Single Amino Acid Substitutions in the Cucumber Mosaic Virus 1a Protein Induce Necrotic Cell Death in Virus-Inoculated Leaves without Affecting Virus Multiplication
Abstract
1. Introduction
2. Materials and Methods
2.1. Plants and Virus
2.2. In Vitro Transcription of Infectious CMV RNA and Production of Reassortant CMV
2.3. Virus Inoculation and Detection
2.4. Virus Quantification by ELISA
2.5. Detection of Cell Death
2.6. RNA-Seq Analysis
2.7. Construction of In Vitro Transcription Vectors Carrying Chimeric cDNA of CMV RNA1
2.8. Single Amino Acid Substitution in 1a Proteins Encoded on RNA1 of CMV(Y)
3. Results
3.1. Response of Arabidopsis thaliana Ecotype Col-0 to a Series of Reassortant CMVs
3.2. Response of A. thaliana Ecotypes to CMV(HYY)
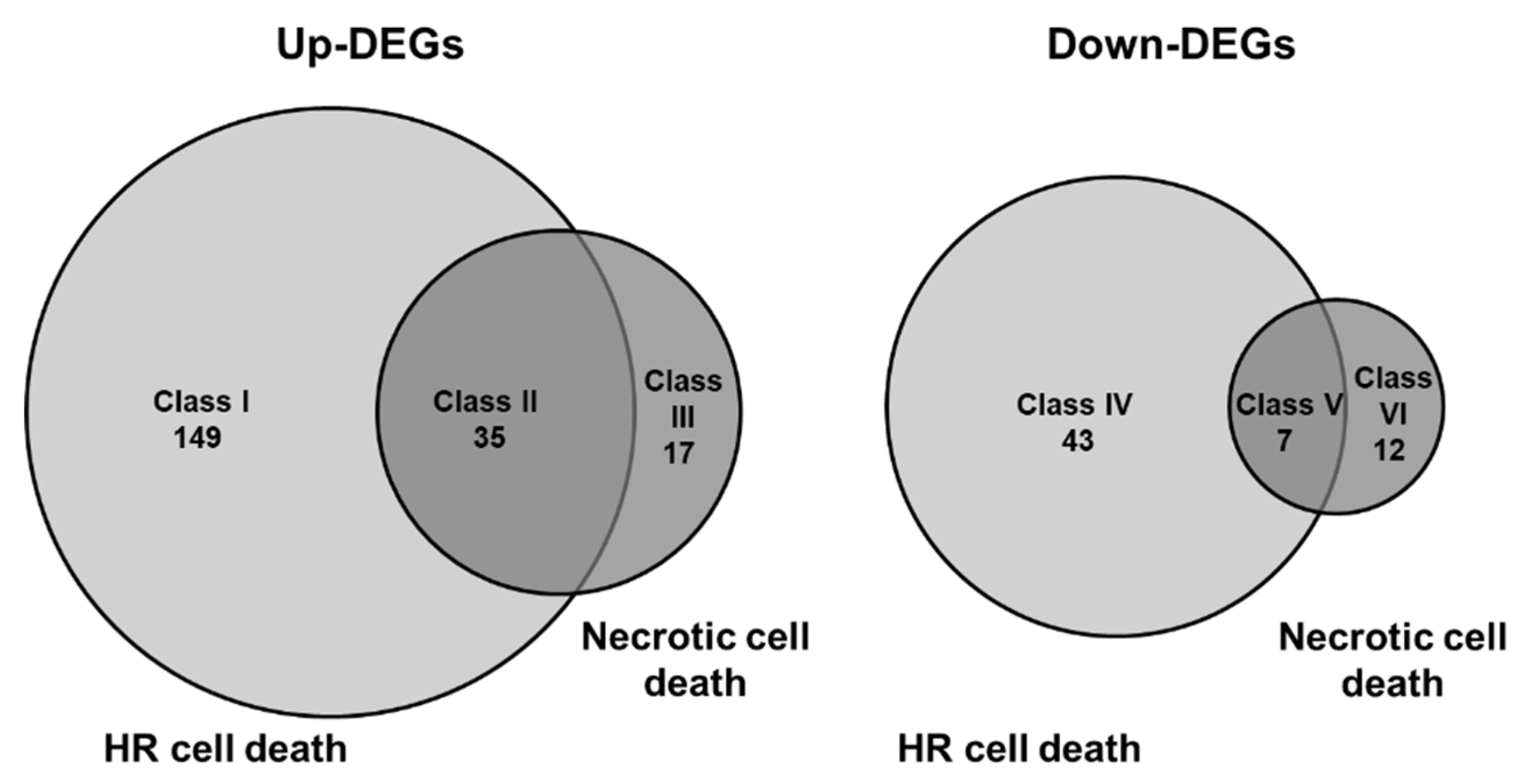
3.3. Comparison of Global Gene Expression Pattern between Two Types of Cell Death in Arabidopsis Leaves
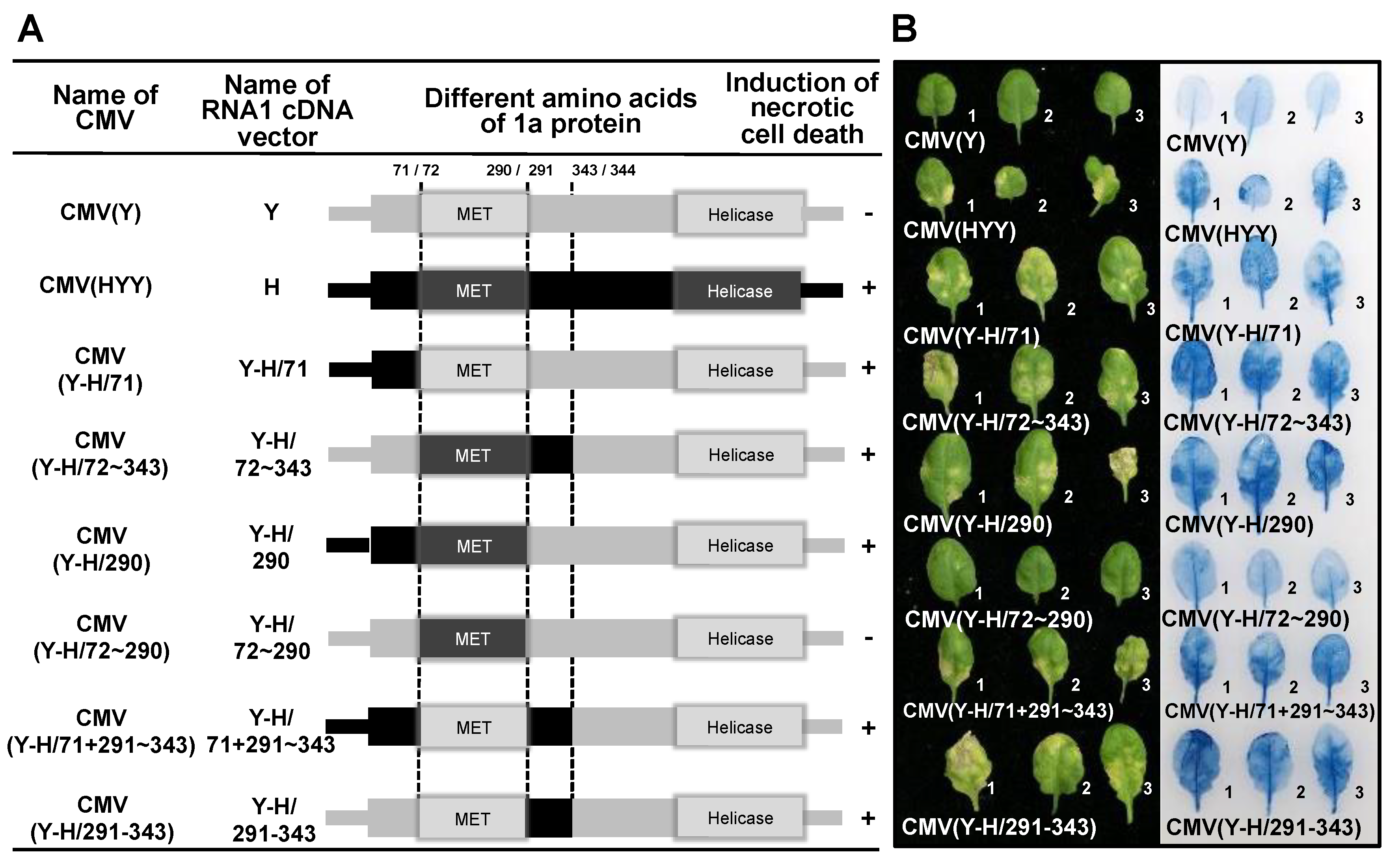
3.4. Analysis of the Viral Sequence in CMV RNA1 Inducing Necrotic Cell Death in Virus-Inoculated Leaves
3.5. Analysis of Single Amino Acid Substitutions in the CMV 1a Protein for Induction of Necrotic Cell Death in Virus-Inoculated Leaves
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Ethical Approval
References
- Mur, L.A.; Kenton, P.; Lloyd, A.J.; Ougham, H.; Prats, E. The hypersensitive response; the centenary is upon us but how much do we know? J. Exp. Bot. 2007, 59, 501–520. [Google Scholar] [CrossRef]
- Künstler, A.; Bacsó, R.; Gullner, G.; Hafez, Y.M.; Király, L. Staying alive—Is cell death dispensable for plant disease resistance during the hypersensitive response? Physiol. Mol. Plant Pathol. 2016, 93, 75–84. [Google Scholar] [CrossRef]
- Hofius, D.; Schultz-Larsen, T.; Joensen, J.; Tsitsigiannis, D.I.; Petersen, N.H.; Mattsson, O.; Jørgensen, L.B.; Jones, J.D.; Mundy, J.; Petersen, M. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 2009, 137, 773–783. [Google Scholar] [CrossRef]
- Hofius, D.; Tsitsigiannis, D.I.; Jones, J.D.; Mundy, J. Inducible cell death in plant immunity. Semin. Cancer Biol. 2007, 17, 166–187. [Google Scholar] [CrossRef] [PubMed]
- Richael, C.; Gilchrist, D. The hypersensitive response: A case of hold or fold? Physiol. Mol. Plant Pathol. 1999, 55, 5–12. [Google Scholar] [CrossRef]
