Taking over Cellular Energy-Metabolism for TBSV Replication: The High ATP Requirement of an RNA Virus within the Viral Replication Organelle
Abstract
1. Introduction
2. TBSV–Host Interactions
3. Budding Yeast as a Surrogate Host to Characterize TBSV–Host Interactions
4. The Expanding Role of Aerobic Glycolysis
5. Exploitation of the Aerobic Glycolytic Pathway by Tombusviruses
6. Exploitation of the Fermentation Pathway by Tombusviruses
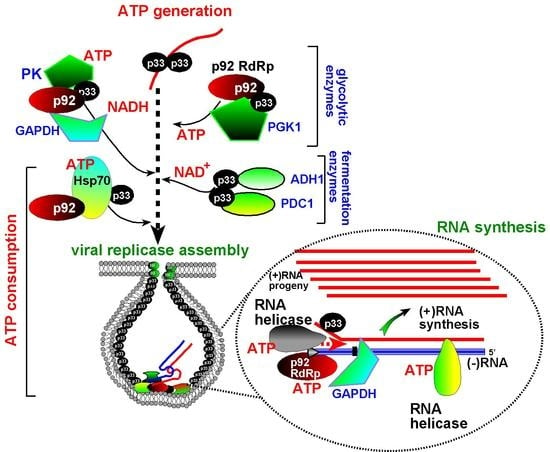
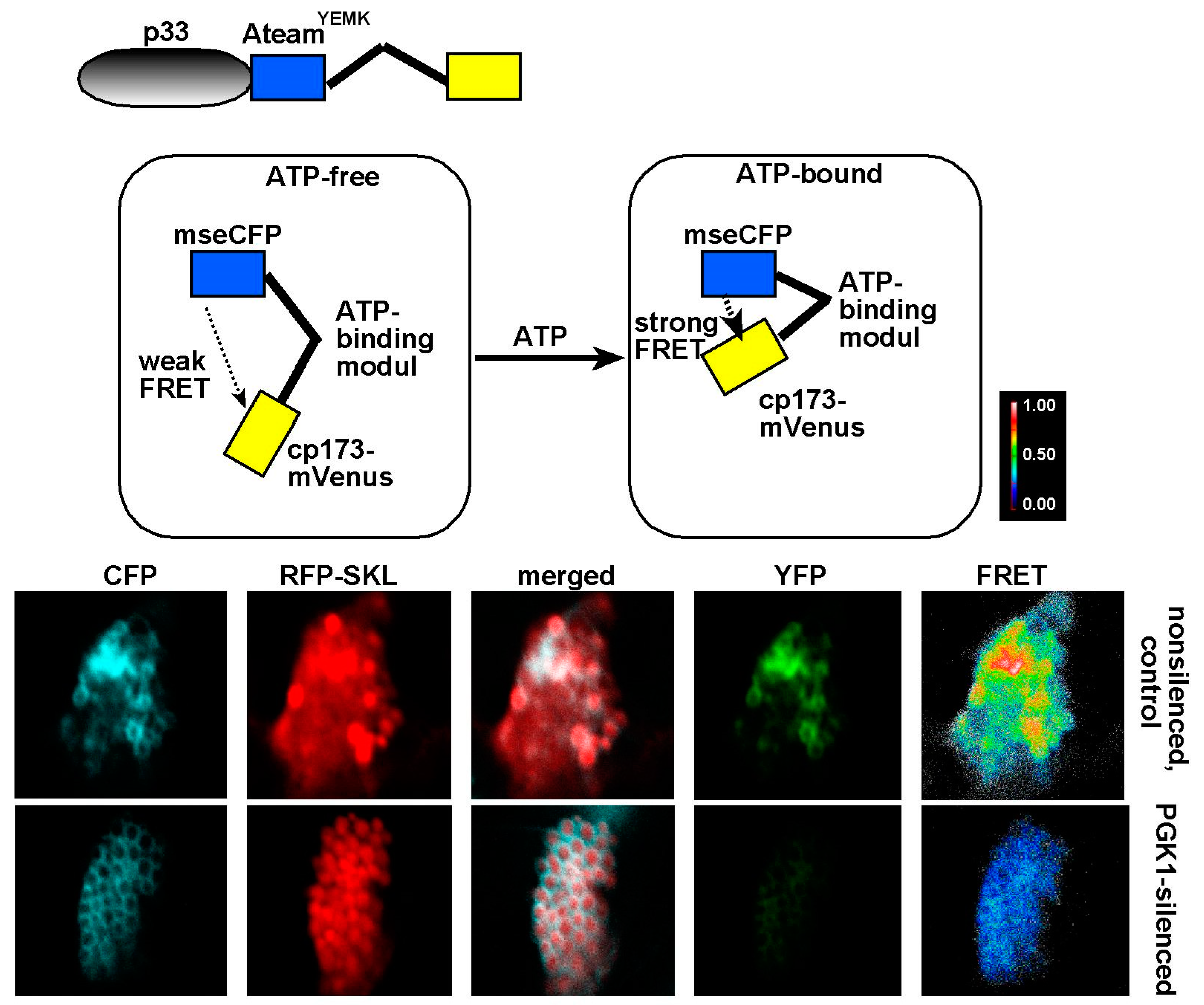
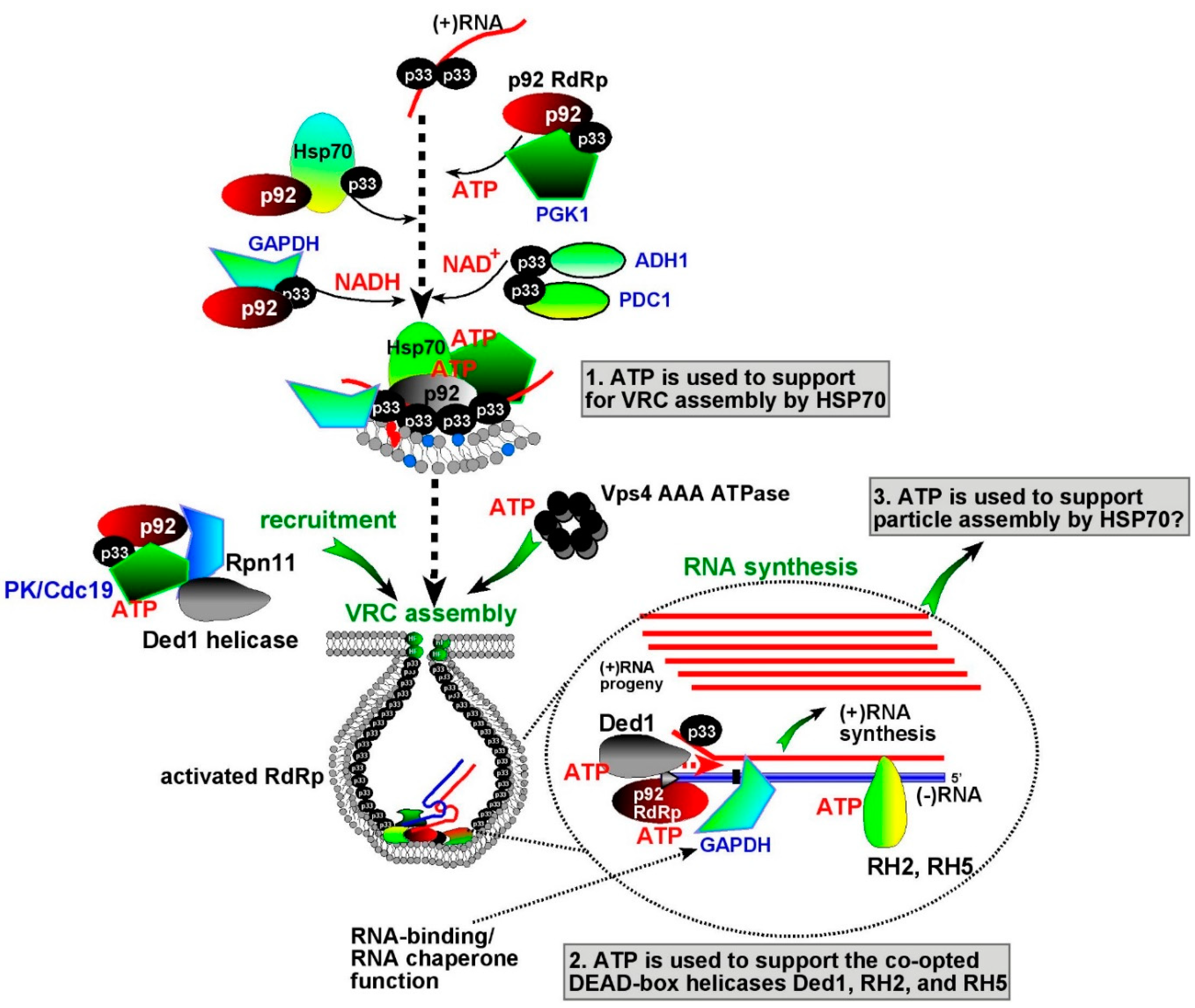
7. The Need for Locally Produced ATP During Tombusvirus Replication
8. Do Glycolytic and Fermentation Enzymes Perform Moonlighting Functions during Virus Replication?
9. Numerous Similarities Between Tombusvirus-Infected Cells and Cancerous Cells in Rewiring Cellular Metabolic Pathways
10. The Role of Glycolysis in Virus–Host Interactions
11. Future Directions
12. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- De Castro, I.F.; Volonte, L.; Risco, C. Virus factories: Biogenesis and structural design. Cell Microbiol. 2013, 15, 24–34. [Google Scholar] [CrossRef]
- Den Boon, J.A.; Ahlquist, P. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu. Rev. Microbiol. 2010, 64, 241–256. [Google Scholar] [CrossRef]
- Nagy, P.D.; Pogany, J. The dependence of viral RNA replication on co-opted host factors. Nat. Rev. Microbiol. 2012, 10, 137–149. [Google Scholar] [CrossRef]
- Wang, A. Dissecting the Molecular Network of Virus-Plant Interactions: The Complex Roles of Host Factors. Annu. Rev. Phytopathol. 2015, 53, 45–66. [Google Scholar] [CrossRef]
- Laliberté, J.-F.; Sanfaçon, H. Cellular Remodeling During Plant Virus Infection. Annu. Rev. Phytopathol. 2010, 48, 69–91. [Google Scholar] [CrossRef]
- Nagy, P.D. Exploitation of a surrogate host, Saccharomyces cerevisiae, to identify cellular targets and develop novel antiviral approaches. Curr. Opin. Virol. 2017, 26, 132–140. [Google Scholar] [CrossRef]
- Nagy, P.D. Tombusvirus-Host Interactions: Co-Opted Evolutionarily Conserved Host Factors Take Center Court. Annu. Rev. Virol. 2016, 3, 491–515. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; He, G.; Filipowicz, N.A.; Randall, G.; Belov, G.A.; Kopek, B.G.; Wang, X. Host Lipids in Positive-Strand RNA Virus Genome Replication. Front. Microbiol. 2019, 10, 286. [Google Scholar] [CrossRef] [PubMed]
