Highly Immunogenic Nanoparticles Based on a Fusion Protein Comprising the M2e of Influenza A Virus and a Lipopeptide
Abstract
1. Introduction
2. Materials and Methods
2.1. Genes for Recombinant Fusion Proteins and Expression Vectors
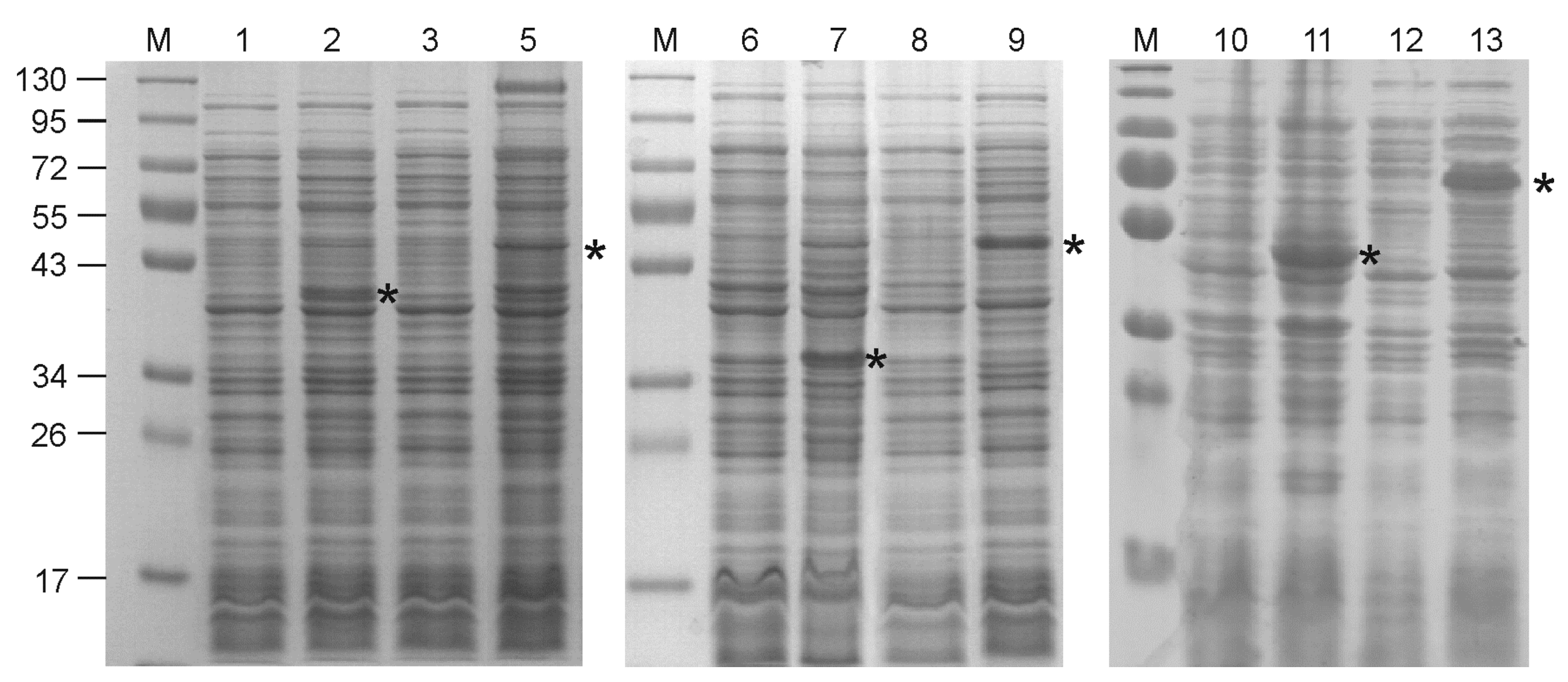
2.2. Expression of Recombinant Proteins
2.3. Purification of Recombinant Proteins
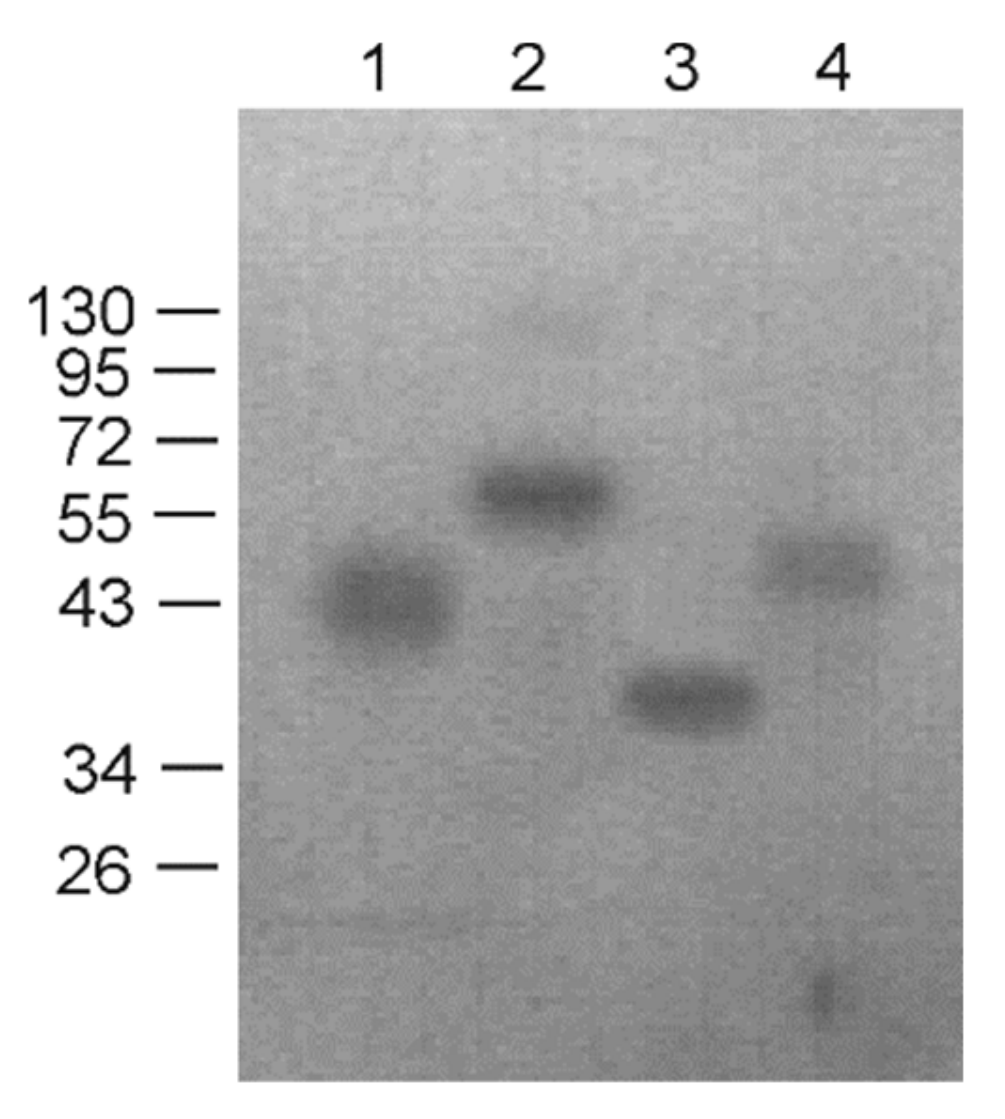
2.4. Western Blotting
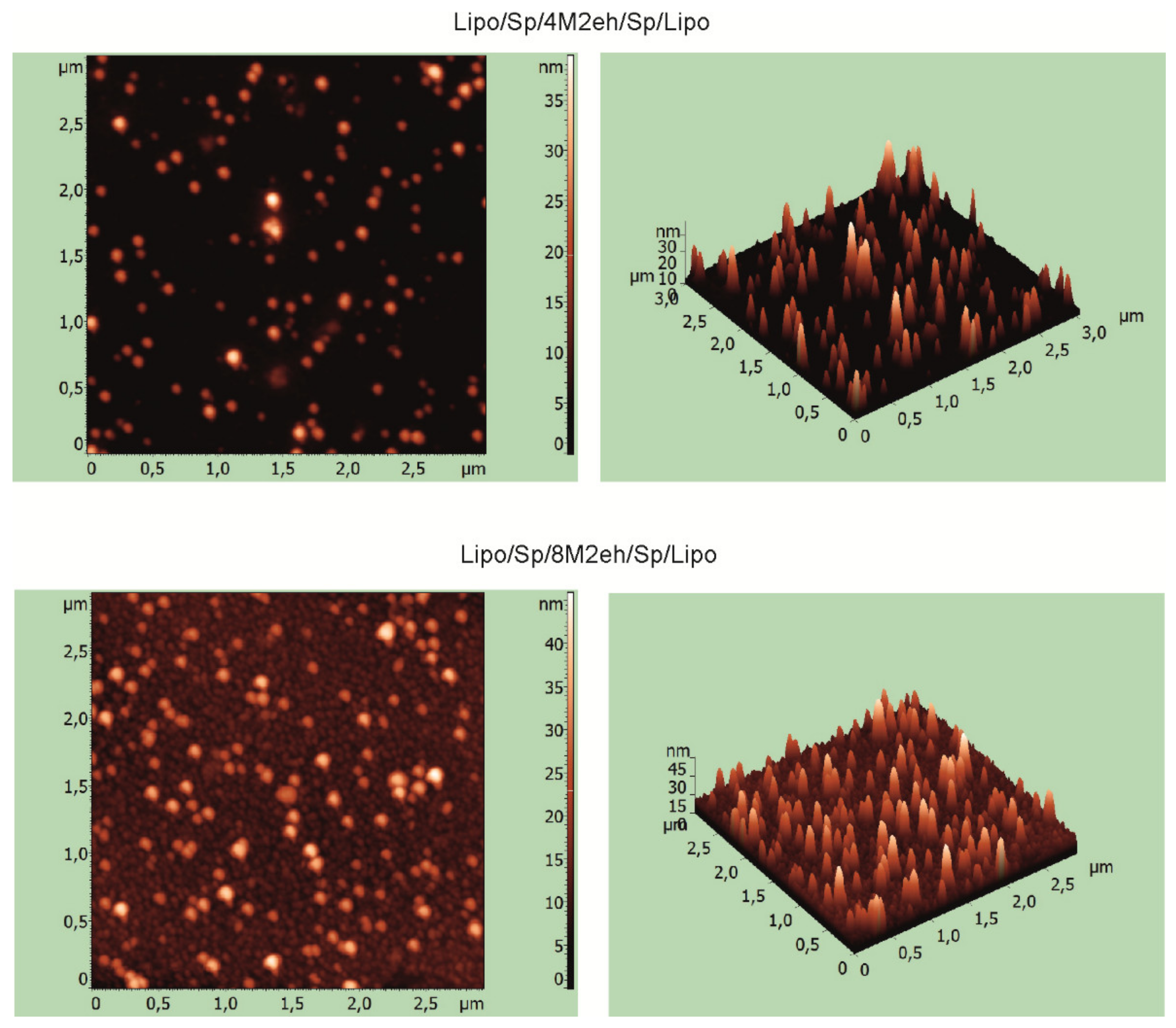
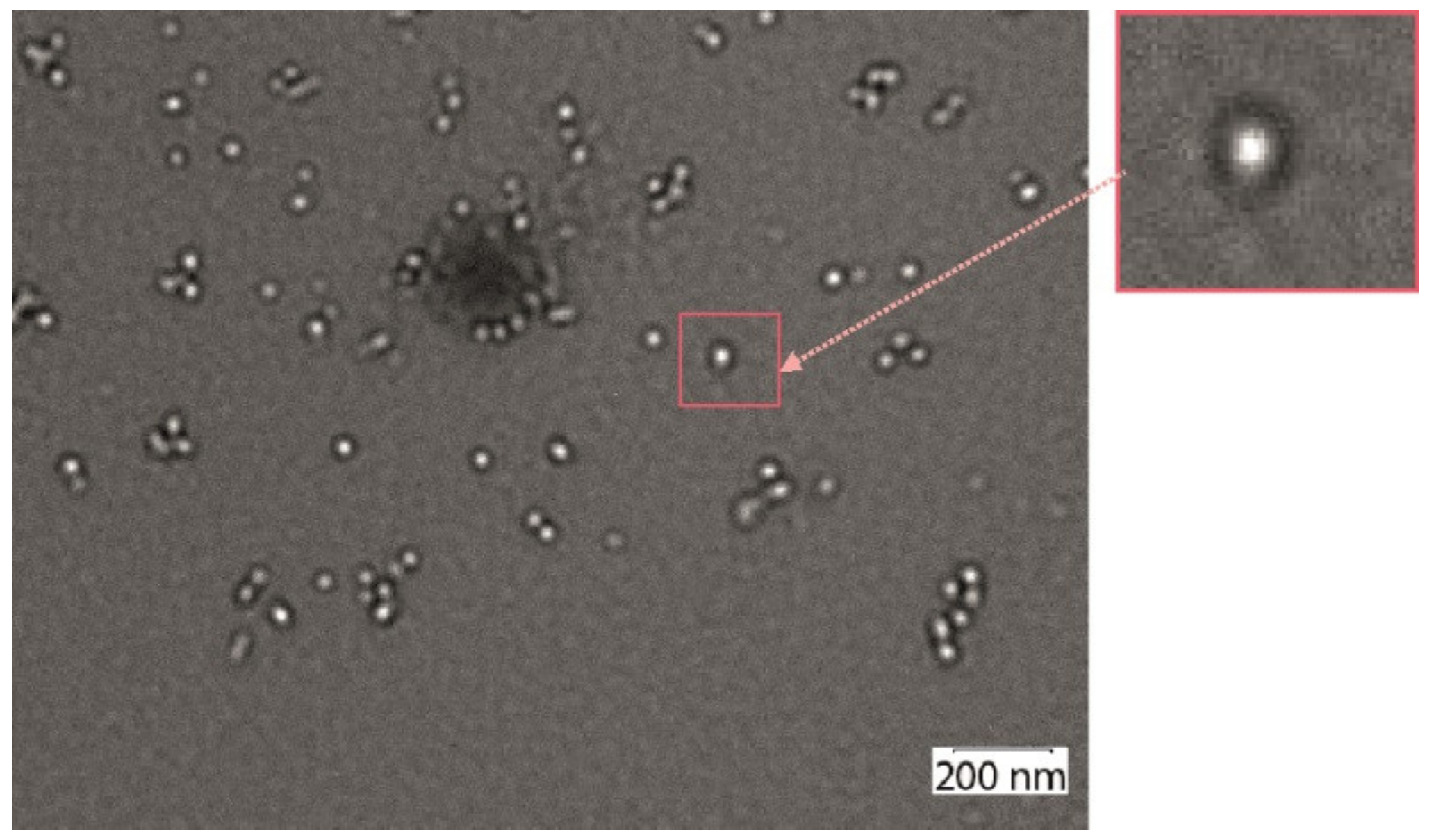
2.5. Electron and Atomic Force Microscopy
2.6. Immunization of Mice
2.7. Antibody Detection in the Sera
3. Results
3.1. Design of Recombinant Proteins, Expression and Purification
3.2. Recombinant Proteins Containing Lipopeptides Form Virus-Like Particles
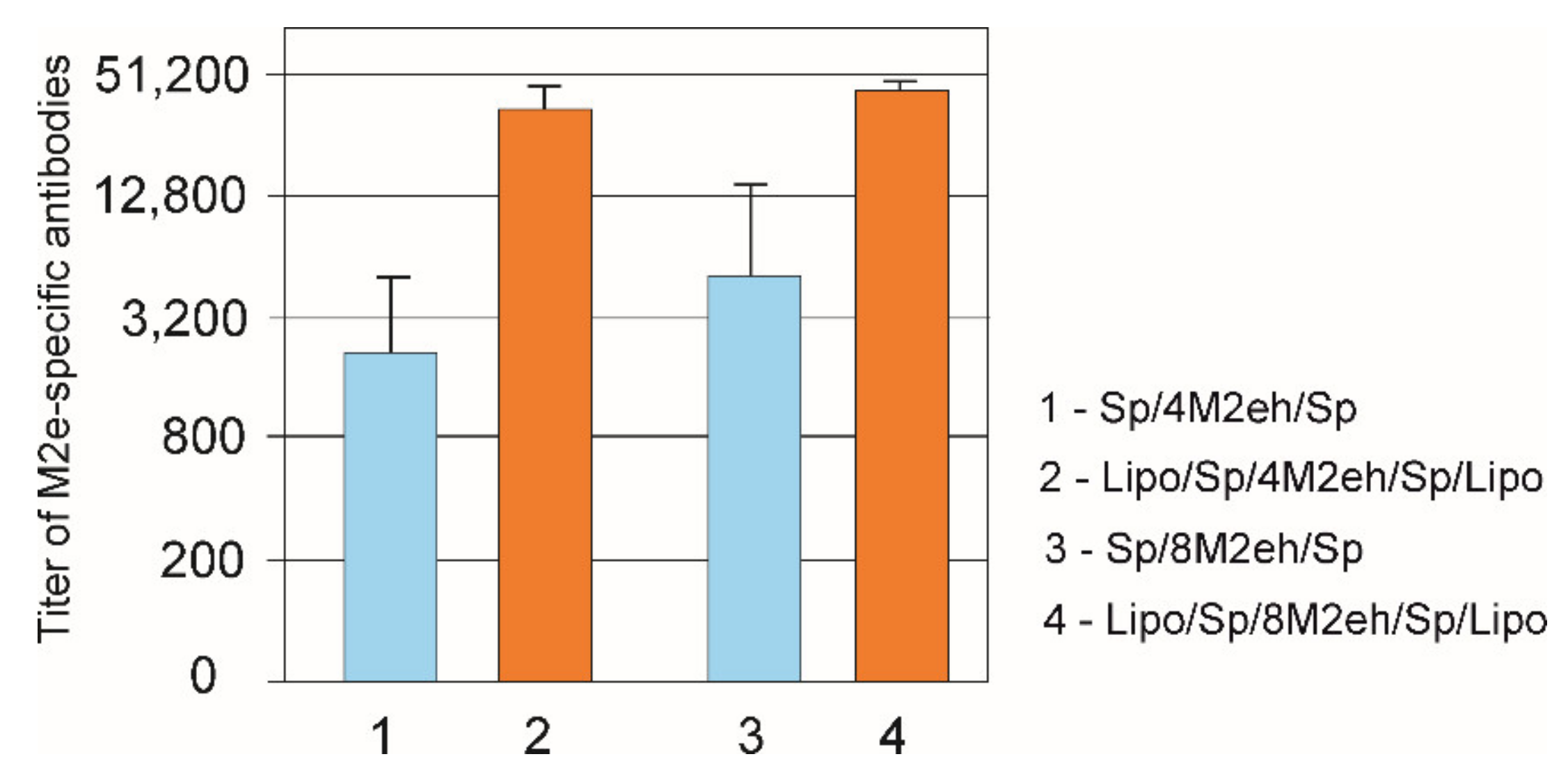
3.3. The Presence of Lipopeptide Increases Immunogenicity of the Fusion Proteins
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Webster, R.G.; Bean, W.J.; Gorman, O.T.; Chambers, T.M.; Kawaoka, Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 1992, 56, 152–179. [Google Scholar] [CrossRef] [PubMed]
- Mezhenskaya, D.; Isakova-Sivak, I.; Rudenko, L. M2e-based universal influenza vaccines: A historical overview and new approaches to development. J. Biomed. Sci. 2019, 26, 76. [Google Scholar] [CrossRef] [PubMed]
- Madsen, A.; Cox, R.J. Prospects and Challenges in the Development of Universal Influenza Vaccines. Vaccines 2020, 8, 361. [Google Scholar]
