Gene Therapy Applications of Non-Human Lentiviral Vectors
Abstract
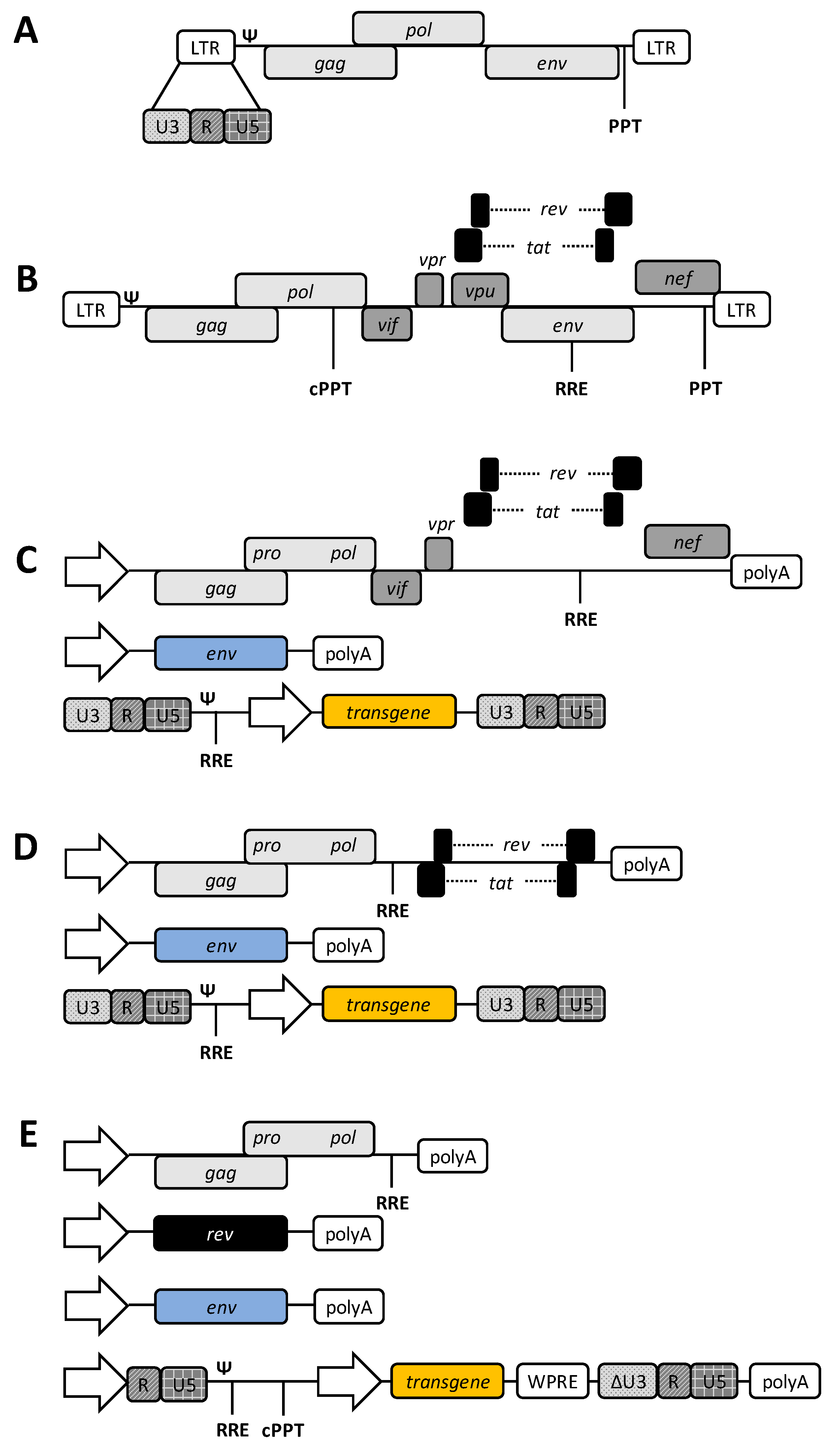
1. Introduction to Lentiviral Vectors
1.1. Retroviral Biology
1.2. HIV-1-Based Lentiviral Vectors
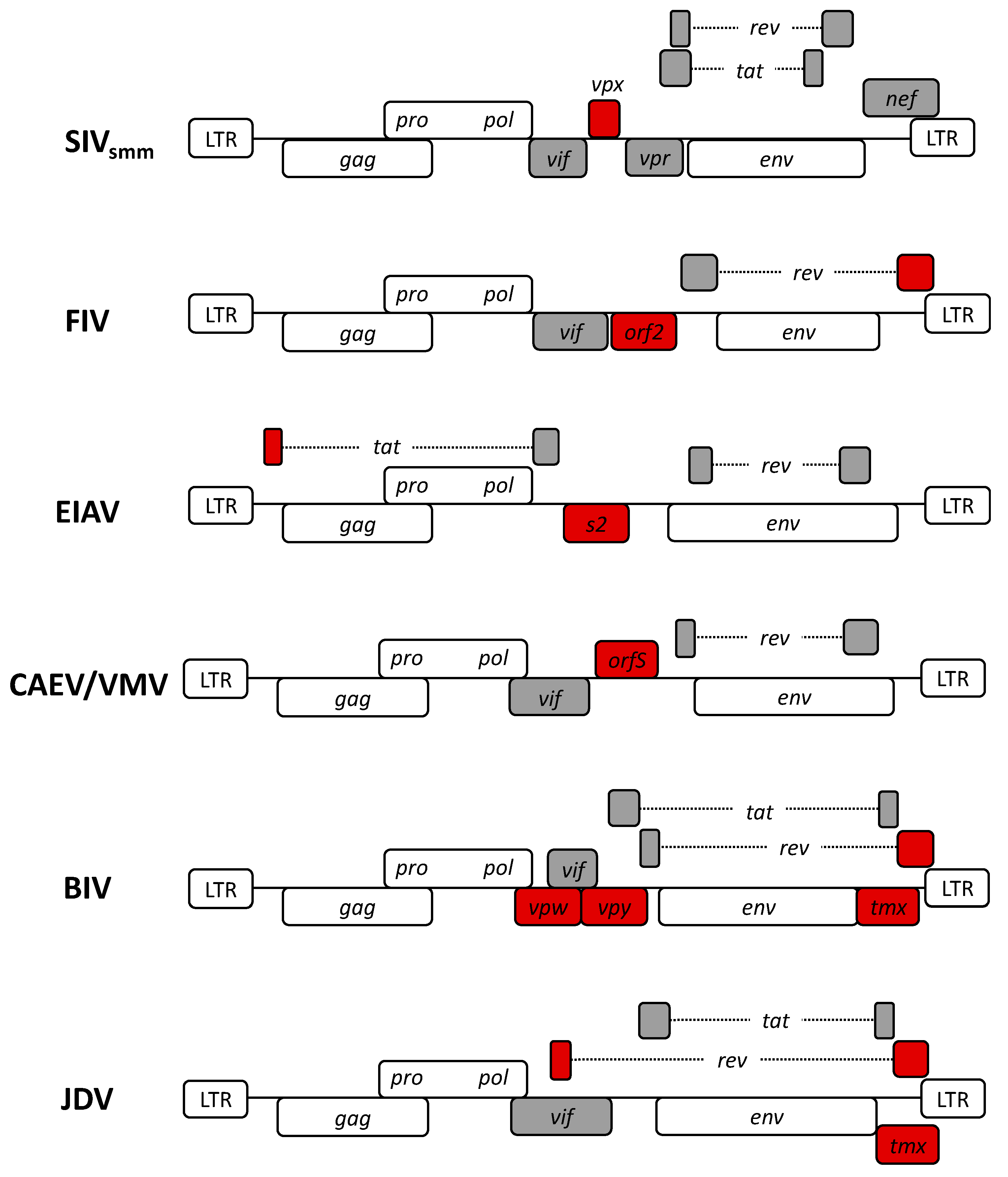
2. Non-Human Lentiviruses
3. Lentiviral Vector Systems Generated Based on Non-Human Lentiviruses
3.1. Simian Lentiviral Vectors
3.2. Feline Lentiviral Vectors
3.3. Equine Lentiviral Vectors
3.4. Caprine and Ovine Lentiviral Vectors
3.5. Bovine Lentiviral Vectors
4. Conclusions and Future Directions
Funding
Acknowledgments
Conflicts of Interest
References
- Weinberg, J.B.; Matthews, T.J.; Cullen, B.R.; Malim, M.H. Productive human immunodeficiency virus type 1 (HIV-1) infection of nonproliferating human monocytes. J. Exp. Med. 1991, 174, 1477–1482. [Google Scholar] [CrossRef]
- Clements, J.E.; Zink, M.C. Molecular biology and pathogenesis of animal lentivirus infections. Clin. Microbiol. Rev. 1996, 9, 100–117. [Google Scholar] [CrossRef] [PubMed]
- Gifford, R.J. Viral evolution in deep time: Lentiviruses and mammals. Trends Genet. 2012, 28, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Narayan, O.; Silverstein, A.M.; Price, D.; Johnson, R.T. Visna Virus Infection of American Lambs. Science 1974, 183, 1202–1203. [Google Scholar] [CrossRef] [PubMed]
- Olmsted, R.A.; Barnes, A.K.; Yamamoto, J.K.; Hirsch, V.M.; Purcell, R.H.; Johnson, P.R. Molecular cloning of feline immunodeficiency virus. Proc. Natl. Acad. Sci. USA 1989, 86, 2448–2452. [Google Scholar] [CrossRef] [PubMed]
- Overbaugh, J.; Donahue, P.; Quackenbush, S.; Hoover, E.; Mullins, J. Molecular cloning of a feline leukemia virus that induces fatal immunodeficiency disease in cats. Science 1988, 239, 906–910. [Google Scholar] [CrossRef] [PubMed]
- Charman, H.P.; Bladen, S.; Gilden, R.V.; Coggins, L. Equine infectious anemia virus: Evidence favoring classification as a retravirus. J. Virol. 1976, 19, 1073–1079. [Google Scholar] [CrossRef]
- Crawford, T.; Adams, D.; Cheevers, W.; Cork, L. Chronic arthritis in goats caused by a retrovirus. Science 1980, 207, 997–999. [Google Scholar] [CrossRef]
- Gonda, M.A.; Braun, M.J.; Carter, S.G.; Kost, T.A.; Bess, J.W.; Arthur, L.O.; Van Der Maaten, M.J. Characterization and molecular cloning of a bovine lentivirus related to human immunodeficiency virus. Nature 1987, 330, 388–391. [Google Scholar] [CrossRef]
- Chadwick, B.J.; Coelen, R.J.; Sammels, L.M.; Kertayadnya, G.; Wilcox, G.E. Genomic sequence analysis identifies Jembrana disease virus as a new bovine lentivirus. J. Gen. Virol. 1995, 76, 189–192. [Google Scholar] [CrossRef]
- Chowdhury, S.; Ikeda, Y. Retroviruses. In Viral Therapy of Cancer; Wiley: Hoboken, NJ, USA, 2008; pp. 69–81. [Google Scholar]
- Baltimore, D. Viral RNA-dependent DNA Polymerase: RNA-dependent DNA Polymerase in Virions of RNA Tumour Viruses. Nature 1970, 226, 1209–1211. [Google Scholar] [CrossRef] [PubMed]
- Telesnitsky, A.; Goff, S.P. Reverse Transcriptase and the Generation of Retroviral DNA. In Retroviruses; Coffin, J.M., Hughes, S.H., Varmus, H.E., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1997; pp. 121–160. [Google Scholar]
- Coffin, J.M.; Hughes, S.H.; Varmus, H.E. (Eds.) The Interactions of Retroviruses and their Hosts. In Retroviruses; Cold Spring Harbor: New York, NY, USA, 1997. [Google Scholar]
- Vogt, V.M. Retroviral virions and genomes. In Retroviruses; Coffin, J.M., Varmus, H.E., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1997; pp. 27–71. [Google Scholar]