- Nagy, P.D.; Pogany, J.; Lin, J.-Y. How yeast can be used as a genetic platform to explore virus–host interactions: From ‘omics’ to functional studies. Trends Microbiol. 2014, 22, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Nagy, P.D.; Pogany, J. Global Genomics and Proteomics Approaches to Identify Host Factors as Targets to Induce Resistance against Tomato Bushy Stunt Virus. Adv. Clin. Chem. 2010, 76, 123–177. [Google Scholar]
- Zhao, R.Y. Yeast for virus research. Microb. Cell 2017, 4, 311–330. [Google Scholar] [CrossRef] [PubMed]
- Romero-Brey, I.; Bartenschlager, R. Membranous Replication Factories Induced by Plus-Strand RNA Viruses. Viruses 2014, 6, 2826–2857. [Google Scholar] [CrossRef] [PubMed]
- Fernandez de Castro, I.; Fernandez, J.J.; Barajas, D.; Nagy, P.D.; Risco, C. Three-dimensional imaging of the intracellular assembly of a functional viral RNA replicase complex. J. Cell Sci. 2017, 130, 260–268. [Google Scholar] [CrossRef] [PubMed]
- Kovalev, N.; Inaba, J.-I.; Li, Z.; Nagy, P.D. The role of co-opted ESCRT proteins and lipid factors in protection of tombusviral double-stranded RNA replication intermediate against reconstituted RNAi in yeast. PLoS Pathog. 2017, 13, e1006520. [Google Scholar] [CrossRef]
- Panaviene, Z.; Panavas, T.; Nagy, P.D. Role of an Internal and Two 3′-Terminal RNA Elements in Assembly of Tombusvirus Replicase. J. Virol. 2005, 79, 10608–10618. [Google Scholar] [CrossRef]
- Panaviene, Z.; Panavas, T.; Serva, S.; Nagy, P.D. Purification of the Cucumber Necrosis Virus Replicase from Yeast Cells: Role of Coexpressed Viral RNA in Stimulation of Replicase Activity. J. Virol. 2004, 78, 8254–8263. [Google Scholar] [CrossRef]
- Gunawardene, C.D.; Donaldson, L.W.; White, K.A. Tombusvirus polymerase: Structure and function. Virus Res. 2017, 234, 74–86. [Google Scholar] [CrossRef]
- Nagy, P.D. Viral Sensing of the Subcellular Environment Regulates the Assembly of New Viral Replicase Complexes during the Course of Infection. J. Virol. 2015, 89, 5196–5199. [Google Scholar] [CrossRef]