- Rajão, D.S.; Pérez, D.R. Universal Vaccines and Vaccine Platforms to Protect against Influenza Viruses in Humans and Agriculture. Front. Microbiol. 2018, 9, 123. [Google Scholar] [CrossRef]
- Neirynck, S.; DeRoo, T.; Saelens, X.; Vanlandschoot, P.; Jou, W.M.; Fiers, W. A universal influenza A vaccine based on the extracellular domain of the M2 protein. Nat. Med. 1999, 5, 1157–1163. [Google Scholar] [CrossRef] [PubMed]
- Fiers, W.; De Filette, M.; El Bakkouri, K.; Schepens, B.; Roose, K.; Schotsaert, M.; Birkett, A.; Saelens, X. M2e-based universal influenza A vaccine. Vaccine 2009, 27, 6280–6283. [Google Scholar] [CrossRef]
- Feng, J.; Zhang, M.; Mozdzanowska, K.; Zharikova, D.; Hoff, H.; Wunner, W.; Couch, R.B.; Gerhard, W. Influenza A virus infection engenders a poor antibody response against the ectodomain of matrix protein 2. Virol. J. 2006, 3, 102. [Google Scholar] [CrossRef] [PubMed]
- Denis, J.; Acosta-Ramirez, E.; Zhao, Y.; Hamelin, M.E.; Koukavica, I.; Baz, M.; Abed, Y.; Savard, C.; Pare, C.; Lopez Macias, C.; et al. Development of a universal influenza A vaccine based on the M2e peptide fused to thepapaya mosaic virus (PapMV) vaccine platform. Vaccine 2008, 26, 3395–3403. [Google Scholar] [CrossRef] [PubMed]
- Petukhova, N.; Gasanova, T.; Stepanova, L.; Rusova, O.; Potapchuk, M.; Korotkov, A.; Skurat, E.V.; Tsybalova, L.M.; Kiselev, O.I.; Ivanov, P.A.; et al. Immunogenicity and Protective Efficacy of Candidate Universal Influenza A Nanovaccines Produced in Plants by Tobacco Mosaic Virus-based Vectors. Curr. Pharm. Des. 2013, 19, 5587–5600. [Google Scholar] [CrossRef] [PubMed]