- Balvay, L.; López-Lastra, M.; Sargueil, B.; Darlix, J.-L.; Ohlmann, T. Translational control of retroviruses. Nat. Rev. Genet. 2007, 5, 128–140. [Google Scholar] [CrossRef]
- Abordo-Adesida, E.; Follenzi, A.; Barcia, C.; Sciascia, S.; Castro, M.G.; Naldini, L.; Lowenstein, P.R. Stability of Lentiviral Vector-Mediated Transgene Expression in the Brain in the Presence of Systemic Antivector Immune Responses. Hum. Gene Ther. 2005, 16, 741–751. [Google Scholar] [CrossRef] [PubMed]
- Reik, W.; Weiher, H.; Jaenisch, R. Replication-competent Moloney murine leukemia virus carrying a bacterial suppressor tRNA gene: Selective cloning of proviral and flanking host sequences. Proc. Natl. Acad. Sci. USA 1985, 82, 1141–1145. [Google Scholar] [CrossRef] [PubMed]
- Hughes, S.; Kosik, E. Mutagenesis of the region between env and src of the SR-A strain of rous sarcoma virus for the purpose of constructing helper-independent vectors. Virology 1984, 136, 89–99. [Google Scholar] [CrossRef]
- Page, K.A.; Liegler, T.; Feinberg, M.B. Use of a Green Fluorescent Protein as a Marker for Human Immunodeficiency Virus Type 1 Infection. AIDS Res. Hum. Retrovir. 1997, 13, 1077–1081. [Google Scholar] [CrossRef]
- Naldini, L.; Blömer, U.; Gallay, P.; Ory, D.; Mulligan, R.; Gage, F.H.; Verma, I.M.; Trono, D. In Vivo Gene Delivery and Stable Transduction of Nondividing Cells by a Lentiviral Vector. Science 1996, 272, 263–267. [Google Scholar] [CrossRef]
- Shimotohno, K.; Temin, H.M. Formation of infectious progeny virus after insertion of herpes simplex thymidine kinase gene into DNA of an avian retrovirus. Cell 1981, 26, 67–77. [Google Scholar] [CrossRef]
- Delenda, C. Lentiviral vectors: Optimization of packaging, transduction and gene expression. J. Gene Med. 2004, 6, S125–S138. [Google Scholar] [CrossRef]
- Miller, A.D. Development and Applications of Retroviral Vectors. In Retroviruses; Coffin, J.M., Hughes, S.H., Varmus, H.E., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1997; pp. 437–474. [Google Scholar]
- Miyoshi, H.; Blömer, U.; Takahashi, M.; Gage, F.H.; Verma, I.M. Development of a Self-Inactivating Lentivirus Vector. J. Virol. 1998, 72, 8150–8157. [Google Scholar] [CrossRef]
- Donello, J.E.; Loeb, J.E.; Hope, T.J. Woodchuck Hepatitis Virus Contains a Tripartite Posttranscriptional Regulatory Element. J. Virol. 1998, 72, 5085–5092. [Google Scholar] [CrossRef] [PubMed]
- Schambach, A.; Galla, M.; Maetzig, T.; Loew, R.; Baum, C. Improving Transcriptional Termination of Self-inactivating Gamma-retroviral and Lentiviral Vectors. Mol. Ther. 2007, 15, 1167–1173. [Google Scholar] [CrossRef] [PubMed]
- Logan, A.C.; Nightingale, S.J.; Haas, D.L.; Cho, G.J.; Pepper, K.A.; Kohn, D.B. Factors Influencing the Titer and Infectivity of Lentiviral Vectors. Hum. Gene Ther. 2004, 15, 976–988. [Google Scholar] [CrossRef] [PubMed]
- Milone, M.C.; O’Doherty, U. Clinical use of lentiviral vectors. Leukemia 2018, 32, 1529–1541. [Google Scholar] [CrossRef]
- Loza, L.I.M.; Yuen, E.C.; McCray, J.P.B. Lentiviral Vectors for the Treatment and Prevention of Cystic Fibrosis Lung Disease. Genes 2019, 10, 218. [Google Scholar] [CrossRef]
- Tisagenlecleucel. Available online: https://www.cancer.gov/about-cancer/treatment/drugs/tisagenlecleucel (accessed on 12 June 2020).
- Axicabtagene Ciloleucel. Available online: https://www.cancer.gov/about-cancer/treatment/drugs/axicabtageneciloleucel (accessed on 12 June 2020).
