Interaction of Human ACE2 to Membrane-Bound SARS-CoV-1 and SARS-CoV-2 S Glycoproteins
Abstract
1. Introduction
2. Material and Methods
2.1. Plasmids
2.2. Cell Lines
2.3. Protein Expression and Purification
2.4. Cell Surface Staining and Flow Cytometry Analysis
2.5. Virus Neutralization Assay
3. Results and Discussion
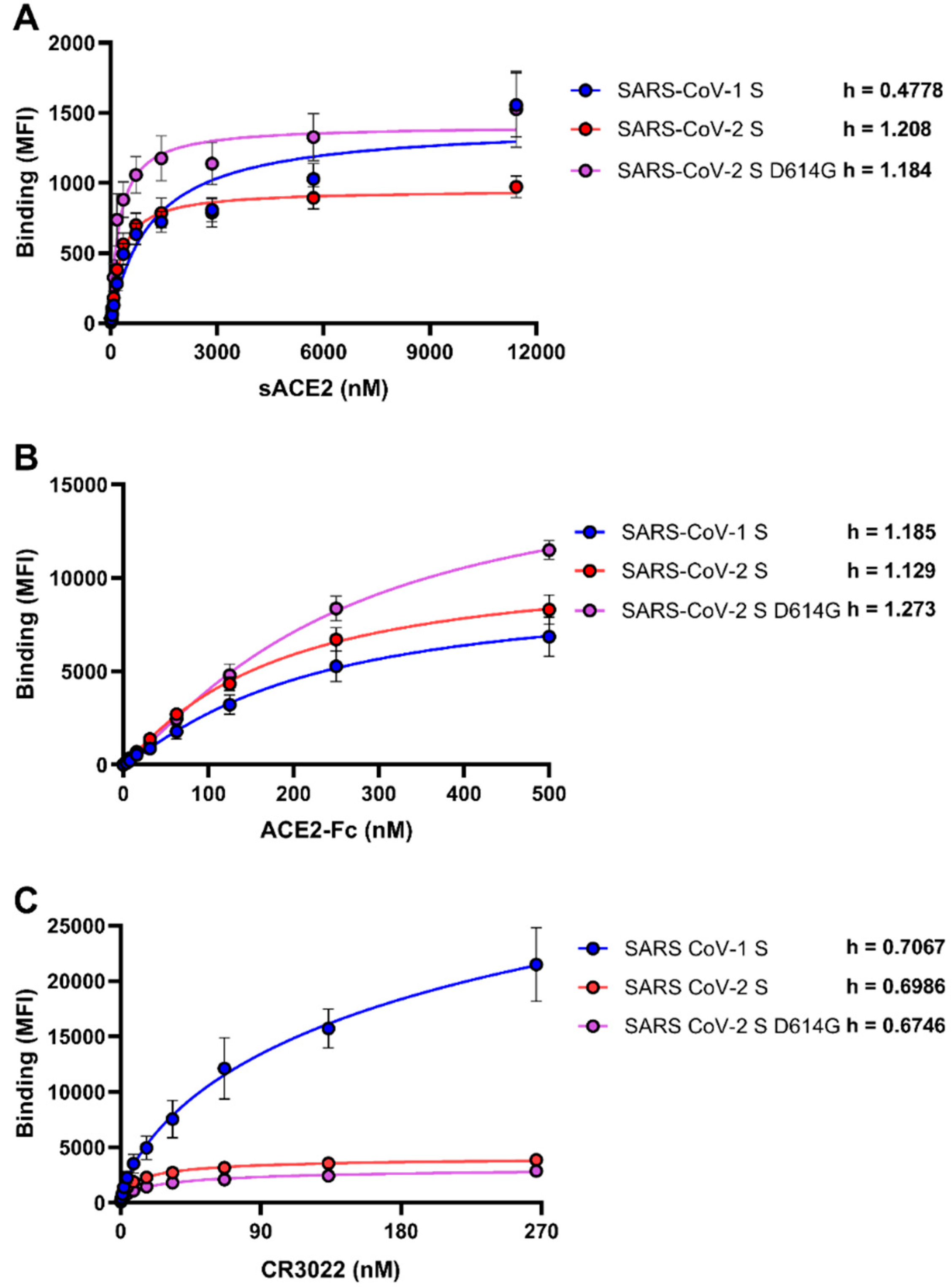
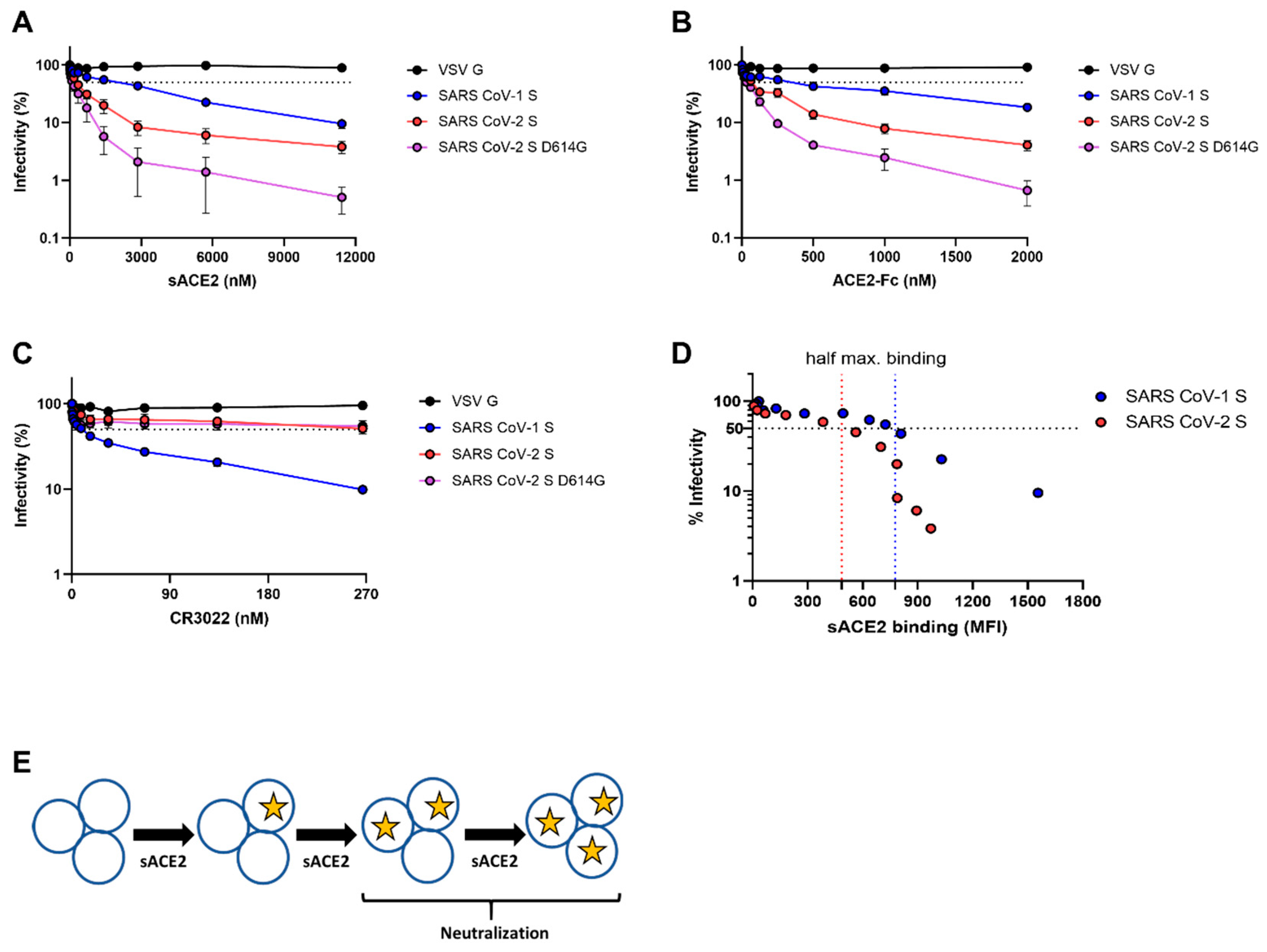
3.1. Differences Between SARS-CoV-1 S and SARS-CoV-2 Spikes in Their Abilities to Engage sACE2, ACE2-Fc, and CR3022
3.2. Sensitivity of Viruses Harboring SARS-CoV-1 S and SARS-CoV-2 Spikes to Neutralization by sACE2, ACE2-Fc, and CR3022
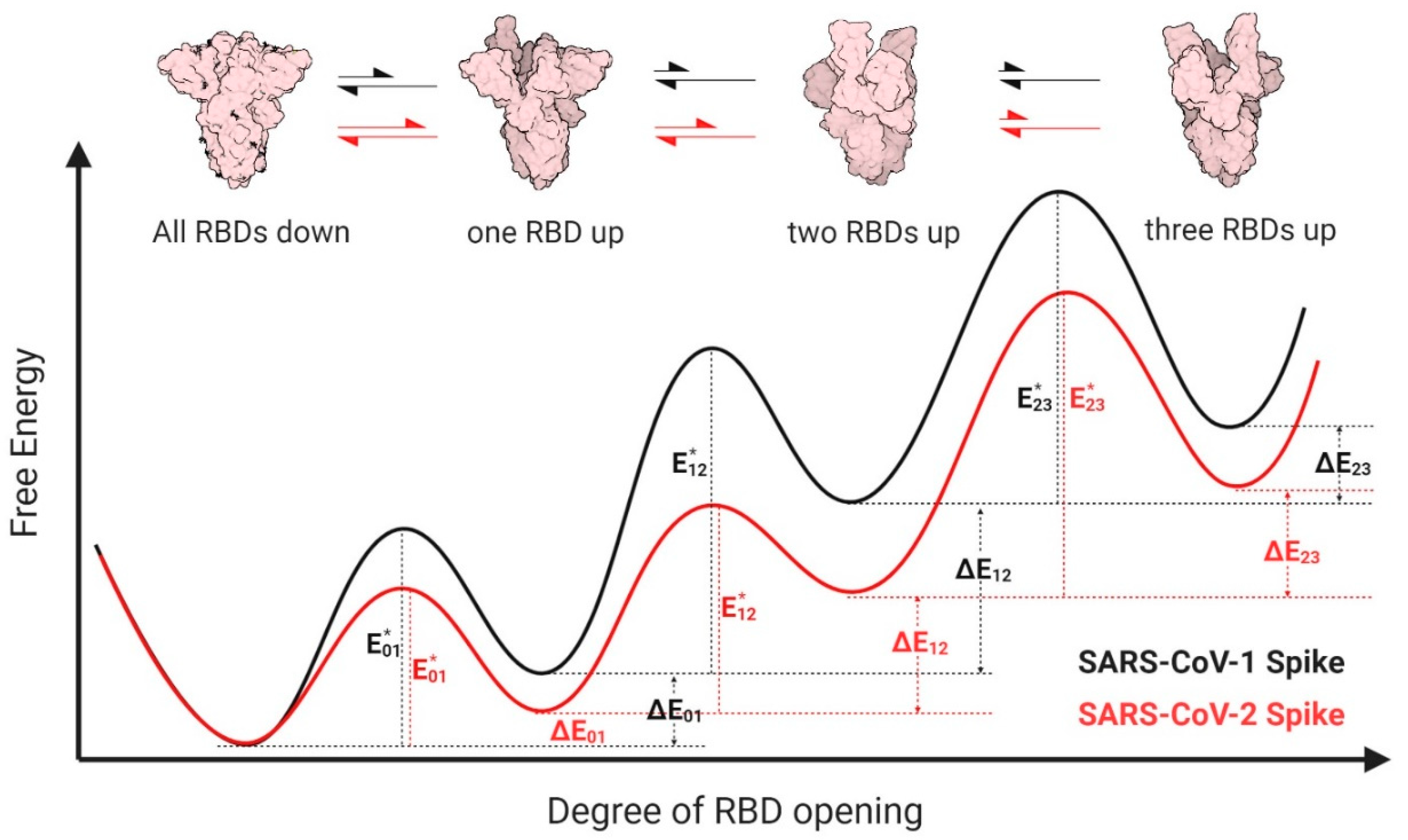
3.3. Proposed Energy Landscapes of Spike Trimer Opening of SARS-CoV-2 and SARS-CoV-1
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Disclaimer
References
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- Yuan, Y.; Cao, D.; Zhang, Y.; Ma, J.; Qi, J.; Wang, Q.; Lu, G.; Wu, Y.; Yan, J.; Shi, Y.; et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017, 8, 15092. [Google Scholar] [CrossRef]
- Gui, M.; Song, W.; Zhou, H.; Xu, J.; Chen, S.; Xiang, Y.; Wang, X. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017, 27, 119–129. [Google Scholar] [CrossRef]
- Cai, Y.; Zhang, J.; Xiao, T.; Peng, H.; Sterling, S.M.; Walsh, R.M., Jr.; Rawson, S.; Rits-Volloch, S.; Chen, B. Distinct conformational states of SARS-CoV-2 spike protein. Science 2020, 369, 1586–1592. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef]
- Crackower, M.A.; Sarao, R.; Oudit, G.Y.; Yagil, C.; Kozieradzki, I.; Scanga, S.E.; Oliveira-dos-Santos, A.J.; da Costa, J.; Zhang, L.; Pei, Y.; et al. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature 2002, 417, 822–828. [Google Scholar] [CrossRef]