- Wibowo, N.; Chuan, Y.P.; Lua, L.H.L.; Middelberg, A.P.J. Modular engineering of amicrobially-produced viral capsomere vaccine for influenza. Chem. Eng. Sci. 2013, 103, 12–20. [Google Scholar] [CrossRef]
- Ravin, N.V.; Blokhina, E.A.; Kuprianov, V.V.; Stepanova, L.A.; Shaldjan, A.A.; Kovaleva, A.A.; Tsybalova, L.M.; Skryabin, K.G. Development of a candidate influenza vaccine based on virus-like particles displaying influenza M2e peptide into the immunodominant loop region of hepatitis B core antigen: Insertion of multiple copies of M2e increases immunogenicity and protective efficiency. Vaccine 2015, 33, 3392–3397. [Google Scholar] [PubMed]
- Huleatt, J.W.; Nakaar, V.; Desai, P.; Huang, Y.; Hewitt, D.; Jacobs, A.; Tang, J.; McDonald, W.; Song, L.; Evans, R.K.; et al. Potent immunogenicity and efficacy of a universal influenza vaccine candidate comprising a recombinant fusion protein linking influenza M2e to the TLR5 ligand flagellin. Vaccine 2008, 26, 201–214. [Google Scholar] [PubMed]
- Turley, C.B.; Rupp, R.E.; Johnson, C.; Taylor, D.N.; Wolfson, J.; Tussey, L.; Kavita, U.; Stanberry, L.; Shaw, A. Safety and immunogenicity of a recombinant M2e–flagellin influenza vaccine (STF2.4xM2e) in healthy adults. Vaccine 2011, 29, 5145–5152. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, L.A.; Mardanova, E.S.; Shuklina, M.A.; Blokhina, E.A.; Kotlyarov, R.; Potapchuk, M.V.; Kovaleva, A.A.; Vidyaeva, I.G.; Korotkov, A.V.; Eletskaya, E.I.; et al. Flagellin-fused protein targeting M2e and HA2 induces potent humoral and T-cell responses and protects mice against various influenza viruses a subtypes. J. Biomed. Sci. 2018, 25, 33. [Google Scholar] [CrossRef]