- Hull, R. Plant Virology, 5th ed.; Academic Press: Salt Lake, UT, USA, 2013. [Google Scholar]
- Goodman, R.N.; Novacky, A.J. The Hypersensitive Reaction in Plants to Pathogens: A Resistance Phenomenon; APS Press: St. Paul, MN, USA, 1994. [Google Scholar]
- Pontier, D.; Balagué, C.; Roby, D. The hypersensitive response. A programmed cell death associated with plant resistance. Comptes Rendus Acad. Sci. Ser. III Sci. 1998, 321, 721–734. [Google Scholar] [CrossRef]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323. [Google Scholar] [CrossRef] [PubMed]
- Lam, E.; Kato, N.; Lawton, M. Programmed cell death, mitochondria and the plant hypersensitive response. Nature 2001, 411, 848. [Google Scholar] [CrossRef] [PubMed]
- Fomicheva, A.S.; Tuzhikov, A.I.; Beloshistov, R.E.; Trusova, S.V.; Galiullina, R.A.; Mochalova, L.V.; Chichkova, N.V.; Vartapetian, A.B. Programmed cell death in plants. Biochemistry (Moscow) 2012, 77, 1452–1464. [Google Scholar] [CrossRef]
- Morel, J.-B.; Dangl, J.L. The hypersensitive response and the induction of cell death in plants. Cell Death Differ. 1997, 4, 671. [Google Scholar] [CrossRef]
- Greenberg, J.T.; Yao, N. The role and regulation of programmed cell death in plant–pathogen interactions. Cell. Microbiol. 2004, 6, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Heath, M.C. Hypersensitive response-related death. In Programmed Cell Death in Higher Plants; Springer: New York, NY, USA, 2000; pp. 77–90. [Google Scholar]
- Canto, T.; Palukaitis, P. The hypersensitive response to cucumber mosaic virus in Chenopodium amaranticolor requires virus movement outside the initially infected cell. Virology 1999, 265, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Wright, K.M.; Duncan, G.H.; Pradel, K.S.; Carr, F.; Wood, S.; Oparka, K.J.; Santa Cruz, S. Analysis of the N gene hypersensitive response induced by a fluorescently tagged tobacco mosaic virus. Plant Physiol. 2000, 123, 1375–1386. [Google Scholar] [CrossRef] [PubMed]
- Murphy, A.M.; Carr, J.P. Salicylic acid has cell-specific effects on tobacco mosaic virus replication and cell-to-cell movement. Plant Physiol. 2002, 128, 552–563. [Google Scholar] [CrossRef]
