MYH9 Key Amino Acid Residues Identified by the Anti-Idiotypic Antibody to Porcine Reproductive and Respiratory Syndrome Virus Glycoprotein 5 Involve in the Virus Internalization by Porcine Alveolar Macrophages
Abstract
1. Introduction
2. Material and Methods
2.1. Cells, Viruses, and Chemicals
2.2. Plasmid Construction and Transfection
2.3. Western Blot Analysis
2.4. Dot Blot Assay
2.5. Enzyme-Linked Immunosorbent Assay (ELISA)
2.6. Immunofluorescence Assays (IFAs)
2.7. Cell Viability Assay
2.8. Quantitative Real-Time PCR (qPCR)
2.9. Homology Modeling and Circular Dichroism Spectroscopy
2.10. PRRSV Inhibition Assay
2.11. Evaluation of Virus Attachment and Internalization
2.12. Expression of PRA, PRA Truncation, and PRA Mutants
2.13. Size-Exclusion Chromatography
2.14. Plasma Membrane Isolation
2.15. Generation of PK-15 Cells Bearing Point Mutation in MYH9
2.16. Statistical Analysis
3. Results
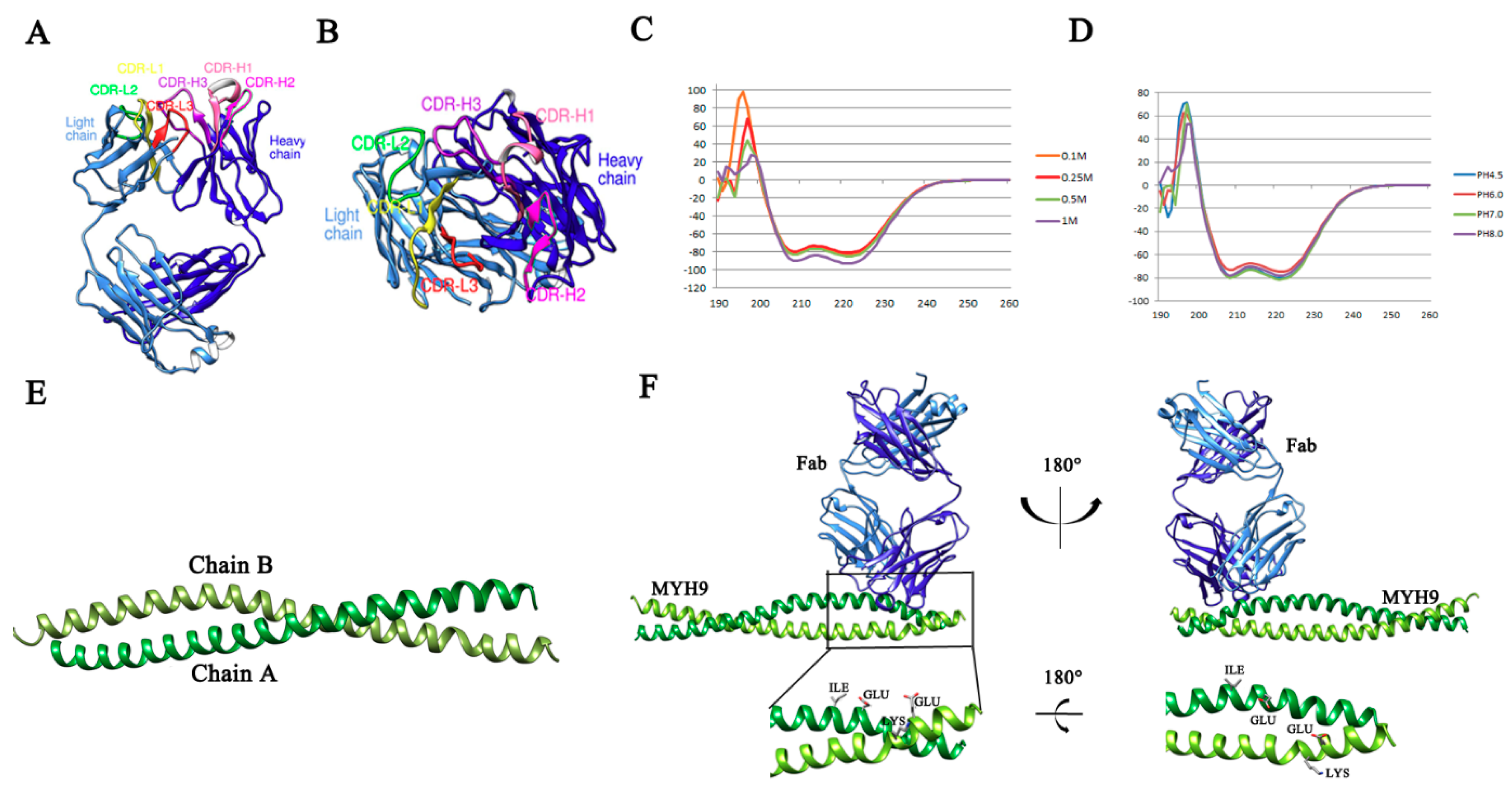
3.1. Mapping of Binding Sites of Mab2-5G2 for MYH9
3.2. Molecular Modeling of the Interaction Region within the Mab2-5G2 Fab-PRA Complex
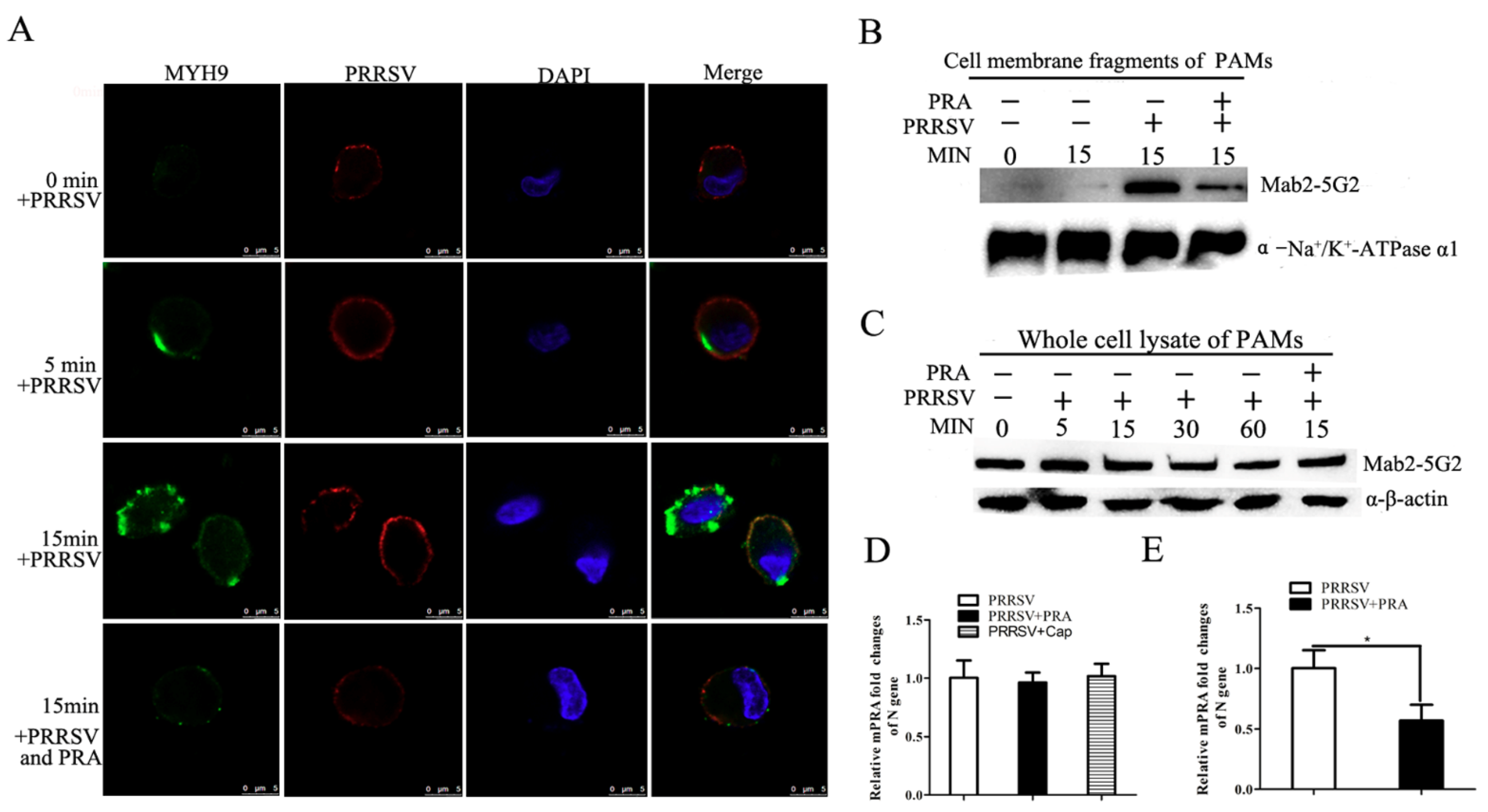
3.3. Mab2-5G2 Blocks PRRSV Internalization by PAMs via Interaction with MYH9
3.4. Interruption of Interaction between PRRSV-GP5 and MYH9 Blocked Internalization of PRRSV to Permissive Cells
3.5. Identification of Key amino acid (aa) Residues Involved in Mab2-5G2 Binding sites within MYH9
3.6. Impaired Binding of Mab2-5G2 to PRA Mutants Correlates with Reduced Inhibition of PRRSV by PRA Mutants
3.7. Porcine Cell Line Bearing Point Mutation in E1670A in MYH9 Demonstrated Reduced Susceptibility for PRRSV Infection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kuhn, J.H.; Lauck, M.; Bailey, A.L.; Shchetinin, A.M.; Vishnevskaya, T.V.; Bao, Y.; Ng, T.F.; LeBreton, M.; Schneider, B.S.; Gillis, A.; et al. Reorganization and expansion of the nidoviral family Arteriviridae. Arch. Virol. 2016, 161, 755–768. [Google Scholar] [CrossRef]