- Hayashi, F.; Smith, K.D.; Ozinsky, A.; Hawn, T.R.; Yi, E.C.; Goodlett, D.R.; Eng, J.K.; Akira, S.; Underhill, D.M.; Aderem, A. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 2001, 410, 1099–1110. [Google Scholar] [CrossRef]
- Ozinsky, A.; Underhill, D.M.; Fontenot, J.D.; Hajjar, A.M.; Smith, K.D.; Wilson, C.B.; Schroeder, L.; Aderem, A. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between Toll-like receptors. Proc. Natl. Acad. Sci. USA 2000, 97, 13766–13771. [Google Scholar] [PubMed]
- Zhang, H.; Niesel, D.W.; Peterson, J.W.; Klimpel, G.R. Lipoprotein Release by Bacteria: Potential Factor in Bacterial Pathogenesis. Infect. Immun. 1998, 66, 5196–5201. [Google Scholar] [CrossRef] [PubMed]
- Ganapamo, F.; Dennis, V.A.; Philipp, M.T. Differential acquired immune responsiveness to bacterial lipoproteins in Lyme disease-resistant and -susceptible mouse strains. Eur. J. Immunol. 2003, 33, 1934–1940. [Google Scholar] [CrossRef]
- Sung, J.W.-C.; Hsieh, S.-Y.; Lin, C.-L.; Leng, C.-H.; Liu, S.-J.; Chou, A.-H.; Lai, L.-W.; Lin, L.-H.; Kwok, Y.; Yang, C.-Y.; et al. Biochemical characterizations of Escherichia coli-expressed protective antigen Ag473 of Neisseria meningitides group B. Vaccine 2010, 28, 8175–8182. [Google Scholar] [CrossRef]