- Lukan, T.; Baebler, Š.; Pompe-Novak, M.; Guček, K.; Zagorščak, M.; Coll, A.; Gruden, K. Cell death is not sufficient for the restriction of potato virus Y spread in hypersensitive response-conferred resistance in potato. Front. Plant Sci. 2018, 9, 168. [Google Scholar] [CrossRef]
- Inaba, J.; Kim, B.M.; Shimura, H.; Masuta, C. Virus-induced necrosis is a consequence of direct protein-protein interaction between a viral RNA-silencing suppressor and a host catalase. Plant Physiol. 2011, 156, 2026–2036. [Google Scholar] [CrossRef]
- Palukaitis, P.; García-Arenal, F. Cucumber Mosaic Virus; APS Press: St. Paul, MN, USA, 2018. [Google Scholar]
- Rozanov, M.N.; Koonin, E.V.; Gorbalenya, A.E. Conservation of the putative methyltransferase domain: A hallmark of the ‘Sindbis-like’ supergroup of positive-strand RNA viruses. J. Gen. Virol. 1992, 73, 2129–2134. [Google Scholar] [CrossRef]
- Habili, N.; Symons, R.H. Evolutionary relationship between luteoviruses and other RNA plant viruses based on sequence motifs in their putative RNA polymerases and nucleic acid helicases. Nucleic Acids Res. 1989, 17, 9543–9555. [Google Scholar] [CrossRef]
- O’Reilly, E.K.; Wang, Z.; French, R.; Kao, C.C. Interactions between the structural domains of the RNA replication proteins of plant-infecting RNA viruses. J. Virol. 1998, 72, 7160–7169. [Google Scholar] [CrossRef]
- Hayes, R.J.; Buck, K.W. Complete replication of a eukaryotic virus RNA in vitro by a purified RNA-dependent RNA polymerase. Cell 1990, 63, 363–368. [Google Scholar] [CrossRef]
- Guo, H.S.; Ding, S.W. A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J. 2002, 21, 398–407. [Google Scholar] [CrossRef] [PubMed]
- Mlotshwa, S.; Voinnet, O.; Mette, M.F.; Matzke, M.; Vaucheret, H.; Ding, S.W.; Pruss, G.; Vance, V.B. RNA silencing and the mobile silencing signal. Plant Cell 2002, 14, S289–S301. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yuan, Y.-R.; Pei, Y.; Lin, S.-S.; Tuschl, T.; Patel, D.J.; Chua, N.-H. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 2006, 20, 3255–3268. [Google Scholar] [CrossRef] [PubMed]
- González, I.; Martínez, L.; Rakitina, D.V.; Lewsey, M.G.; Atencio, F.A.; Llave, C.; Kalinina, N.O.; Carr, J.P.; Palukaitis, P.; Canto, T. Cucumber mosaic virus 2b protein subcellular targets and interactions: Their significance to RNA silencing suppressor activity. Mol. Plant Microbe Interact. 2010, 23, 294–303. [Google Scholar] [CrossRef]