- Noueiry, A.O.; Ahlquist, P. Brome mosaic virus RNA replication: Revealing the role of the host in RNA virus replication. Annu. Rev. Phytopathol. 2003, 41, 77–98. [Google Scholar] [CrossRef]
- Boon, J.A.D.; Diaz, A.; Ahlquist, P. Cytoplasmic viral replication complexes. Cell Host Microbe 2010, 8, 77–85. [Google Scholar] [CrossRef]
- Jones, W.; Bianchi, K. Aerobic Glycolysis: Beyond Proliferation. Front. Immunol. 2015, 6, 227. [Google Scholar] [CrossRef] [PubMed]
- Heiden, M.G.V.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Lunt, S.Y.; Heiden, M.G.V. Aerobic Glycolysis: Meeting the Metabolic Requirements of Cell Proliferation. Annu. Rev. Cell Dev. Boil. 2011, 27, 441–464. [Google Scholar] [CrossRef] [PubMed]
- Kürsteiner, O.; Dupuis, I.; Kuhlemeier, C. The Pyruvate decarboxylase1 Gene of Arabidopsis Is Required during Anoxia But Not Other Environmental Stresses. Plant. Physiol. 2003, 132, 968–978. [Google Scholar] [CrossRef]
- Gándara, L.; Wappner, P. Metabo-Devo: A metabolic perspective of development. Mech. Dev. 2018, 154, 12–23. [Google Scholar] [CrossRef]
- Krejcova, G.; Danielova, A.; Nedbalova, P.; Kazek, M.; Strych, L.; Chawla, G.; Tennessen, J.M.; Lieskovska, J.; Jindra, M.; Dolezal, T.; et al. Drosophila macrophages switch to aerobic glycolysis to mount effective antibacterial defense. Elife 2019, 8, e50414. [Google Scholar] [CrossRef]
- Ghosh, S.; Castillo, E.; Frias, E.S.; Swanson, R.A. Bioenergetic regulation of microglia. Glia 2018, 66, 1200–1212. [Google Scholar] [CrossRef]
- Palm, W.; Thompson, C.B. Nutrient acquisition strategies of mammalian cells. Nature 2017, 546, 234–242. [Google Scholar] [CrossRef]
- Shannon, B.J.; Vaishnavi, S.N.; Vlassenko, A.G.; Shimony, J.S.; Rutlin, J.; Raichle, M.E. Brain aerobic glycolysis and motor adaptation learning. Proc. Natl. Acad. Sci. USA 2016, 113, E3782–E3791. [Google Scholar] [CrossRef]
- Yu, L.; Chen, X.; Wang, L.; Chen, S. Oncogenic virus-induced aerobic glycolysis and tumorigenesis. J. Cancer 2018, 9, 3699–3706. [Google Scholar] [CrossRef]