- Monteil, V.; Kwon, H.; Prado, P.; Hagelkruys, A.; Wimmer, R.A.; Stahl, M.; Leopoldi, A.; Garreta, E.; Hurtado Del Pozo, C.; Prosper, F.; et al. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell 2020, 181, 905–913.e7. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.A.; Xie, X.; Kalveram, B.; Lokugamage, K.G.; Muruato, A.; Zou, J.; Zhang, X.; Juelich, T.; Smith, J.K.; Zhang, L.; et al. Furin Cleavage Site Is Key to SARS-CoV-2 Pathogenesis. BioRxiv 2020. [Google Scholar] [CrossRef]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Tsybovsky, Y.; Olia, A.S.; Gorman, J.; Rapp, M.A.; Cerutti, G.; Katsamba, P.S.; Nazzari, A.; Schon, A.; Wang, P.D.; et al. A pH-dependent switch mediates conformational masking of SARS-CoV-2 spike. BioRxiv 2020. [Google Scholar] [CrossRef]
- Ke, Z.; Oton, J.; Qu, K.; Cortese, M.; Zila, V.; McKeane, L.; Nakane, T.; Zivanov, J.; Neufeldt, C.J.; Cerikan, B.; et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 2020, 1–7. [Google Scholar] [CrossRef]
- Tai, W.; He, L.; Zhang, X.; Pu, J.; Voronin, D.; Jiang, S.; Zhou, Y.; Du, L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: Implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol. Immunol. 2020, 17, 613–620. [Google Scholar] [CrossRef]
- Lui, I.; Zhou, X.X.; Lim, S.A.; Elledge, S.K.; Solomon, P.; Rettko, N.J.; Zha, B.S.; Kirkemo, L.L.; Gramespacher, J.A.; Liu, J.; et al. Trimeric SARS-CoV-2 Spike interacts with dimeric ACE2 with limited intra-Spike avidity. BioRxiv 2020. [Google Scholar] [CrossRef]