- Schwinghamer, M.W.; Symons, R.H. Fractionation of cucumber mosaic virus RNA and its translation in a wheat embryo cell-free system. Virology 1975, 63, 252–262. [Google Scholar] [CrossRef]
- Ding, B.; Li, Q.; Nguyen, L.; Palukaitis, P.; Lucas, W.J. Cucumber mosaic virus 3a protein potentiates cell-to-cell trafficking of CMV RNA in tobacco plants. Virology 1995, 207, 345–353. [Google Scholar] [CrossRef][Green Version]
- Ding, S.W.; Anderson, B.J.; Haase, H.R.; Symons, R.H. New overlapping gene encoded by the cucumber mosaic virus genome. Virology 1994, 198, 593–601. [Google Scholar] [CrossRef]
- Ando, S.; Miyashita, S.; Takahashi, H. Plant defense systems against cucumber mosaic virus: Lessons learned from CMV–Arabidopsis interactions. J. Gen. Plant Pathol. 2019, 85, 174–181. [Google Scholar] [CrossRef]
- Sekine, K.; Kawakami, S.; Hase, S.; Kubota, M.; Ichinose, Y.; Shah, J.; Kang, H.G.; Klessig, D.F.; Takahashi, H. High level expression of a virus resistance gene, RCY1, confers extreme resistance to cucumber mosaic virus in Arabidopsis thaliana. Mol. Plant Microbe Interact. 2008, 21, 1398–1407. [Google Scholar] [CrossRef]
- Takahashi, H.; Goto, N.; Ehara, Y. Hypersensitive response in cucumber mosaic virus-inoculated Arabidopsis thaliana. Plant J. 1994, 6, 369–377. [Google Scholar] [CrossRef]
- Tomaru, K.; Hidaka, Z. Strains of cucumber mosaic virus isolated from tobacco plants. II. A mild strain. Bull. Hatano Tob. Exp. Stn. 1960, 46, 143–149. [Google Scholar] [CrossRef]
- Suzuki, M.; Kuwata, S.; Kataoka, J.; Masuta, C.; Nitta, N.; Takanami, Y. Functional analysis of deletion mutants of cucumber mosaic virus RNA3 using an in vitro transcription system. Virology 1991, 183, 106–113. [Google Scholar] [CrossRef]
- Sambrook, J.; Russel, D. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Takahashi, H.; Ehara, Y. Severe chlorotic spot symptoms in cucumber mosaic virus strain Y-infected tobaccos are induced by a combination of the virus coat protein gene and two host recessive genes. Mol. Plant Microbe Interact. 1993, 6, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Koenig, R. Indirect ELISA methods for the broad specificity detection of plant viruses. J. Gen. Virol. 1981, 55, 53–62. [Google Scholar] [CrossRef]