- Chen, H.-W.; Liu, S.-J.; Liu, H.-H.; Kwok, Y.; Lin, C.-L.; Lin, L.-H.; Chen, M.-Y.; Tsai, J.-P.; Chang, L.-S.; Chiu, F.-F.; et al. A novel technology for the production of a heterologous lipoprotein immunogen in high yield has implications for the field of vaccine design. Vaccine 2009, 27, 1400–1409. [Google Scholar] [CrossRef] [PubMed]
- Zom, G.G.; Khan, S.; Filippov, D.V.; Ossendorp, F. TLR Ligand–Peptide Conjugate Vaccines. Adv. Immunol. 2012, 114, 177–201. [Google Scholar]
- Pejoski, D.; Zeng, W.; Rockman, S.; Brown, L.E.; Jackson, D.C. A lipopeptide based on the M2 and HA proteins of influenza A viruses induces protective antibody. Immunol. Cell Biol. 2010, 88, 605–611. [Google Scholar] [CrossRef]
- Zeng, W.; Tan, A.C.; Horrocks, K.; Jackson, D. A lipidated form of the extracellular domain of influenza M2 protein as a self-adjuvanting vaccine candidate. Vaccine 2015, 33, 3526–3532. [Google Scholar] [CrossRef] [PubMed]
- Blokhina, E.A.; Kuprianov, V.V.; Stepanova, L.A.; Tsybalova, L.M.; Kiselev, O.I.; Ravin, N.V.; Skryabin, S.K. A molecular assembly system for presentation of antigens on the surface of HBc virus-like particles. Virology 2013, 435, 293–300. [Google Scholar] [CrossRef]
- Liu, W.; Peng, Z.; Liu, Z.; Lu, Y.; Ding, J.; Chen, Y.-H. High epitope density in a single recombinant protein molecule of the extracellular domain of influenza A virus M2 protein significantly enhances protective immunity. Vaccine 2004, 23, 366–371. [Google Scholar] [CrossRef]
- De Filette, M.; Jou, W.M.; Birkett, A.; Lyons, K.; Schultz, B.; Tonkyro, A.; Resch, S.; Fiers, W. Universal influenza A vaccine: Optimization of M2-based constructs. Virology 2005, 337, 149–161. [Google Scholar] [CrossRef]