- Lentiviral Vector. Available online: https://clinicaltrials.gov/ct2/results?cond=&term=lentiviral+vector&cntry=&state=&city=&dist= (accessed on 12 June 2020).
- Locatelli, S.; Peeters, M. Cross-species transmission of simian retroviruses. AIDS 2012, 26, 659–673. [Google Scholar] [CrossRef]
- Aghokeng, A.F.; Ayouba, A.; Mpoudi-Ngolé, E.; Loul, S.; Liegeois, F.; Delaporte, E.; Peeters, M. Extensive survey on the prevalence and genetic diversity of SIVs in primate bushmeat provides insights into risks for potential new cross-species transmissions. Infect. Genet. Evol. 2010, 10, 386–396. [Google Scholar] [CrossRef]
- Gao, F.; Bailes, E.; Robertson, D.L.; Chen, Y.; Rodenburg, C.M.; Michael, S.F.; Cummins, L.B.; Arthur, L.O.; Peeters, M.; Shaw, G.M.; et al. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 1999, 397, 436–441. [Google Scholar] [CrossRef]
- Hirsch, V.M.; Olmsted, R.A.; Murphey-Corb, M.; Purcell, R.H.; Johnson, P.R. An African primate lentivirus (SIVsmclosely related to HIV-2. Nature 1989, 339, 389–392. [Google Scholar] [CrossRef]
- Lauck, M.; Switzer, W.M.; Sibley, S.D.; Hyeroba, D.; Tumukunde, A.; Weny, G.; Taylor, B.; Shankar, A.; Ting, N.; Chapman, C.A.; et al. Discovery and full genome characterization of two highly divergent simian immunodeficiency viruses infecting black-and-white colobus monkeys (Colobus guereza) in Kibale National Park, Uganda. Retrovirology 2013, 10, 107. [Google Scholar] [CrossRef]
- Sauter, D.; Kirchhoff, F. Chapter 4-Properties of Human and Simian Immunodeficiency Viruses. In Natural Hosts of SIV; Ansari, A.A., Silvestri, G., Eds.; Elsevier: Amsterdam, The Netherlands, 2014; Chapter 4; pp. 69–84. Available online: https://doi.org/10.1016/B978-0-12-404734-1.00004-8 (accessed on 20 August 2020).
- Pandrea, I.; Apetrei, C. Where the Wild Things Are: Pathogenesis of SIV Infection in African Nonhuman Primate Hosts. Curr. HIV/AIDS Rep. 2010, 7, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.A. SIV pathogenicity. Fast-acting slow viruses. Nature 1990, 345, 572–573. [Google Scholar] [CrossRef] [PubMed]
- Pisoni, G.; Bertoni, G.; Boettcher, P.; Ponti, W.; Moroni, P. Phylogenetic analysis of the gag region encoding the matrix protein of small ruminant lentiviruses: Comparative analysis and molecular epidemiological applications. Virus Res. 2006, 116, 159–167. [Google Scholar] [CrossRef] [PubMed]
- VandenDriessche, T.; Thorrez, L.; Naldini, L.; Follenzi, A.; Moons, L.; Berneman, Z.; Collen, D.; Chuah, M.K. Lentiviral vectors containing the human immunodeficiency virus type-1 central polypurine tract can efficiently transduce nondividing hepatocytes and antigen-presenting cells in vivo. Blood 2002, 100, 813–822. [Google Scholar] [CrossRef]
- Frech, K.; Brack-Werner, R.; Werner, T. Common Modular Structure of Lentivirus LTRs. Virology 1996, 224, 256–267. [Google Scholar] [CrossRef]
- Kim, S.H.; Jun, H.J.; Jang, S.I.; You, J.C. The Determination of Importance of Sequences Neighboring the Psi Sequence in Lentiviral Vector Transduction and Packaging Efficiency. PLoS ONE 2012, 7, e50148. [Google Scholar] [CrossRef]
- Naldini, L.; Trono, D.; Verma, I.M. Lentiviral vectors, two decades later. Science 2016, 353, 1101–1102. [Google Scholar] [CrossRef]
- Naldini, L.; Verma, I.M. Lentiviral Vectors; Elsevier: Amsterdam, The Netherlands, 2000; Volume 55, pp. 599–609. [Google Scholar]
- Clements, J.E.; Zink, M.C.; Narayan, O.; Gabuzda, D.H. Lentivirus infection of macrophages. Immunol. Ser. 1994, 60, 589–600. [Google Scholar]
- Shimojima, M.; Miyazawa, T.; Ikeda, Y.; McMonagle, E.; Haining, H.; Akashi, H.; Takeuchi, Y.; Hosie, M.J.; Willett, B.J. Use of CD134 As a Primary Receptor by the Feline Immunodeficiency Virus. Science 2004, 303, 1192–1195. [Google Scholar] [CrossRef]
- Phillips, T.R.; Lamont, C.; A Konings, D.; Shacklett, B.L.; A Hamson, C.; A Luciw, P.; Elder, J.H. Identification of the Rev transactivation and Rev-responsive elements of feline immunodeficiency virus. J. Virol. 1992, 66, 5464–5471. [Google Scholar] [CrossRef]
- Chadwick, B.J.; Coelen, R.J.; Wilcox, G.E.; Sammels, L.M.; Kertayadnya, G. Nucleotide sequence analysis of Jembrana disease virus: A bovine lentivirus associated with an acute disease syndrome. J. Gen. Virol. 1995, 76, 1637–1650. [Google Scholar] [CrossRef] [PubMed]
- Curran, M.A.; Nolan, G.P. Nonprimate Lentiviral Vectors; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2002; Volume 261, pp. 75–105. [Google Scholar]
- Villet, S.; Faure, C.; Bouzar, B.A.; Morin, T.; Verdier, G.; Chebloune, Y.; Legras, C. Lack of trans-activation function for Maedi Visna virus and Caprine arthritis encephalitis virus Tat proteins. Virology 2003, 307, 317–327. [Google Scholar] [CrossRef]
- Schiltz, R.L.; Shih, D.S.; Rasty, S.; Montelaro, R.C.; E Rushlow, K. Equine infectious anemia virus gene expression: Characterization of the RNA splicing pattern and the protein products encoded by open reading frames S1 and S2. J. Virol. 1992, 66, 3455–3465. [Google Scholar] [CrossRef] [PubMed]
- Villet, S.; Bouzar, B.A.; Morin, T.; Verdier, G.; Legras, C.; Chebloune, Y. Maedi-Visna Virus and Caprine Arthritis Encephalitis Virus Genomes Encode a Vpr-Like but No Tat Protein. J. Virol. 2003, 77, 9632–9638. [Google Scholar] [CrossRef]
- De Parseval, A.; Elder, J.H. Demonstration that orf2 Encodes the Feline Immunodeficiency Virus Transactivating (Tat) Protein and Characterization of a Unique Gene Product with Partial Rev Activity. J. Virol. 1999, 73, 608–617. [Google Scholar] [CrossRef]
- Leroux, C.; Montelaro, R.C. Equine Infectious Anemia Virus (EIAV): What has HIV?s country cousin got to tell us? Vet. Res. 2004, 35, 485–512. [Google Scholar] [CrossRef]
- Stopak, K.; De Noronha, C.; Yonemoto, W.; Greene, W.C. HIV-1 Vif Blocks the Antiviral Activity of APOBEC3G by Impairing Both Its Translation and Intracellular Stability. Mol. Cell 2003, 12, 591–601. [Google Scholar] [CrossRef]
- Li, F.; Leroux, C.; Craigo, J.K.; Cook, S.J.; Issel, C.J.; Montelaro, R.C. The S2 Gene of Equine Infectious Anemia Virus Is a Highly Conserved Determinant of Viral Replication and Virulence Properties in Experimentally Infected Ponies. J. Virol. 2000, 74, 573–579. [Google Scholar] [CrossRef]
- Fagerness, A.J.; Flaherty, M.T.; Perry, S.T.; Jia, B.; Payne, S.L.; Fuller, F. The S2 accessory gene of equine infectious anemia virus is essential for expression of disease in ponies. Virology 2006, 349, 22–30. [Google Scholar] [CrossRef]
