, 9, 129. [Google Scholar] [CrossRef]
- Netea, M.G.; Quintin, J.; van der Meer, J.W. Trained immunity: A memory for innate host defense. Cell Host Microbe 2011, 9, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Vita, R.; Mahajan, S.; Overton, J.A.; Dhanda, S.K.; Martini, S.; Cantrell, J.R.; Wheeler, D.K.; Sette, A.; Peters, B. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 2019, 47, D339–D343. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
| 1. Intestinal dysbiosis: Differences in microbiota composition of vaccine responders and nonresponders among different populations. | ||
| Vaccine Type | Microbial Richness Responsible for Vaccine Effectiveness | Tested Population (Ref) |
| Pentavalent diphtheria–tetanus–acellular pertussis-inactivated poliomyelitis–Haemophilus influenzae type B vaccine (DTaP-IPV-Hib) | Existence of intestinal Bifidobacterium sp. especially B. longum, B. infantis and B. breve under phylum Actinobacteria enhanced anti-poliovirus IgA antibodies. | Infants (France) [76] |
| OPV, Parenteral tetanus toxoid (TT), Bacillus Calmette–Guérin (BCG) and hepatitis B vaccine (HBV) | Abundance of Bifidobacterium longum of phylum Actinobacteria augmented oral and parenteral vaccine avidities among infants. Amplified frequencies of Enterobacteriales and Pseudomonadales encumbered vaccine specific immune responses. | Infants (Bangladesh) [61] |
| RVV | i. Streptococcus bovis under Firmicutes correlated with oral RVV response. Increased Bacteroides sp. and Prevotella sp. of phylum Bacteroidetes were associated with lack of RVV response. | Infants (Ghana) [74] |
| ii. Abundance of bacteria related to Clostridium cluster IX of phylum Firmicutes and Serratia sp. and E. coli of phylum Proteobacteria augmented RVV efficiency in a population. | Infants (Pakistan) [73] | |
| Trivalent inactivated influenza vaccine (TIV) and OPV | Activation of toll-like receptor 5 (TLR5) enriched antibody response against TIV and OPV. Impaired antibody response in both GF mice and antibiotic-treated mice was restored by oral reconstitution with flagellated strain of E. coli. | Mice [99,100] |
| Live attenuated oral typhoid (Ty21a) vaccine | Cell-mediated immune response was found to be associated with SFB under phylum Firmicutes, while humoral response was independent of any microbial dysbiosis. | Healthy adults [72] |
| 2. EE: SBBO disturbing the normal gut microbiota is responsible for suboptimal response of many vaccines in low-income settings and developing countries | ||
| Live cholera vaccine CVD-103-HgR | SBBO could blunt the immunological response to live cholera vaccine candidate CVD 103-HgR | Children from high- and low-income countries [60,89] |
| Live oral Shigella flexneri 2a candidate SC602 vaccine | Lower dose of 104 CFU was adequate to elicit a protective response in north American cohort while the ingestion of 104, 105 or 106 CFU of SC602 was not adequate for Bangladeshi toddlers because of SBBO. | [94,95] |
| Oral RVV, OPV | i. EE was found to be associated with refuted efficacy of oral RVV but had no impact on the parenteral administered vaccines– tetanus, pertussis, diphtheria, Haemophilus influenzae type B and measles. | [96,97] |
| ii. Enhanced frequencies of Campylobacter sp. under phylum Proteobacteria and enteroviruses at the time of immunization was negatively correlated with immunogenicity of OPV and diminished rotavirus immunoglobulin A titer (RVI). | Urban slum area of Bangladesh [98] | |
| 3. Probiotics and prebiotics: Several studies mentioned about the role of both probiotics and prebiotics on vaccine efficacy which further substantiates the significance of residential gut microbiota on vaccine effectiveness | ||