- Bowling, S.A.; Guo, A.; Cao, H.; Gordon, A.S.; Klessig, D.F.; Dong, X. A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 1994, 6, 1845–1857. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Chen, H.; Boutros, P.C. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Diveki, Z.; Salanki, K.; Balazs, E. The necrotic pathotype of the cucumber mosaic virus (CMV) ns strain is solely determined by amino acid 461 of the 1a protein. Mol. Plant Microbe Interact. 2004, 17, 837–845. [Google Scholar] [CrossRef]
- Salánki, K.; Gellért, Á.; Náray-Szabó, G.; Balázs, E. Modeling-based characterization of the elicitor function of amino acid 461 of cucumber mosaic virus 1a protein in the hypersensitive response. Virology 2007, 358, 109–118. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kang, W.H.; Seo, J.K.; Chung, B.N.; Kim, K.H.; Kang, B.C. Helicase domain encoded by cucumber mosaic virus RNA1 determines systemic infection of Cmr1 in pepper. PLoS ONE 2012, 7, e43136. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.K.; Kwon, S.J.; Choi, H.S.; Kim, K.H. Evidence for alternate states of cucumber mosaic virus replicase assembly in positive-and negative-strand RNA synthesis. Virology 2009, 383, 248–260. [Google Scholar] [CrossRef] [PubMed]
- Fujisaki, K.; Hagihara, F.; Azukawa, Y.; Kaido, M.; Okuno, T.; Mise, K. Identification and characterization of the SSB1 locus involved in symptom development by spring beauty latent virus infection in Arabidopsis thaliana. Mol. Plant Microbe Interact. 2004, 17, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Adam Arsovski, A.; Yu, K.; Wang, A. Deep sequencing leads to the identification of eukaryotic translation initiation factor 5A as a key element in Rsv1-mediated lethal systemic hypersensitive response to soybean mosaic virus infection in soybean. Mol. Plant Pathol. 2017, 18, 391–404. [Google Scholar] [CrossRef]
- Kim, B.M.; Suehiro, N.; Natsuaki, T.; Inukai, T.; Masuta, C. The P3 protein of turnip mosaic virus can alone induce hypersensitive response-like cell death in Arabidopsis thaliana carrying TuNI. Mol. Plant Microbe Interact. 2010, 23, 144–152. [Google Scholar] [CrossRef]