- Li, Y.; Wang, H.; Tang, X.; Fang, S.; Ma, D.; Du, C.; Wang, Y.; Pan, H.; Yao, W.; Zhang, R.; et al. SARS-CoV-2 and three related coronaviruses utilize multiple ACE2 orthologs and are potently blocked by an improved ACE2-Ig. J. Virol. 2020. [Google Scholar] [CrossRef]
- Lei, C.; Qian, K.; Li, T.; Zhang, S.; Fu, W.; Ding, M.; Hu, S. Neutralization of SARS-CoV-2 spike pseudotyped virus by recombinant ACE2-Ig. Nat. Commun. 2020, 11, 2070. [Google Scholar] [CrossRef]
- Lodge, R.; Lalonde, J.P.; Lemay, G.; Cohen, E.A. The membrane-proximal intracytoplasmic tyrosine residue of HIV-1 envelope glycoprotein is critical for basolateral targeting of viral budding in MDCK cells. EMBO J. 1997, 16, 695–705. [Google Scholar] [CrossRef]
- Prevost, J.; Gasser, R.; Beaudoin-Bussieres, G.; Richard, J.; Duerr, R.; Laumaea, A.; Anand, S.P.; Goyette, G.; Ding, S.; Medjahed, H.; et al. Cross-sectional evaluation of humoral responses against SARS-CoV-2 Spike. BioRxiv 2020. [Google Scholar] [CrossRef]
- Weiss, J.N. The Hill equation revisited: Uses and misuses. FASEB J. 1997, 11, 835–841. [Google Scholar] [CrossRef] [PubMed]
- Cattoni, D.I.; Chara, O.; Kaufman, S.B.; Gonzalez Flecha, F.L. Cooperativity in Binding Processes: New Insights from Phenomenological Modeling. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e9. [Google Scholar] [CrossRef] [PubMed]
- Beaudoin-Bussières, G.; Laumaea, A.; Anand, S.P.; Prévost, J.; Gasser, R.; Goyette, G.; Medjahed, H.; Perreault, J.; Tremblay, T.; Lewin, A.; et al. Decline of humoral responses against SARS-CoV-2 Spike in convalescent individuals. BioRxiv 2020. [Google Scholar] [CrossRef]