- Arai, R.; Ueda, H.; Kitayama, A.; Kamiya, N.; Nagamune, T. Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein Eng. 2001, 14, 529–532. [Google Scholar] [CrossRef] [PubMed]
- Robinson, C.R.; Sauer, R.T. Optimizing the stability of single-chain proteins by linker length and composition mutagenesis. Proc. Natl. Acad. Sci. USA 1998, 95, 5929–5934. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, M.F.; Jennings, G.T. Vaccine delivery: A matter of size, geometry, kinetics and molecular patterns. Nat. Rev. Immunol. 2010, 10, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Marqusee, S.; Baldwin, R.L. Helix stabilization by Glu-. Lys+ salt bridges in short peptides of de novo design. Proc. Natl. Acad. Sci. USA 1987, 84, 8898–8902. [Google Scholar] [CrossRef]
- Chen, X.; Zaro, J.L.; Shen, W.-C. Fusion protein linkers: Property, design and functionality. Adv. Drug Deliv. Rev. 2013, 65, 1357–1369. [Google Scholar] [CrossRef]
- Fujita, Y.; Taguchi, H. Nanoparticle-Based Peptide Vaccines. Micro Nanotechnol. Vaccine Dev. 2017, 149–170. [Google Scholar] [CrossRef]
- Mason, J.M.; Arndt, K.M. Coiled coil domains: Stability, specificity, and biological implications. ChemBioChem 2004, 5, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Tamborrini, M.; Geib, N.; Marrero-Nodarse, A.; Jud, M.; Hauser, J.; Aho, C.; Lamelas, A.; Zúñiga, A.; Pluschke, G.; Ghasparian, A.; et al. A Synthetic Virus-Like Particle Streptococcal Vaccine Candidate Using B-Cell Epitopes from the Proline-Rich Region of Pneumococcal Surface Protein A. Vaccines 2015, 3, 850–874. [Google Scholar] [CrossRef] [PubMed]