- Vaishnavi, S.N.; Vlassenko, A.G.; Rundle, M.M.; Snyder, A.Z.; Mintun, M.A.; Raichle, M.E. Regional aerobic glycolysis in the human brain. Proc. Natl. Acad. Sci. USA 2010, 107, 17757–17762. [Google Scholar] [CrossRef] [PubMed]
- Vlassenko, A.G.; Vaishnavi, S.N.; Couture, L.; Sacco, D.; Shannon, B.J.; Mach, R.H.; Morris, J.C.; Raichle, M.E.; Mintun, M.A. Spatial correlation between brain aerobic glycolysis and amyloid-beta (Abeta) deposition. Proc. Natl. Acad. Sci. USA 2010, 107, 17763–17767. [Google Scholar] [CrossRef] [PubMed]
- Prasanth, K.R.; Chuang, C.; Nagy, P.D. Co-opting ATP-generating glycolytic enzyme PGK1 phosphoglycerate kinase facilitates the assembly of viral replicase complexes. PLoS Pathog. 2017, 13, e1006689. [Google Scholar] [CrossRef] [PubMed]
- Chuang, C.; Prasanth, K.R.; Nagy, P.D. The Glycolytic Pyruvate Kinase Is Recruited Directly into the Viral Replicase Complex to Generate ATP for RNA Synthesis. Cell Host Microbe 2017, 22, 639–652e7. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Nagy, P.D. RNA virus replication depends on enrichment of phosphatidylethanolamine at replication sites in subcellular membranes. Proc. Natl. Acad. Sci. USA 2015, 112, E1782–E1791. [Google Scholar] [CrossRef]
- Inaba, J.-I.; Nagy, P.D. Tombusvirus RNA replication depends on the TOR pathway in yeast and plants. Virology 2018, 519, 207–222. [Google Scholar] [CrossRef]
- Olson, K.A.; Schell, J.C.; Rutter, J. Pyruvate and Metabolic Flexibility: Illuminating a Path toward Selective Cancer Therapies. Trends Biochem. Sci. 2016, 41, 219–230. [Google Scholar] [CrossRef]
- Lin, W.; Liu, Y.; Molho, M.; Zhang, S.; Wang, L.; Xie, L.; Nagy, P.D. Co-opting the fermentation pathway for tombusvirus replication: Compartmentalization of cellular metabolic pathways for rapid ATP generation. PLoS Pathog. 2019, 15, e1008092. [Google Scholar] [CrossRef]
- Knight, M.; Stanley, S. HIF-1alpha as a central mediator of cellular resistance to intracellular pathogens. Curr. Opin. Immunol. 2019, 60, 111–116. [Google Scholar] [CrossRef]
- Verdone, J.E.; Zarif, J.C.; Pienta, K.J. Aerobic glycolysis, motility, and cytoskeletal remodeling. Cell Cycle 2015, 14, 169–170. [Google Scholar] [CrossRef][Green Version]