- Chande, A.; Cuccurullo, E.C.; Rosa, A.; Ziglio, S.; Carpenter, S.; Pizzato, M. S2 from equine infectious anemia virus is an infectivity factor which counteracts the retroviral inhibitors SERINC5 and SERINC3. Proc. Natl. Acad. Sci. USA 2016, 113, 13197–13202. [Google Scholar] [CrossRef]
- Stivahtis, G.L.; A Soares, M.; A Vodicka, M.; Hahn, B.H.; Emerman, M. Conservation and host specificity of Vpr-mediated cell cycle arrest suggest a fundamental role in primate lentivirus evolution and biology. J. Virol. 1997, 71, 4331–4338. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, T.M.; Brichacek, B.; Sharova, N.; Newman, M.A.; Stivahtis, G.; Sharp, P.M.; Emerman, M.; Hahn, B.H.; Stevenson, M. Nuclear import and cell cycle arrest functions of the HIV-1 Vpr protein are encoded by two separate genes in HIV-2/SIV(SM). EMBO J. 1996, 15, 6155–6165. [Google Scholar] [CrossRef] [PubMed]
- Gonda, M.A.; Luther, D.G.; Fong, S.E.; Tobin, G.J. Bovine immunodeficiency virus: Molecular biology and virus-host interactions. Virus Res. 1994, 32, 155–181. [Google Scholar] [CrossRef] [PubMed]
- St-Louis, M.-C.; Cojocariu, M.; Archambault, D. The molecular biology of bovine immunodeficiency virus: A comparison with other lentiviruses. Anim. Health Res. Rev. 2004, 5, 125–143. [Google Scholar] [CrossRef] [PubMed]
- Elder, J.H.; Lerner, D.L.; Hasselkus-Light, C.S.; Fontenot, D.J.; Hunter, E.; A Luciw, P.; Montelaro, R.C.; Phillips, T.R. Distinct subsets of retroviruses encode dUTPase. J. Virol. 1992, 66, 1791–1794. [Google Scholar] [CrossRef]
- CaraDonna, S.J.; Cheng, Y.C. Induction of uracil-DNA glycosylase and dUTP nucleotidohydrolase activity in herpes simplex virus-infected human cells. J. Boil. Chem. 1981, 256, 9834–9837. [Google Scholar]
- Broyles, S.S. Vaccinia Virus Encodes a Functional dUTPase. Virology 1993, 195, 863–865. [Google Scholar] [CrossRef]
- Bergman, A.-C.; Bjornberg, O.; Nord, J.; Nyman, P.; Rosengren, A. The Protein p30, Encoded at the gag-pro Junction of Mouse Mammary Tumor Virus, is a dUTPase Fused with a Nucleocapsid Protein. Virology 1994, 204, 420–421. [Google Scholar] [CrossRef]
- Köppe, B.; Menéndez-Arias, L.; Oroszlan, S. Expression and purification of the mouse mammary tumor virus gag-pro transframe protein p30 and characterization of its dUTPase activity. J. Virol. 1994, 68, 2313–2319. [Google Scholar] [CrossRef]
- Vassylyev, D.G.; Morikawa, K. Precluding uracil from DNA. Structure 1996, 4, 1381–1385. [Google Scholar] [CrossRef]
- Hizi, A.; Herzig, E. dUTPase: The frequently overlooked enzyme encoded by many retroviruses. Retrovirology 2015, 12, 70. [Google Scholar] [CrossRef] [PubMed]
- Podolny, Y.; Herzig, E.; Hizi, A. Insights into the molecular and biological features of the dUTPase-related gene of bovine immunodeficiency virus. Virology 2017, 506, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Steagall, W.K.; Robek, M.D.; Perry, S.T.; Fuller, F.J.; Payne, S.L. Incorporation of Uracil into Viral DNA Correlates with Reduced Replication of EIAV in Macrophages. Virology 1995, 210, 302–313. [Google Scholar] [CrossRef]
- Wagaman, P.C.; Hasselkus-Light, C.S.; Henson, M.; Lerner, D.L.; Phillips, T.R.; Elder, J.H. Molecular Cloning and Characterization of Deoxyuridine Triphosphatase from Feline Immunodeficiency Virus (FIV). Virology 1993, 196, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Threadgill, D.S.; Steagall, W.K.; Flaherty, M.T.; Fuller, F.J.; Perry, S.T.; E Rushlow, K.; Le Grice, S.F.; Payne, S.L. Characterization of equine infectious anemia virus dUTPase: Growth properties of a dUTPase-deficient mutant. J. Virol. 1993, 67, 2592–2600. [Google Scholar] [CrossRef]
- Turelli, P.; Pétursson, G.; Guiguen, F.; Mornex, J.F.; Vigne, R.; Quérat, G. Replication properties of dUTPase-deficient mutants of caprine and ovine lentiviruses. J. Virol. 1996, 70, 1213–1217. [Google Scholar] [CrossRef]
- Lichtenstein, D.L.; E Rushlow, K.; Cook, R.F.; Raabe, M.L.; Swardson, C.J.; Kociba, G.J.; Issel, C.J.; Montelaro, R.C. Replication in vitro and in vivo of an equine infectious anemia virus mutant deficient in dUTPase activity. J. Virol. 1995, 69, 2881–2888. [Google Scholar] [CrossRef]
- Pétursson, G.; Turelli, P.; Matthíasdóttir, S.; Georgsson, G.; Andrésson, O.S.; Torsteinsdóttir, S.; Vigne, R.; Andrésdóttir, V.; Gunnarsson, E.; Agnarsdóttir, G.; et al. Visna Virus dUTPase Is Dispensable for Neuropathogenicity. J. Virol. 1998, 72, 1657–1661. [Google Scholar] [CrossRef]
- Turelli, P.; Guiguen, F.; Mornex, J.F.; Vigne, R.; Quérat, G. dUTPase-minus caprine arthritis-encephalitis virus is attenuated for pathogenesis and accumulates G-to-A substitutions. J. Virol. 1997, 71, 4522–4530. [Google Scholar] [CrossRef]
- Lerner, D.L.; Wagaman, P.C.; Phillips, T.R.; Prospéro-García, O.; Henriksen, S.J.; Fox, H.S.; Bloom, F.E.; Elder, J.H. Increased mutation frequency of feline immunodeficiency virus lacking functional deoxyuridine-triphosphatase. Proc. Natl. Acad. Sci. USA 1995, 92, 7480–7484. [Google Scholar] [CrossRef]
- Giavedoni, L.D.; Yilma, T. Construction and characterization of replication-competent simian immunodeficiency virus vectors that express gamma interferon. J. Virol. 1996, 70, 2247–2251. [Google Scholar] [CrossRef] [PubMed]
- Alexander, L.; Veazey, R.S.; Czajak, S.; DeMaria, M.; Rosenzweig, M.; Lackner, A.A.; Desrosiers, R.C.; Sasseville, V.G. Recombinant Simian Immunodeficiency Virus Expressing Green Fluorescent Protein Identifies Infected Cells in Rhesus Monkeys. AIDS Res. Hum. Retrovir. 1999, 15, 11–21. [Google Scholar] [CrossRef]
- Ruprecht, R.M. Live attenuated AIDS viruses as vaccines: Promise or peril? Immunol. Rev. 1999, 170, 135–149. [Google Scholar] [CrossRef] [PubMed]
- Koff, W.C.; Johnson, P.R.; Watkins, D.I.; Burton, D.R.; Lifson, J.D.; Hasenkrug, K.J.; McDermott, A.B.; Schultz, A.; Zamb, T.J.; Boyle, R.; et al. HIV vaccine design: Insights from live attenuated SIV vaccines. Nat. Immunol. 2005, 7, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, T.; Nakamaru, K.; Ido, E.; Terao, K.; Hayami, M.; Hasegawa, M. Development of Novel Simian Immunodeficiency Virus Vectors Carrying a Dual Gene Expression System. Hum. Gene Ther. 2000, 11, 1863–1874. [Google Scholar] [CrossRef]