- Zhang, L.; Jackson, C.B.; Mou, H.; Ojha, A.; Rangarajan, E.S.; Izard, T.; Farzan, M.; Choe, H. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. BioRxiv 2020. [Google Scholar] [CrossRef]
- Tian, X.; Li, C.; Huang, A.; Xia, S.; Lu, S.; Shi, Z.; Lu, L.; Jiang, S.; Yang, Z.; Wu, Y.; et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020, 9, 382–385. [Google Scholar] [CrossRef]
- ter Meulen, J.; van den Brink, E.N.; Poon, L.L.; Marissen, W.E.; Leung, C.S.; Cox, F.; Cheung, C.Y.; Bakker, A.Q.; Bogaards, J.A.; van Deventer, E.; et al. Human monoclonal antibody combination against SARS coronavirus: Synergy and coverage of escape mutants. PLoS Med. 2006, 3, e237. [Google Scholar] [CrossRef]
- Huo, J.; Zhao, Y.; Ren, J.; Zhou, D.; Duyvesteyn, H.M.E.; Ginn, H.M.; Carrique, L.; Malinauskas, T.; Ruza, R.R.; Shah, P.N.M.; et al. Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike. Cell Host Microbe 2020, 28, 445–454. [Google Scholar] [CrossRef]
- Procko, E. The sequence of human ACE2 is suboptimal for binding the S spike protein of SARS coronavirus 2. BioRxiv 2020. [Google Scholar] [CrossRef]
- Chan, K.K.; Dorosky, D.; Sharma, P.; Abbasi, S.A.; Dye, J.M.; Kranz, D.M.; Herbert, A.S.; Procko, E. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science 2020, 369, 1261–1265. [Google Scholar] [CrossRef] [PubMed]
- Henderson, R.; Edwards, R.J.; Mansouri, K.; Janowska, K.; Stalls, V.; Gobeil, S.M.C.; Kopp, M.; Li, D.; Parks, R.; Hsu, A.L.; et al. Controlling the SARS-CoV-2 spike glycoprotein conformation. Nat. Struct. Mol. Biol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Gui, M.; Wang, X.; Xiang, Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018, 14, e1007236. [Google Scholar] [CrossRef] [PubMed]