- Shiraishi, T.; Verdone, J.E.; Huang, J.; Kahlert, U.D.; Hernandez, J.R.; Torga, G.; Zarif, J.C.; Epstein, T.; Gatenby, R.; McCartney, A.; et al. Glycolysis is the primary bioenergetic pathway for cell motility and cytoskeletal remodeling in human prostate and breast cancer cells. Oncotarget 2015, 6, 130–143. [Google Scholar] [CrossRef] [PubMed]
- Araiza-Olivera, D.; Chiquete-Felix, N.; Rosas-Lemus, M.; Sampedro, J.G.; Pena, A.; Mujica, A.; Uribe-Carvajal, S. A glycolytic metabolon in Saccharomyces cerevisiae is stabilized by F-actin. FEBS J. 2013, 280, 3887–3905. [Google Scholar] [CrossRef] [PubMed]
- Sweetlove, L.J.; Fernie, A.R. The role of dynamic enzyme assemblies and substrate channelling in metabolic regulation. Nat. Commun. 2018, 9, 2136. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.Y.-L.; Stork, J.; Pogany, J.; Nagy, P.D. A temperature sensitive mutant of heat shock protein 70 reveals an essential role during the early steps of tombusvirus replication. Virology 2009, 394, 28–38. [Google Scholar] [CrossRef]
- Wang, R.Y.-L.; Stork, J.; Nagy, P.D. A Key Role for Heat Shock Protein 70 in the Localization and Insertion of Tombusvirus Replication Proteins to Intracellular Membranes. J. Virol. 2009, 83, 3276–3287. [Google Scholar] [CrossRef]
- Pogany, J.; Stork, J.; Li, Z.; Nagy, P.D. In vitro assembly of the Tomato bushy stunt virus replicase requires the host Heat shock protein 70. Proc. Natl. Acad. Sci. USA 2008, 105, 19956–19961. [Google Scholar] [CrossRef]
- Barajas, D.; Martin, I.F.; Pogany, J.; Risco, C.; Nagy, P.D. Noncanonical Role for the Host Vps4 AAA+ ATPase ESCRT Protein in the Formation of Tomato Bushy Stunt Virus Replicase. PLoS Pathog. 2014, 10, e1004087. [Google Scholar] [CrossRef]
- Xu, K.; Nagy, P.D. Enrichment of Phosphatidylethanolamine in Viral Replication Compartments via Co-opting the Endosomal Rab5 Small GTPase by a Positive-Strand RNA Virus. PLoS Biol. 2016, 14, e2000128. [Google Scholar] [CrossRef]
- Nawaz-Ul-Rehman, M.S.; Prasanth, K.R.; Xu, K.; Sasvari, Z.; Kovalev, N.; de Castro Martin, I.F.; Barajas, D.; Risco, C.; Nagy, P.D. Viral Replication Protein Inhibits Cellular Cofilin Actin Depolymerization Factor to Regulate the Actin Network and Promote Viral Replicase Assembly. PLoS Pathog. 2016, 12, e1005440. [Google Scholar] [CrossRef]