- Stitz, J.; Mühlebach, M.; Blömer, U.; Scherr, M.; Selbert, M.; Wehner, P.; Steidl, S.; Schmitt, I.; König, R.; Schweizer, M.; et al. A Novel Lentivirus Vector Derived from Apathogenic Simian Immunodeficiency Virus. Virology 2001, 291, 191–197. [Google Scholar] [CrossRef]
- Pandya, S.; Boris-Lawrie, K.; Leung, N.J.; Akkina, R.; Planelles, V. Development of an Rev-Independent, Minimal Simian Immunodeficiency Virus-Derived Vector System. Hum. Gene Ther. 2001, 12, 847–857. [Google Scholar] [CrossRef]
- White, S.M.; Renda, M.; Nam, N.-Y.; Klimatcheva, E.; Zhu, Y.; Fisk, J.; Halterman, M.; Rimel, B.J.; Federoff, H.; Pandya, S.; et al. Lentivirus Vectors Using Human and Simian Immunodeficiency Virus Elements. J. Virol. 1999, 73, 2832–2840. [Google Scholar] [CrossRef]
- A Rizvi, T.; Panganiban, A.T. Simian immunodeficiency virus RNA is efficiently encapsidated by human immunodeficiency virus type 1 particles. J. Virol. 1993, 67, 2681–2688. [Google Scholar] [CrossRef]
- Schnell, T.; Foley, P.; Wirth, M.; Münch, J.; Überla, K. Development of a Self-Inactivating, Minimal Lentivirus Vector Based on Simian Immunodeficiency Virus. Hum. Gene Ther. 2000, 11, 439–447. [Google Scholar] [CrossRef]
- Hanawa, H.; Hematti, P.; Keyvanfar, K.; Metzger, M.E.; Krouse, A.; Donahue, R.E.; Kepes, S.; Gray, J.; Dunbar, C.E.; Persons, D.A.; et al. Efficient gene transfer into rhesus repopulating hematopoietic stem cells using a simian immunodeficiency virus–based lentiviral vector system. Blood 2004, 103, 4062–4069. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Iida, A.; Ueda, Y.; Hasegawa, M. Pseudotyped Lentivirus Vectors Derived from Simian Immunodeficiency Virus SIVagm with Envelope Glycoproteins from Paramyxovirus. J. Virol. 2003, 77, 2607–2614. [Google Scholar] [CrossRef] [PubMed]
- Naumann, N.; De Ravin, S.S.; Choi, U.; Moayeri, M.; Whiting-Theobald, N.; Linton, G.F.; Ikeda, Y.; Malech, H.L. Simian immunodeficiency virus lentivector corrects human X-linked chronic granulomatous disease in the NOD/SCID mouse xenograft. Gene Ther. 2007, 14, 1513–1524. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kitagawa, R.; Miyachi, S.; Hanawa, H.; Takada, M.; Shimada, T. Differential characteristics of HIV-based versus SIV-based lentiviral vector systems: Gene delivery to neurons and axonal transport of expressed gene. Neurosci. Res. 2007, 57, 550–558. [Google Scholar] [CrossRef]
- Ikeda, Y.; Yonemitsu, Y.; Miyazaki, M.; Kohno, R.-I.; Murakami, Y.; Murata, T.; Tabata, T.; Ueda, Y.; Ono, F.; Suzuki, T.; et al. Stable Retinal Gene Expression in Nonhuman Primates via Subretinal Injection of SIVagm-Based Lentiviral Vectors. Hum. Gene Ther. 2009, 20, 573–579. [Google Scholar] [CrossRef]
- Alton, E.W.F.W.; Beekman, J.M.; Boyd, A.C.; Brand, J.; Carlon, M.S.; Connolly, M.M.; Chan, M.; Conlon, S.; E Davidson, H.; Davies, J.C.; et al. Preparation for a first-in-man lentivirus trial in patients with cystic fibrosis. Thorax 2016, 72, 137–147. [Google Scholar] [CrossRef]
- Sandrin, V.; Boson, B.; Salmon, P.; Gay, W.; Nègre, D.; Le Grand, R.; Trono, D.; Cosset, F.-L.; Kaudewitz, P.; Steinhoff, M.; et al. Lentiviral vectors pseudotyped with a modified RD114 envelope glycoprotein show increased stability in sera and augmented transduction of primary lymphocytes and CD34+ cells derived from human and nonhuman primates. Blood 2002, 100, 823–832. [Google Scholar] [CrossRef]
- Mangeot, P.E.; Nègre, D.; Dubois, B.; Winter, A.J.; Leissner, P.; Mehtali, M.; Kaiserlian, D.; Cosset, F.-L.; Darlix, J.-L. Development of Minimal Lentivirus Vectors Derived from Simian Immunodeficiency Virus (SIVmac251) and Their Use for Gene Transfer into Human Dendritic Cells. J. Virol. 2000, 74, 8307–8315. [Google Scholar] [CrossRef]
- Stitz, J.; Buchholz, C.; Engelstädter, M.; Uckert, W.; Bloemer, U.; Schmitt, I.; Cichutek, K. Lentiviral Vectors Pseudotyped with Envelope Glycoproteins Derived from Gibbon Ape Leukemia Virus and Murine Leukemia Virus 10A1. Virology 2000, 273, 16–20. [Google Scholar] [CrossRef]
- Mitomo, K.; Griesenbach, U.; Inoue, M.; Somerton, L.; Meng, C.; Akiba, E.; Tabata, T.; Ueda, Y.; Frankel, G.M.; Farley, R.; et al. Toward Gene Therapy for Cystic Fibrosis Using a Lentivirus Pseudotyped With Sendai Virus Envelopes. Mol. Ther. 2010, 18, 1173–1182. [Google Scholar] [CrossRef]
- Hlavatý, J.; Tonar, Z.; Renner, M.; Panitz, S.; Petznek, H.; Schweizer, M.; Schüle, S.; Kloke, B.-P.; Moldzio, R.; Witter, K. Tropism, intracerebral distribution, and transduction efficiency of HIV- and SIV-based lentiviral vectors after injection into the mouse brain: A qualitative and quantitative in vivo study. Histochem. Cell Boil. 2017, 148, 313–329. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Goujon, C.; Rivière, L.; Jarrosson-Wuilleme, L.; Bernaud, J.; Rigal, D.; Darlix, J.-L.; Cimarelli, A. SIVSM/HIV-2 Vpx proteins promote retroviral escape from a proteasome-dependent restriction pathway present in human dendritic cells. Retrovirology 2007, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Sharova, N.; Wu, Y.; Zhu, X.; Stránská, R.; Kaushik, R.; Sharkey, M.; Stevenson, M. Primate Lentiviral Vpx Commandeers DDB1 to Counteract a Macrophage Restriction. PLoS Pathog. 2008, 4, e1000057. [Google Scholar] [CrossRef] [PubMed]
- Negri, D.; Rossi, A.; Blasi, M.; Michelini, Z.; Leone, P.; Chiantore, M.V.; Baroncelli, S.; Perretta, G.; Cimarelli, A.; E Klotman, M.; et al. Simian immunodeficiency virus-Vpx for improving integrase defective lentiviral vector-based vaccines. Retrovirology 2012, 9, 69. [Google Scholar] [CrossRef]
- Goujon, C.; Jarrosson-Wuillème, L.; Bernaud, J.; Rigal, D.; Darlix, J.-L.; Cimarelli, A. With a little help from a friend: Increasing HIV transduction of monocyte-derived dendritic cells with virion-like particles of SIVMAC. Gene Ther. 2006, 13, 991–994. [Google Scholar] [CrossRef] [PubMed]
- Berger, G.; Goujon, C.; Darlix, J.-L.; Cimarelli, A. SIVMAC Vpx improves the transduction of dendritic cells with nonintegrative HIV-1-derived vectors. Gene Ther. 2008, 16, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Sheehy, A.M.; Gaddis, N.; Choi, J.D.; Malim, M.H. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 2002, 418, 646–650. [Google Scholar] [CrossRef]
- Stremlau, M.; Owens, C.M.; Perron, M.J.; Kiessling, M.; Autissier, P.; Sodroski, J. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 2004, 427, 848–853. [Google Scholar] [CrossRef]