- Hajimorad, M.; Eggenberger, A.; Hill, J. Loss and gain of elicitor function of soybean mosaic virus G7 provoking Rsv1-mediated lethal systemic hypersensitive response maps to P3. J. Virol. 2005, 79, 1215–1222. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, K.; Hashimoto, M.; Ozeki, J.; Yamaji, Y.; Maejima, K.; Senshu, H.; Himeno, M.; Okano, Y.; Kagiwada, S.; Namba, S. Viral-induced systemic necrosis in plants involves both programmed cell death and the inhibition of viral multiplication, which are regulated by independent pathways. Mol. Plant Microbe Interact. 2010, 23, 283–293. [Google Scholar] [CrossRef]
- Hashimoto, M.; Komatsu, K.; Iwai, R.; Keima, T.; Maejima, K.; Shiraishi, T.; Ishikawa, K.; Yoshida, T.; Kitazawa, Y.; Okano, Y. Cell death triggered by a putative amphipathic helix of radish mosaic virus helicase protein is tightly correlated with host membrane modification. Mol. Plant Microbe Interact. 2015, 28, 675–688. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Buss, G.; Roane, C.; Tolin, S. Inheritance in soybean of resistant and necrotic reactions to soybean mosaic virus strains. Crop Sci. 1994, 34, 414–422. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, A.; Miyashita, S.; Ando, S.; Takahashi, H. Single Amino Acid Substitutions in the Cucumber Mosaic Virus 1a Protein Induce Necrotic Cell Death in Virus-Inoculated Leaves without Affecting Virus Multiplication. Viruses 2020, 12, 91. https://doi.org/10.3390/v12010091
Tian A, Miyashita S, Ando S, Takahashi H. Single Amino Acid Substitutions in the Cucumber Mosaic Virus 1a Protein Induce Necrotic Cell Death in Virus-Inoculated Leaves without Affecting Virus Multiplication. Viruses. 2020; 12(1):91. https://doi.org/10.3390/v12010091
Chicago/Turabian StyleTian, Ainan, Shuhei Miyashita, Sugihiro Ando, and Hideki Takahashi. 2020. "Single Amino Acid Substitutions in the Cucumber Mosaic Virus 1a Protein Induce Necrotic Cell Death in Virus-Inoculated Leaves without Affecting Virus Multiplication" Viruses 12, no. 1: 91. https://doi.org/10.3390/v12010091
APA StyleTian, A., Miyashita, S., Ando, S., & Takahashi, H. (2020). Single Amino Acid Substitutions in the Cucumber Mosaic Virus 1a Protein Induce Necrotic Cell Death in Virus-Inoculated Leaves without Affecting Virus Multiplication. Viruses, 12(1), 91. https://doi.org/10.3390/v12010091