- Chuang, C.; Prasanth, K.R.; Nagy, P.D. Coordinated Function of Cellular DEAD-Box Helicases in Suppression of Viral RNA Recombination and Maintenance of Viral Genome Integrity. PLoS Pathog. 2015, 11, e1004680. [Google Scholar] [CrossRef]
- Kovalev, N.; Nagy, P.D. The Expanding Functions of Cellular Helicases: The Tombusvirus RNA Replication Enhancer Co-opts the Plant eIF4AIII-Like AtRH2 and the DDX5-Like AtRH5 DEAD-Box RNA Helicases to Promote Viral Asymmetric RNA Replication. PLoS Pathog. 2014, 10, e1004051. [Google Scholar] [CrossRef] [PubMed]
- Kovalev, N.; Pogány, J.; Nagy, P.D. A Co-Opted DEAD-Box RNA Helicase Enhances Tombusvirus Plus-Strand Synthesis. PLoS Pathog. 2012, 8, e1002537. [Google Scholar] [CrossRef] [PubMed]
- Kovalev, N.; Pogany, J.; Nagy, P.D. Template Role of Double-Stranded RNA in Tombusvirus Replication. J. Virol. 2014, 88, 5638–5651. [Google Scholar] [CrossRef] [PubMed]
- Alam, S.B.; Rochon, D. Cucumber necrosis virus recruits cellular heat shock protein 70 homologs at several stages of infection. J. Virol. 2015, 90, 3302–3317. [Google Scholar] [CrossRef] [PubMed]
- Garcin, E.D. GAPDH as a model non-canonical AU-rich RNA binding protein. Semin. Cell Dev. Boil. 2019, 86, 162–173. [Google Scholar] [CrossRef] [PubMed]
- White, M.R.; Khan, M.M.; Deredge, D.; Ross, C.R.; Quintyn, R.; Zucconi, B.E.; Wysocki, V.H.; Wintrode, P.L.; Wilson, G.M.; Garcin, E.D. A dimer interface mutation in glyceraldehyde 3-phosphate dehydrogenase regulates its binding to AU-rich RNA. J. Boil. Chem. 2015, 290, 4129. [Google Scholar] [CrossRef]
- White, M.R.; Garcin, E.D. The sweet side of RNA regulation: Glyceraldehyde-3-phosphate dehydrogenase as a noncanonical RNA-binding protein. Wiley Interdiscip. Rev. RNA 2016, 7, 53–70. [Google Scholar] [CrossRef]
- Beckmann, B.M.; Horos, R.; Fischer, B.; Castelló, A.; Eichelbaum, K.; Alleaume, A.-M.; Schwarzl, T.; Curk, T.; Foehr, S.; Huber, W.; et al. The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nat. Commun. 2015, 6, 10127. [Google Scholar] [CrossRef]