- Neil, S.J.; Zang, T.; Bieniasz, P. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 2008, 451, 425–430. [Google Scholar] [CrossRef]
- Van Damme, N.; Goff, D.; Katsura, C.; Jorgenson, R.L.; Mitchell, R.; Johnson, M.C.; Stephens, E.B.; Guatelli, J. The Interferon-Induced Protein BST-2 Restricts HIV-1 Release and Is Downregulated from the Cell Surface by the Viral Vpu Protein. Cell Host Microbe 2008, 3, 245–252. [Google Scholar] [CrossRef]
- Lahouassa, H.; Daddacha, W.; Hofmann, H.; Ayinde, D.; Logue, E.C.; Dragin, L.; Bloch, N.; Maudet, C.; Bertrand, M.; Gramberg, T.; et al. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 2012, 13, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Rosa, A.; Chande, A.; Ziglio, S.; De Sanctis, V.; Bertorelli, R.; Goh, S.L.; McCauley, S.M.; Nowosielska, A.; Antonarakis, S.E.; Luban, J.; et al. HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature 2015, 526, 212–217. [Google Scholar] [CrossRef]
- Usami, Y.; Wu, Y.; Göttlinger, H. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature 2015, 526, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.S.; Fregoso, O.I.; McCoy, C.O.; Matsen, F.A.; Malik, H.S.; Emerman, M. The Ability of Primate Lentiviruses to Degrade the Monocyte Restriction Factor SAMHD1 Preceded the Birth of the Viral Accessory Protein Vpx. Cell Host Microbe 2012, 11, 194–204. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Serra-Moreno, R.; Neidermyer, W.; Rahmberg, A.; Mackey, J.; Ben Fofana, I.; Johnson, W.E.; Westmoreland, S.; Evans, D.T. Species-Specific Activity of SIV Nef and HIV-1 Vpu in Overcoming Restriction by Tetherin/BST2. PLoS Pathog. 2009, 5, e1000429. [Google Scholar] [CrossRef] [PubMed]
- Heigele, A.; Kmiec, D.; Regensburger, K.; Langer, S.; Peiffer, L.; Stürzel, C.M.; Sauter, D.; Peeters, M.; Pizzato, M.; Learn, G.H.; et al. The Potency of Nef-Mediated SERINC5 Antagonism Correlates with the Prevalence of Primate Lentiviruses in the Wild. Cell Host Microbe 2016, 20, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Kmiec, D.; Akbil, B.; Ananth, S.; Hotter, D.; Sparrer, K.M.J.; Stürzel, C.M.; Trautz, B.; Ayouba, A.; Peeters, M.; Yao, Z.; et al. SIVcol Nef counteracts SERINC5 by promoting its proteasomal degradation but does not efficiently enhance HIV-1 replication in human CD4+ T cells and lymphoid tissue. PLoS Pathog. 2018, 14, e1007269. [Google Scholar] [CrossRef]
- Gaddis, N.; Sheehy, A.M.; Ahmad, K.M.; Swanson, C.M.; Bishop, K.N.; Beer, B.E.; Marx, P.A.; Gao, F.; Bibollet-Ruche, F.; Hahn, B.H.; et al. Further Investigation of Simian Immunodeficiency Virus Vif Function in Human Cells. J. Virol. 2004, 78, 12041–12046. [Google Scholar] [CrossRef]
- Ylinen, L.M.J.; Keckesova, Z.; Wilson, S.J.; Ranasinghe, S.; Towers, G.J. Differential Restriction of Human Immunodeficiency Virus Type 2 and Simian Immunodeficiency Virus SIVmac by TRIM5α Alleles. J. Virol. 2005, 79, 11580–11587. [Google Scholar] [CrossRef]
- OhAinle, M.; Kim, K.; Keceli, S.K.; Felton, A.; Campbell, E.; Luban, J.; Emerman, M. TRIM34 restricts HIV-1 and SIV capsids in a TRIM5α-dependent manner. PLoS Pathog. 2020, 16, e1008507. [Google Scholar] [CrossRef]
- Pizzato, M.; McCauley, S.M.; Neagu, M.R.; Pertel, T.; Firrito, C.; Ziglio, S.; Dauphin, A.; Zufferey, M.; Berthoux, L.; Luban, J. Lv4 Is a Capsid-Specific Antiviral Activity in Human Blood Cells That Restricts Viruses of the SIVMAC/SIVSM/HIV-2 Lineage Prior to Integration. PLoS Pathog. 2015, 11, e1005050. [Google Scholar] [CrossRef] [PubMed]
- Kahl, C.A.; Cannon, P.M.; Oldenburg, J.; Tarantal, A.F.; Kohn, D.B. Tissue-specific restriction of cyclophilin A-independent HIV-1- and SIV-derived lentiviral vectors. Gene Ther. 2008, 15, 1079–1089. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-J.; Kim, Y.-S.; LaRochelle, A.; Renaud, G.; Wolfsberg, T.G.; Adler, R.; Donahue, R.E.; Hematti, P.; Hong, B.; Roayaei, J.; et al. Sustained high-level polyclonal hematopoietic marking and transgene expression 4 years after autologous transplantation of rhesus macaques with SIV lentiviral vector–transduced CD34+ cells. Blood 2009, 113, 5434–5443. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Poeschla, E.M.; Wong-Staal, F.; Looney, D.J. Efficient transduction of nondividing human cells by feline immunodeficiency virus lentiviral vectors. Nat. Med. 1998, 4, 354–357. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, K. Clinical Aspects of Feline Retroviruses: A Review. Viruses 2012, 4, 2684–2710. [Google Scholar] [CrossRef]
- Curran, M.A.; Kaiser, S.M.; Achacoso, P.L.; Nolan, G.P. Efficient Transduction of Nondividing Cells by Optimized Feline Immunodeficiency Virus Vectors. Mol. Ther. 2000, 1, 31–38. [Google Scholar] [CrossRef]
- Johnston, J.C.; Gasmi, M.; Lim, L.E.; Elder, J.H.; Yee, J.-K.; Jolly, D.J.; Campbell, K.P.; Davidson, B.L.; Sauter, S.L. Minimum Requirements for Efficient Transduction of Dividing and Nondividing Cells by Feline Immunodeficiency Virus Vectors. J. Virol. 1999, 73, 4991–5000. [Google Scholar] [CrossRef]
- Khare, P.D.; A Loewen, N.; Teo, W.; A Barraza, R.; Saenz, D.T.; Johnson, D.H.; Poeschla, E.M. Durable, Safe, Multi-gene Lentiviral Vector Expression in Feline Trabecular Meshwork. Mol. Ther. 2008, 16, 97–106. [Google Scholar] [CrossRef]
- Yee, J.-K.; Friedmann, T.; Burns, J.C. Chapter 5 Generation of High-Titer Pseudotyped Retroviral Vectors with Very Broad Host Range. Method. Cell Biol. 1994, 43, 99–112. [Google Scholar] [CrossRef]
- Song, J.J.; Lee, B.; Chang, J.W.; Kim, J.-H.; Kwon, Y.K.; Lee, H. Optimization of vesicular stomatitis virus-G pseudotyped feline immunodeficiency virus vector for minimized cytotoxicity with efficient gene transfer. Virus Res. 2003, 93, 25–30. [Google Scholar] [CrossRef]
- Stein, C.S.; Martins, I.; Davidson, B.L. In The Lymphocytic Choriomeningitis Virus Envelope Glycoprotein Targets Lentiviral Gene Transfer Vector to Neural Progenitors in the Murine Brain. Mol. Ther. 2005, 11, 382–389. [Google Scholar] [CrossRef]
- Shai, E.; Palmon, A.; Panet, A.; Marmary, Y.; Sherman, Y.; Curran, M.A.; Galun, E.; Condiotti, R. Prolonged transgene expression in murine salivary glands following non-primate lentiviral vector transduction. Mol. Ther. 2005, 12, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Stein, C.S.; Heth, J.A.; Sinn, P.L.; Penisten, A.K.; Staber, P.D.; Ratliff, K.L.; Shen, H.; Barker, C.K.; Martins, I.; et al. In Vivo Gene Transfer Using a Nonprimate Lentiviral Vector Pseudotyped with Ross River Virus Glycoproteins. J. Virol. 2002, 76, 9378–9388. [Google Scholar] [CrossRef] [PubMed]