- De Filette, M.; Fiers, W.; Martens, W.; Birkett, A.; Ramne, A.; Löwenadler, B.; Lycke, N.; Jou, W.M.; Saelens, X. Improved design and intranasal delivery of an M2e-based human influenza A vaccine. Vaccine 2006, 24, 6597–6601. [Google Scholar] [CrossRef]
- Mardanova, E.S.; Kotlyarov, R.; Kuprianov, V.V.; Stepanova, L.A.; Tsybalova, L.M.; Lomonossoff, G.P.; Ravin, N.V. Rapid high-yield expression of a candidate influenza vaccine based on the ectodomain of M2 protein linked to flagellin in plants using viral vectors. BMC Biotechnol. 2015, 15, 42. [Google Scholar] [CrossRef] [PubMed]
- Shim, B.-S.; Choi, Y.K.; Yun, C.-H.; Lee, E.-G.; Jeon, Y.S.; Park, S.-M.; Cheon, I.S.; Joo, D.-H.; Cho, C.H.; Song, M.-S.; et al. Sublingual Immunization with M2-Based Vaccine Induces Broad Protective Immunity against Influenza. PLoS ONE 2011, 6, e27953. [Google Scholar] [CrossRef] [PubMed]
- De Filette, M.; Ramne, A.; Birkett, A.; Lycke, N.; Löwenadler, B.; Jou, W.M.; Saelens, X.; Fiers, W. The universal influenza vaccine M2e-HBc administered intranasally in combination with the adjuvant CTA1-DD provides complete protection. Vaccine 2006, 24, 544–551. [Google Scholar] [CrossRef] [PubMed]
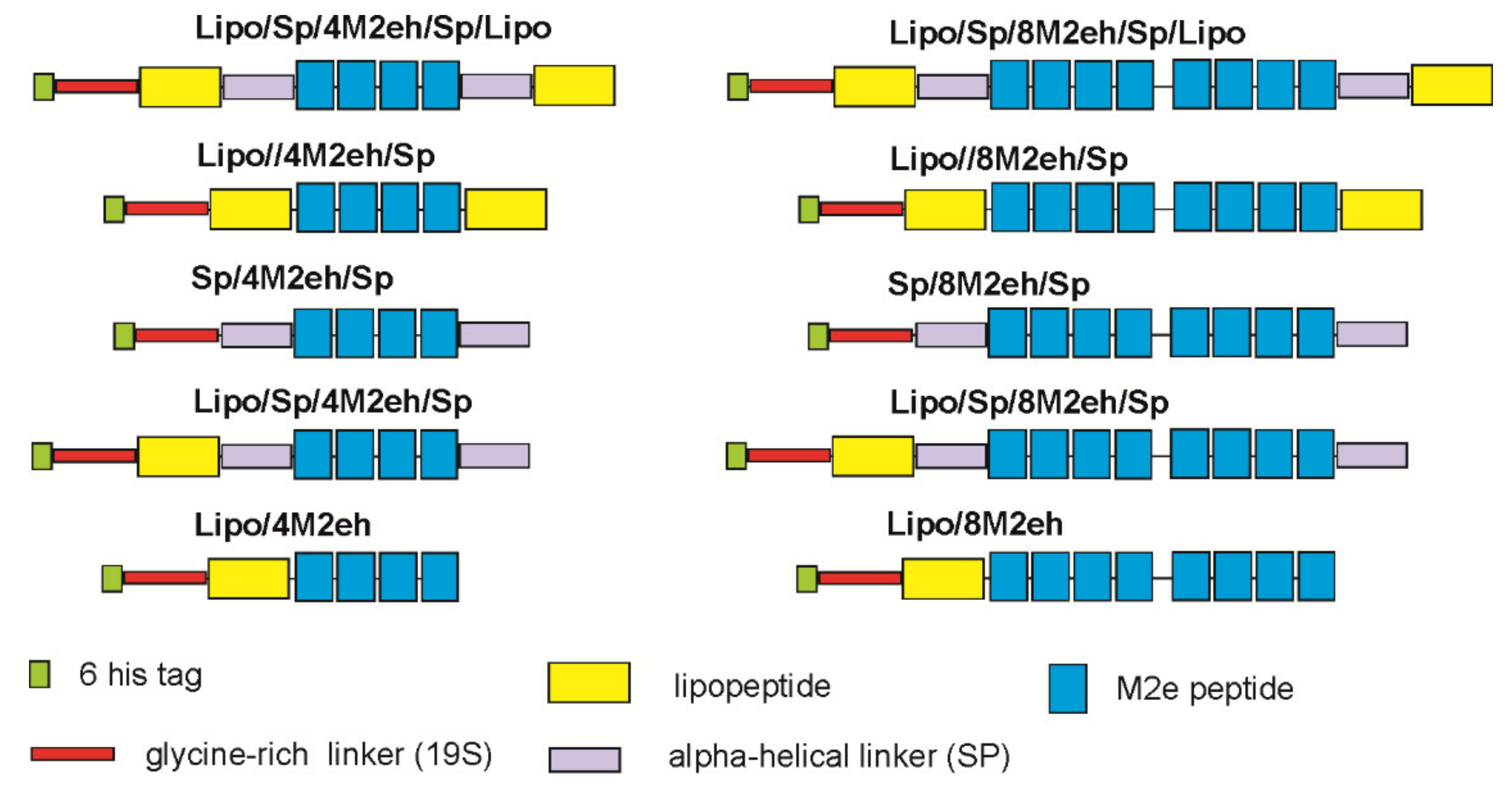
| Protein | Growth Medium | Induction Conditions for Expression of the Target Gene | ||
|---|---|---|---|---|
| Time (h) | Temperature | IPTG | ||
| Lipo/Sp/4M2eh/Sp/Lipo | 2× TY | 4 h | 37 °C | 0.5 mM |