- Adams, M.J.; Lefkowitz, E.J.; King, A.M.; Harrach, B.; Harrison, R.L.; Knowles, N.J.; Kropinski, A.M.; Krupovic, M.; Kuhn, J.H.; Mushegian, A.R.; et al. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2016). Arch. Virol. 2016, 161, 2921–2949. [Google Scholar] [CrossRef]
- Forsberg, R. Divergence time of porcine reproductive and respiratory syndrome virus subtypes. Mol. Biol. Evol. 2005, 22, 2131–2134. [Google Scholar] [CrossRef]
- Van Woensel, P.A.; Liefkens, K.; Demaret, S. Effect on viraemia of an American and a European serotype PRRSV vaccine after challenge with European wild-type strains of the virus. Vet. Rec. 1998, 142, 510–512. [Google Scholar] [CrossRef] [PubMed]
- Morgan, S.B.; Frossard, J.P.; Pallares, F.J.; Gough, J.; Stadejek, T.; Graham, S.P.; Steinbach, F.; Drew, T.W.; Salguero, F.J. Pathology and Virus Distribution in the Lung and Lymphoid Tissues of Pigs Experimentally Inoculated with Three Distinct Type 1 PRRS Virus Isolates of Varying Pathogenicity. Transbound. Emerg. Dis. 2016, 63, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Albina, E.; Carrat, C.; Charley, B. Interferon-alpha response to swine arterivirus (PoAV), the porcine reproductive and respiratory syndrome virus. J. Interferon Cytokine Res. 1998, 18, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Liu, Y.; Ding, Y.; Zhang, Y.; Zhang, J. PRRSV receptors and their roles in virus infection. Arch. Microbiol. 2015, 197, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Delputte, P.L.; Vanderheijden, N.; Nauwynck, H.J.; Pensaert, M.B. Involvement of the matrix protein in attachment of porcine reproductive and respiratory syndrome virus to a heparinlike receptor on porcine alveolar macrophages. J. Virol. 2002, 76, 4312–4320. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Fahad, A.M.; Shanmukhappa, K.; Kapil, S. Defining the cellular target(s) of porcine reproductive and respiratory syndrome virus blocking monoclonal antibody 7G10. J. Virol. 2006, 80, 689–696. [Google Scholar] [CrossRef]