- Sinn, P.L.; Burnight, E.R.; Hickey, M.A.; Blissard, G.W.; McCray, P.B. Persistent Gene Expression in Mouse Nasal Epithelia following Feline Immunodeficiency Virus-Based Vector Gene Transfer. J. Virol. 2005, 79, 12818–12827. [Google Scholar] [CrossRef] [PubMed]
- Sinn, P.L.; Goreham-Voss, J.D.; Arias, A.C.; Hickey, M.A.; Maury, W.J.; Chikkanna-Gowda, C.; McCray, P.B. Enhanced Gene Expression Conferred by Stepwise Modification of a Nonprimate Lentiviral Vector. Hum. Gene Ther. 2007, 18, 1244–1252. [Google Scholar] [CrossRef]
- Sinn, P.L.; Cooney, A.L.; Oakland, M.; E Dylla, D.; Wallen, T.J.; A Pezzulo, A.; Chang, E.H.; McCray, P.B. Lentiviral Vector Gene Transfer to Porcine Airways. Mol. Ther. Nucleic Acids 2012, 1, e56. [Google Scholar] [CrossRef]
- Oakland, M.; Maury, W.J.; McCray, P.B.; Sinn, P.L. Intrapulmonary Versus Nasal Transduction of Murine Airways With GP64-pseudotyped Viral Vectors. Mol. Ther. Nucleic Acids 2013, 2, e69. [Google Scholar] [CrossRef]
- Loewen, N.A.; Fautsch, M.P.; Peretz, M.; Bahler, C.K.; Cameron, J.D.; Johnson, D.H.; Poeschla, E.M. Genetic Modification of Human Trabecular Meshwork with Lentiviral Vectors. Hum. Gene Ther. 2001, 12, 2109–2119. [Google Scholar] [CrossRef]
- Loewen, N.; Bahler, C.; Teo, W.-L.; Whitwam, T.; Peretz, M.; Xu, R.; Fautsch, M.P.; Johnson, D.H.; Poeschla, E.M. Preservation of aqueous outflow facility after second-generation FIV vector-mediated expression of marker genes in anterior segments of human eyes. Investig. Ophthalmol. Vis. Sci. 2002, 43, 3686–3690. [Google Scholar]
- Lotery, A.; Derksen, T.A.; Russell, S.; Mullins, R.F.; Sauter, S.; Affatigato, L.M.; Stone, E.M.; Davidson, B.L. Gene Transfer to the Nonhuman Primate Retina with Recombinant Feline Immunodeficiency Virus Vectors. Hum. Gene Ther. 2002, 13, 689–696. [Google Scholar] [CrossRef]
- Barraza, R.A.; Rasmussen, C.A.; A Loewen, N.; Cameron, J.D.; Gabelt, B.T.; Teo, W.-L.; Kaufman, P.L.; Poeschla, E.M.E.M. Prolonged Transgene Expression with Lentiviral Vectors in the Aqueous Humor Outflow Pathway of Nonhuman Primates. Hum. Gene Ther. 2009, 20, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Derksen, T.A.; Sauter, S.L.; Davidson, B.L. Feline immunodeficiency virus vectors. Gene transfer to mouse retina following intravitreal injection. J. Gene Med. 2002, 4, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Janic, B.; Zhang, X.; Li, W. Feline immunodeficiency virus-mediated long-term transgene expression in undifferentiated retinal progenitor cells and its downregulation in differentiated cells. Mol. Vis. 2008, 14, 2117–2125. [Google Scholar] [PubMed]
- Cheng, L.; Toyoguchi, M.; Looney, D.J.; Lee, J.; Davidson, M.C.; Freeman, W.R. Efficient gene transfer to retinal pigment epithelium cells with long-term expression. Retin. J. Retin. Vit. Dis. 2005, 25, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Cheng, Q.; Nguyen, T.; A Bonanno, J. Knockdown of NBCe1 in vivo compromises the corneal endothelial pump. Investig. Opthalmology Vis. Sci. 2010, 51, 5190–5197. [Google Scholar] [CrossRef]
- Hatziioannou, T.; Perez-Caballero, D.; Yang, A.; Cowan, S.; Bieniasz, P.D. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5. Proc. Natl. Acad. Sci. USA 2004, 101, 10774–10779. [Google Scholar] [CrossRef]
- Saenz, D.T.; Teo, W.; Olsen, J.C.; Poeschla, E.M. Restriction of Feline Immunodeficiency Virus by Ref1, Lv1, and Primate TRIM5α Proteins. J. Virol. 2005, 79, 15175–15188. [Google Scholar] [CrossRef]
- Towers, G.J.; Collins, M.; Takeuchi, Y. Abrogation of Ref1 Retrovirus Restriction in Human Cells. J. Virol. 2002, 76, 2548–2550. [Google Scholar] [CrossRef]
- Olsen, J.C. Gene transfer vectors derived from equine infectious anemia virus. Gene Ther. 1998, 5, 1481–1487. [Google Scholar] [CrossRef]
- O’Rourke, J.P.; Olsen, J.C.; A Bunnell, B. Optimization of equine infectious anemia derived vectors for hematopoietic cell lineage gene transfer. Gene Ther. 2004, 12, 22–29. [Google Scholar] [CrossRef]
- Mitrophanous, K.; Yoon, S.; Rohll, J.B.; Patil, D.; Wilkes, F.J.; Kim, V.N.; Kingsman, S.M.; Kingsman, A.J.; Mazarakis, N.D. Stable gene transfer to the nervous system using a non-primate lentiviral vector. Gene Ther. 1999, 6, 1808–1818. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.-F.; Azzouz, M.; Walmsley, L.E.; Askham, Z.; Wilkes, F.J.; Mitrophanous, K.A.; Kingsman, S.M.; Mazarakis, N.D. Erratum to “Transduction patterns of pseudotyped lentiviral vectors in the nervous system”. Mol. Ther. 2004, 9, 765. [Google Scholar] [CrossRef]
- Ikeda, Y.; Collins, M.; Radcliffe, P.; Mitrophanous, K.; Takeuchi, Y. Gene transduction efficiency in cells of different species by HIV and EIAV vectors. Gene Ther. 2002, 9, 932–938. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Olsen, J.C.; Patel, M.; Rao, K.W.; Walsh, C.E. Functional Correction of Fanconi Anemia Group C Hematopoietic Cells by the Use of a Novel Lentiviral Vector. Mol. Ther. 2001, 3, 485–490. [Google Scholar] [CrossRef] [PubMed]
- O’Rourke, J.P.; Newbound, G.C.; Kohn, D.B.; Olsen, J.C.; Bunnell, B.A. Comparison of Gene Transfer Efficiencies and Gene Expression Levels Achieved with Equine Infectious Anemia Virus- and Human Immunodeficiency Virus Type 1-Derived Lentivirus Vectors. J. Virol. 2002, 76, 1510–1515. [Google Scholar] [CrossRef]
- Beutelspacher, S.C.; Ardjomand, N.; Tan, P.H.; Patton, G.S.; Larkin, D.F.P.; George, A.J.; O McClure, M. Comparison of HIV-1 and EIAV-based lentiviral vectors in corneal transduction. Exp. Eye Res. 2005, 80, 787–794. [Google Scholar] [CrossRef] [PubMed]
- Stewart, H.J.; A Leroux-Carlucci, M.; Sion, C.J.M.; A Mitrophanous, K.; A Radcliffe, P. Development of inducible EIAV-based lentiviral vector packaging and producer cell lines. Gene Ther. 2009, 16, 805–814. [Google Scholar] [CrossRef]
- Rohll, J.B.; Mitrophanous, K.A.; Martin-Rendon, E.; Ellard, F.M.; Radcliffe, P.A.; Mazarakis, N.D.; Kingsman, S.M. [27] Design, production, safety, evaluation, and clinical applications of nonprimate lentiviral vectors. Methods Enzymol. 2002, 346, 466–500. [Google Scholar] [CrossRef]
- Campochiaro, P.A.; Lauer, A.K.; Sohn, E.H.; Mir, T.A.; Naylor, S.; Anderton, M.C.; Kelleher, M.; Harrop, R.; Ellis, S.; Mitrophanous, K.A. Lentiviral Vector Gene Transfer of Endostatin/Angiostatin for Macular Degeneration (GEM) Study. Hum. Gene Ther. 2016, 28, 99–111. [Google Scholar] [CrossRef]