| Pseudotype | Neutralization (IC50; nM) | |||
|---|---|---|---|---|
| sACE2 | ACE2-Fc | CR3022 | Fold Change | |
| (sACE2/ACE2-Fc) | ||||
| VSV G | >11500 | >2000 | >270 | - |
| SARS CoV-1 S | 1359 | 218.5 | 7.481 | 6.22 |
| SARS CoV-2 S | 245.4 | 56.82 | 131.1 | 4.32 |
| SARS CoV-2 S D614G | 103.8 | 28.73 | 80.25 | 3.61 |
| Legend | ||||
| sACE2 | ACE2-Fc | CR3022 | ||
| IC50 < 1000 | IC50 < 100 | IC50 < 10 | ||
| 1000 < IC50 < 11500 | 100 < IC50 < 2000 | 10 < IC50 < 270 | ||
| IC50 > 11500 | IC50 > 2000 | IC50 > 270 | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anand, S.P.; Chen, Y.; Prévost, J.; Gasser, R.; Beaudoin-Bussières, G.; Abrams, C.F.; Pazgier, M.; Finzi, A. Interaction of Human ACE2 to Membrane-Bound SARS-CoV-1 and SARS-CoV-2 S Glycoproteins. Viruses 2020, 12, 1104. https://doi.org/10.3390/v12101104
Anand SP, Chen Y, Prévost J, Gasser R, Beaudoin-Bussières G, Abrams CF, Pazgier M, Finzi A. Interaction of Human ACE2 to Membrane-Bound SARS-CoV-1 and SARS-CoV-2 S Glycoproteins. Viruses. 2020; 12(10):1104. https://doi.org/10.3390/v12101104
Chicago/Turabian StyleAnand, Sai Priya, Yaozong Chen, Jérémie Prévost, Romain Gasser, Guillaume Beaudoin-Bussières, Cameron F. Abrams, Marzena Pazgier, and Andrés Finzi. 2020. "Interaction of Human ACE2 to Membrane-Bound SARS-CoV-1 and SARS-CoV-2 S Glycoproteins" Viruses 12, no. 10: 1104. https://doi.org/10.3390/v12101104
APA StyleAnand, S. P., Chen, Y., Prévost, J., Gasser, R., Beaudoin-Bussières, G., Abrams, C. F., Pazgier, M., & Finzi, A. (2020). Interaction of Human ACE2 to Membrane-Bound SARS-CoV-1 and SARS-CoV-2 S Glycoproteins. Viruses, 12(10), 1104. https://doi.org/10.3390/v12101104