| The impact of several probiotic strains on the efficacy of 17 different vaccines, e.g., diphtheria, tetanus toxoids and pertussis (DTP), whole-cell DTP vaccine (DTwP), diphtheria, tetanus, acellular pertussis, with Haemophilus influenzae type B (DTaP-Hib), DTaP-IPV-Hib, hepatitis A vaccine (HAV), hepatitis A vaccine (HBV), Hib, live attenuated influenza vaccine (LAIV), attenuated virus MMR vaccine with chickenpox vaccine or varicella vaccine (MMRV), oral cholera vaccine (OCV), OPV, oral rabies vaccines (ORV), pneumococcal conjugate vaccine (PCV7), pneumococcal polysaccharide vaccine (PPV23), polio, trivalent inactivated influenza (TIV) and Ty21a | Lactobacillus sp., Bifidobacterium sp. and Saccharomyces boulardii were the most frequently used microorganisms in probiotics. The beneficial effects of probiotics were found to be strongest in both oral and parenteral vaccines | [71] |
| S. typhimurium SL1479 vaccination | The administration of a prebiotic (a nondigestible food component that promotes the growth of beneficial microorganisms), e.g., fructo-oligosaccharide/inulin mix was shown to enhance efficacy of S. Typhimurium SL1479 vaccination | Mice [101] |
| 4. Antibiotics: Several studies in both mice and humans have shown the effect of antibiotic-mediated dysbiosis on vaccine responses | ||
| BCG vaccine, Bexsero meningococcal serogroup B vaccine (MenB), the meningococcal serogroup C vaccine-NeisVac-C (MenC); the Prevenar 13-valent pneumococcal conjugate vaccine (PVC13); the hexavalent combination vaccine against hepatitis B, diphtheria, tetanus, pertussis, Hemophilus influenzae type B, inactivated poliomyelitis virus (INFANRIX Hexa) | Antibiotic-mediated dysbiosis in early life impaired antibody response against these vaccines. Restoration of commensal microbiota retrieved vaccine efficiencies. | Newborn mice [13] |
| RVV | Positive correlation was observed in antibiotic-driven microbiota modulation and increased immunological response | Adult cohort [73] |
| Alternation of Microbiome Frequencies Due to SARS-CoV-2 Infection | |
|---|---|
| Sensitive Patients | Protective Patients |
| Phylum Bacteroidetes | Phylum Bacteroidetes |
| Capnocytophaga gingivalis | Alistipes onderdonkii |
| Prevotella melaninogenica | Parabacteroides merdae |
| Bacteroides nordi | Bacteroides stercoris |
| Capnocytophaga sp. | Alistipes onderdonkii |
| Phylum Firmicutes | Bacteroides ovatus |
| Clostridium ramosum | Bacteroides dorei |
| Clostridium hathewayi | Bacteroides thetaiotaomicron |
| Erysipelotrichaceae bacterium | Bacteroides massiliensis |
| Ruthenibacterium lactatiformans | Bacteroides stercoris |
| Veillonella sp. | Phylum Firmicutes |
| Veillonella parvula | Faecalibacterium prausnitzii |
| Streptococcus infantis | Lachnospiraceae bacteria |
| Ruminococcus gnavus | Eubacterium rectale |
| Enterococcus avium | Ruminococcus obeum |
| Phylum Actinobacteria | Dorea formicigenerans |
| Actinomyces viscosus | Lactobacillus sp. |
| Corynebacterium sp. | Lachnospiraceae bacteria |
| Collinsella aerofaciens | Phylum Actinobacteria |
| Collinsella tanakaei | Bifidobacterium sp. |
| Phylum Fusobacteria | Phylum Fusobacteria |
| Leptotrichia buccalis | Fusobacterium periodonticum |
| Phylum Proteobacteria | |
| Acinetobacter baumannii | |
| Klebsiella pneumoniae | |
| Morganella morganii | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shelly, A.; Gupta, P.; Ahuja, R.; Srichandan, S.; Meena, J.; Majumdar, T. Impact of Microbiota: A Paradigm for Evolving Herd Immunity against Viral Diseases. Viruses 2020, 12, 1150. https://doi.org/10.3390/v12101150
Shelly A, Gupta P, Ahuja R, Srichandan S, Meena J, Majumdar T. Impact of Microbiota: A Paradigm for Evolving Herd Immunity against Viral Diseases. Viruses. 2020; 12(10):1150. https://doi.org/10.3390/v12101150
Chicago/Turabian StyleShelly, Asha, Priya Gupta, Rahul Ahuja, Sudeepa Srichandan, Jairam Meena, and Tanmay Majumdar. 2020. "Impact of Microbiota: A Paradigm for Evolving Herd Immunity against Viral Diseases" Viruses 12, no. 10: 1150. https://doi.org/10.3390/v12101150
APA StyleShelly, A., Gupta, P., Ahuja, R., Srichandan, S., Meena, J., & Majumdar, T. (2020). Impact of Microbiota: A Paradigm for Evolving Herd Immunity against Viral Diseases. Viruses, 12(10), 1150. https://doi.org/10.3390/v12101150