- Palfi, S.; Gurruchaga, J.M.; Ralph, G.S.; Lepetit, H.; Lavisse, S.; Buttery, P.C.; Watts, C.; Miskin, J.; Kelleher, M.; Deeley, S.; et al. Long-term safety and tolerability of ProSavin, a lentiviral vector-based gene therapy for Parkinson’s disease: A dose escalation, open-label, phase 1/2 trial. Lancet 2014, 383, 1138–1146. [Google Scholar] [CrossRef]
- Mselli-Lakhal, L.; Favier, C.; Teixeira, M.F.D.S.; Chettab, K.; Legras, C.; Ronfort, C.; Verdier, G.; Mornex, J.F.; Chebloune, Y. Defective RNA packaging is responsible for low transduction efficiency of CAEV-based vectors. Arch. Virol. 1998, 143, 681–695. [Google Scholar] [CrossRef] [PubMed]
- Mselli-Lakhal, L.; Favier, C.; Leung, K.; Guiguen, F.; Grézel, D.; Miossec, P.; Mornex, J.-F.; Narayan, O.; Quérat, G.; Chebloune, Y. Lack of Functional Receptors Is the Only Barrier That Prevents Caprine Arthritis-Encephalitis Virus from Infecting Human Cells. J. Virol. 2000, 74, 8343–8348. [Google Scholar] [CrossRef] [PubMed]
- Mselli-Lakhal, L.; Guiguen, F.; Greenland, T.; Mornex, J.-F.; Chebloune, Y. Gene transfer system derived from the caprine arthritis–encephalitis lentivirus. J. Virol. Methods 2006, 136, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Berkowitz, R.D.; Ilves, H.; Plavec, I.; Veres, G. Gene Transfer Systems Derived from Visna Virus: Analysis of Virus Production and Infectivity. Virology 2001, 279, 116–129. [Google Scholar] [CrossRef] [PubMed]
- Metharom, P.; Takyar, S.; Xia, H.H.; Ellem, K.A.O.; Macmillan, J.; Shepherd, R.W.; Wilcox, G.E.; Wei, M.Q. Novel bovine lentiviral vectors based on Jembrana disease virus. J. Gene Med. 2000, 2, 176–185. [Google Scholar] [CrossRef]
- Berkowitz, R.; Ilves, H.; Lin, W.Y.; Eckert, K.; Coward, A.; Tamaki, S.; Veres, G.; Plavec, I. Construction and Molecular Analysis of Gene Transfer Systems Derived from Bovine Immunodeficiency Virus. J. Virol. 2001, 75, 3371–3382. [Google Scholar] [CrossRef]
- Takahashi, K.; Luo, T.; Saishin, Y.; Saishin, Y.; Sung, J.; Hackett, S.; Brazzell, R.; Kaleko, M.; Campochiaro, P.A. Sustained Transduction of Ocular Cells with a Bovine Immunodeficiency Viral Vector. Hum. Gene Ther. 2002, 13, 1305–1316. [Google Scholar] [CrossRef]
- Matukonis, M.; Li, M.; Molina, R.P.; Paszkiet, B.; Kaleko, M.; Luo, T. Development of Second- and Third-Generation Bovine Immunodeficiency Virus-Based Gene Transfer Systems. Hum. Gene Ther. 2002, 13, 1293–1303. [Google Scholar] [CrossRef]
- Hacein-Bey-Abina, S.; Von Kalle, C.; Schmidt, M.; McCormack, M.P.; Wulffraat, N.; Leboulch, P.; Lim, A.; Osborne, C.S.; Pawliuk, R.; Morillon, E.; et al. LMO2-Associated Clonal T Cell Proliferation in Two Patients after Gene Therapy for SCID-X1. Science 2003, 302, 415–419. [Google Scholar] [CrossRef]
- Hacein-Bey-Abina, S.; Garrigue, A.; Wang, G.P.; Soulier, J.; Lim, A.; Morillon, E.; Clappier, E.; Caccavelli, L.; Delabesse, E.; Beldjord, K.; et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Investig. 2008, 118, 3132–3142. [Google Scholar] [CrossRef]
- Wang, G.P.; Ciuffi, A.; Leipzig, J.; Berry, C.C.; Bushman, F.D. HIV integration site selection: Analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007, 17, 1186–1194. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Li, Y.; Crise, B.; Burgess, S.M. Transcription Start Regions in the Human Genome Are Favored Targets for MLV Integration. Science 2003, 300, 1749–1751. [Google Scholar] [CrossRef] [PubMed]
- Biffi, A.; Bartolomae, C.C.; Cesana, D.; Cartier, N.; Aubourg, P.; Ranzani, M.; Cesani, M.; Benedicenti, F.; Plati, T.; Rubagotti, E.; et al. Lentiviral vector common integration sites in preclinical models and a clinical trial reflect a benign integration bias and not oncogenic selection. Blood 2011, 117, 5332–5339. [Google Scholar] [CrossRef] [PubMed]
- Goldstone, D.C.; Ennis-Adeniran, V.; Hedden, J.J.; Groom, H.C.T.; Rice, G.I.; Christodoulou, E.; Walker, P.A.; Kelly, G.; Haire, L.F.; Yap, M.W.; et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 2011, 480, 379–382. [Google Scholar] [CrossRef]
- E Mangeot, P.; Duperrier, K.; Nègre, D.; Boson, B.; Rigal, D.; Cosset, F.-L.; Darlix, J.-L. High Levels of Transduction of Human Dendritic Cells with Optimized SIV Vectors. Mol. Ther. 2002, 5, 283–290. [Google Scholar] [CrossRef]
- Mereby, S.A.; Maehigashi, T.; Holler, J.M.; Kim, N.-H.; Schinazi, R.F.; Kim, B. Interplay of ancestral non-primate lentiviruses with the virus-restricting SAMHD1 proteins of their hosts. J. Boil. Chem. 2018, 293, 16402–16412. [Google Scholar] [CrossRef]
- Laguette, N.; Sobhian, B.; Casartelli, N.; Ringeard, M.; Chable-Bessia, C.; Ségéral, E.; Yatim, A.; Emiliani, S.; Schwartz, O.; Benkirane, M. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 2011, 474, 654–657. [Google Scholar] [CrossRef]
| HIV-1 | HIV-2 | SIVcpz | SIVsmm | SIVagm | SIVlho | SIVsyk | SIVcol | |
|---|---|---|---|---|---|---|---|---|
| rev | + | + | + | + | + | + | + | + |
| vif | + | + | + | + | + | + | + | + |
| tat | + | + | + | + | + | + | + | + |
| vpr | + | + | + | + | + | + | + | + |
| vpx | − | + | − | + | − | − | − | − |
| vpu | + | − | + | − | − | − | − | − |
| nef | + | + | + | + | + | + | + | + |
| SIVsmm | FIV | EIAV | CAEV/VMV | BIV | JDV | |
|---|---|---|---|---|---|---|
| rev | + | + | + | + | + | + |
| vif | + | + | − | + | + | + |
| tat | + | − | + | − | + | + |
| vpr | + | − | − | − | − | − |
| vpx | + | − | − | − | − | − |
| nef | + | − | − | − | − | − |
| orf2 | − | + | − | − | − | − |
| orfS | − | − | − | + | − | − |
| s2 | − | − | + | − | − | − |
| vpw | − | − | - | − | + | − |
| vpy | − | − | - | − | + | − |
| tmx | − | − | - | − | + | + |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Munis, A.M. Gene Therapy Applications of Non-Human Lentiviral Vectors. Viruses 2020, 12, 1106. https://doi.org/10.3390/v12101106
Munis AM. Gene Therapy Applications of Non-Human Lentiviral Vectors. Viruses. 2020; 12(10):1106. https://doi.org/10.3390/v12101106
Chicago/Turabian StyleMunis, Altar M. 2020. "Gene Therapy Applications of Non-Human Lentiviral Vectors" Viruses 12, no. 10: 1106. https://doi.org/10.3390/v12101106
APA StyleMunis, A. M. (2020). Gene Therapy Applications of Non-Human Lentiviral Vectors. Viruses, 12(10), 1106. https://doi.org/10.3390/v12101106