| Lipo/Sp/8M2eh/Sp/Lipo | 2× TY | 4 h | 37 °C | 0.5 mM |
| Lipo/4M2eh/Lipo | 2× TY | 4 h | 37 °C | 0.5 mM |
| Lipo/8M2eh/Lipo | 2× TY | 4 h | 37 °C | 0.5 mM |
| Sp/4M2eh/Sp | LB | 12–14 h | 28 °C | 1 mM |
| Sp/8M2eh/Sp | LB | 12–14 h | 28 °C | 1 mM |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zykova, A.A.; Blokhina, E.A.; Kotlyarov, R.Y.; Stepanova, L.A.; Tsybalova, L.M.; Kuprianov, V.V.; Ravin, N.V. Highly Immunogenic Nanoparticles Based on a Fusion Protein Comprising the M2e of Influenza A Virus and a Lipopeptide. Viruses 2020, 12, 1133. https://doi.org/10.3390/v12101133
Zykova AA, Blokhina EA, Kotlyarov RY, Stepanova LA, Tsybalova LM, Kuprianov VV, Ravin NV. Highly Immunogenic Nanoparticles Based on a Fusion Protein Comprising the M2e of Influenza A Virus and a Lipopeptide. Viruses. 2020; 12(10):1133. https://doi.org/10.3390/v12101133
Chicago/Turabian StyleZykova, Anna A., Elena A. Blokhina, Roman Y. Kotlyarov, Liudmila A. Stepanova, Liudmila M. Tsybalova, Victor V. Kuprianov, and Nikolai V. Ravin. 2020. "Highly Immunogenic Nanoparticles Based on a Fusion Protein Comprising the M2e of Influenza A Virus and a Lipopeptide" Viruses 12, no. 10: 1133. https://doi.org/10.3390/v12101133
APA StyleZykova, A. A., Blokhina, E. A., Kotlyarov, R. Y., Stepanova, L. A., Tsybalova, L. M., Kuprianov, V. V., & Ravin, N. V. (2020). Highly Immunogenic Nanoparticles Based on a Fusion Protein Comprising the M2e of Influenza A Virus and a Lipopeptide. Viruses, 12(10), 1133. https://doi.org/10.3390/v12101133