- Huang, T.-S.; Nagy, P.D. Direct Inhibition of Tombusvirus Plus-Strand RNA Synthesis by a Dominant Negative Mutant of a Host Metabolic Enzyme, Glyceraldehyde-3-Phosphate Dehydrogenase, in Yeast and Plants. J. Virol. 2011, 85, 9090–9102. [Google Scholar] [CrossRef][Green Version]
- Wang, R.Y.-L.; Nagy, P.D. Tomato bushy stunt virus Co-Opts the RNA-Binding Function of a Host Metabolic Enzyme for Viral Genomic RNA Synthesis. Cell Host Microbe 2008, 3, 178–187. [Google Scholar] [CrossRef]
- Sanchez, E.L.; Lagunoff, M. Viral activation of cellular metabolism. Virology 2015, 479, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, K.A.; Sanchez, E.L.; Camarda, R.; Lagunoff, M. Dengue virus induces and requires glycolysis for optimal replication. J. Virol. 2015, 89, 2358–2366. [Google Scholar] [CrossRef] [PubMed]
- Delgado, T.; Sanchez, E.L.; Camarda, R.; Lagunoff, M. Global Metabolic Profiling of Infection by an Oncogenic Virus: KSHV Induces and Requires Lipogenesis for Survival of Latent Infection. PLoS Pathog. 2012, 8, 1002866. [Google Scholar] [CrossRef] [PubMed]
- Diamond, D.L.; Syder, A.J.; Jacobs, J.M.; Sorensen, C.M.; Walters, K.-A.; Proll, S.C.; McDermott, J.E.; Gritsenko, M.A.; Zhang, Q.; Zhao, R.; et al. Temporal Proteome and Lipidome Profiles Reveal Hepatitis C Virus-Associated Reprogramming of Hepatocellular Metabolism and Bioenergetics. PLoS Pathog. 2010, 6, e1000719. [Google Scholar] [CrossRef] [PubMed]
- Ramière, C.; Rodriguez, J.; Enache, L.S.; Lotteau, V.; André, P.; Diaz, O. Activity of Hexokinase Is Increased by Its Interaction with Hepatitis C Virus Protein NS5A. J. Virol. 2014, 88, 3246–3254. [Google Scholar] [CrossRef]
- Ripoli, M.; D’Aprile, A.; Quarato, G.; Sarasin-Filipowicz, M.; Gouttenoire, J.; Scrima, R.; Cela, O.; Boffoli, D.; Heim, M.H.; Moradpour, D.; et al. Hepatitis C virus-linked mitochondrial dysfunction promotes hypoxia-inducible factor 1 alpha-mediated glycolytic adaptation. J. Virol. 2010, 84, 647–660. [Google Scholar] [CrossRef]
- Carroll, K.D.; Bu, W.; Palmeri, D.; Spadavecchia, S.; Lynch, S.J.; Marras, S.A.; Tyagi, S.; Lukac, D.M. Kaposi’s Sarcoma-associated herpesvirus lytic switch protein stimulates DNA binding of RBP-Jk/CSL to activate the Notch pathway. J. Virol. 2006, 80, 9697–9709. [Google Scholar] [CrossRef]
- Mazzon, M.; Peters, N.E.; Loenarz, C.; Krysztofinska, E.M.; Ember, S.W.J.; Ferguson, B.J.; Smith, G.L. A mechanism for induction of a hypoxic response by vaccinia virus. Proc. Natl. Acad. Sci. USA 2013, 110, 12444–12449. [Google Scholar] [CrossRef]
- Darekar, S.; Georgiou, K.; Yurchenko, M.; Yenamandra, S.P.; Chachami, G.; Simos, G.; Klein, G.; Kashuba, E. Epstein-Barr Virus Immortalization of Human B-Cells Leads to Stabilization of Hypoxia-Induced Factor 1 Alpha, Congruent with the Warburg Effect. PLoS ONE 2012, 7, e42072. [Google Scholar] [CrossRef]
- Presek, P.; Reinacher, M.; Eigenbrodt, E. Pyruvate kinase type M2 is phosphorylated at tyrosine residues in cells transformed by Rous sarcoma virus. FEBS Lett. 1988, 242, 194–198. [Google Scholar] [CrossRef]
- Teng, C.-F.; Hsieh, W.-C.; Wu, H.-C.; Lin, Y.-J.; Tsai, H.-W.; Huang, W.; Su, I.-J. Hepatitis B Virus Pre-S2 Mutant Induces Aerobic Glycolysis through Mammalian Target of Rapamycin Signal Cascade. PLoS ONE 2015, 10, e0122373. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.-T.; Han, C.; Xu, D.-Q.; Fu, X.-W.; Wang, J.-S.; Kong, L.-Y. 4-Octyl itaconate inhibits aerobic glycolysis by targeting GAPDH to exert anti-inflammatory effects. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nagy, P.D.; Lin, W. Taking over Cellular Energy-Metabolism for TBSV Replication: The High ATP Requirement of an RNA Virus within the Viral Replication Organelle. Viruses 2020, 12, 56. https://doi.org/10.3390/v12010056
Nagy PD, Lin W. Taking over Cellular Energy-Metabolism for TBSV Replication: The High ATP Requirement of an RNA Virus within the Viral Replication Organelle. Viruses. 2020; 12(1):56. https://doi.org/10.3390/v12010056
Chicago/Turabian StyleNagy, Peter D., and Wenwu Lin. 2020. "Taking over Cellular Energy-Metabolism for TBSV Replication: The High ATP Requirement of an RNA Virus within the Viral Replication Organelle" Viruses 12, no. 1: 56. https://doi.org/10.3390/v12010056
APA StyleNagy, P. D., & Lin, W. (2020). Taking over Cellular Energy-Metabolism for TBSV Replication: The High ATP Requirement of an RNA Virus within the Viral Replication Organelle. Viruses, 12(1), 56. https://doi.org/10.3390/v12010056