- Wu, J.; Peng, X.; Zhou, A.; Qiao, M.; Wu, H.; Xiao, H.; Liu, G.; Zheng, X.; Zhang, S.; Mei, S. MiR-506 inhibits PRRSV replication in MARC-145 cells via CD151. Mol. Cell Biochem. 2014, 394, 275–281. [Google Scholar] [CrossRef]
- Guo, L.; Niu, J.; Yu, H.; Gu, W.; Li, R.; Luo, X.; Huang, M.; Tian, Z.; Feng, L.; Wang, Y. Modulation of CD163 expression by metalloprotease ADAM17 regulates porcine reproductive and respiratory syndrome virus entry. J. Virol. 2014, 88, 10448–10458. [Google Scholar] [CrossRef] [PubMed]
- Delputte, P.L.; Van Breedam, W.; Delrue, I.; Oetke, C.; Crocker, P.R.; Nauwynck, H.J. Porcine arterivirus attachment to the macrophage-specific receptor sialoadhesin is dependent on the sialic acid-binding activity of the N-terminal immunoglobulin domain of sialoadhesin. J. Virol. 2007, 81, 9546–9550. [Google Scholar] [CrossRef] [PubMed]
- Pineyro, P.E.; Subramaniam, S.; Kenney, S.P.; Heffron, C.L.; Gimenez-Lirola, L.G.; Meng, X.J. Modulation of Proinflammatory Cytokines in Monocyte-Derived Dendritic Cells by Porcine Reproductive and Respiratory Syndrome Virus Through Interaction with the Porcine Intercellular-Adhesion-Molecule-3-Grabbing Nonintegrin. Viral Immunol. 2016, 29, 546–556. [Google Scholar] [CrossRef] [PubMed]
- Hou, G.; Xue, B.; Li, L.; Nan, Y.; Zhang, L.; Li, K.; Zhao, Q.; Hiscox, J.A.; Stewart, J.P.; Wu, C.; et al. Direct Interaction Between CD163 N-Terminal Domain and MYH9 C-Terminal Domain Contributes to Porcine Reproductive and Respiratory Syndrome Virus Internalization by Permissive Cells. Front. Microbiol. 2019, 10, 1815. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Hou, G.; Zhang, G.; Huang, J.; Li, L.; Nan, Y.; Mu, Y.; Wang, L.; Zhang, L.; Han, X.; et al. MYH9 Aggregation Induced by Direct Interaction With PRRSV GP5 Ectodomain Facilitates Viral Internalization by Permissive Cells. Front. Microbiol. 2019, 10, 2313. [Google Scholar] [CrossRef]
- Sellers, J.R.; Umemoto, S. Effect of multiple phosphorylations on movement of smooth muscle and cytoplasmic myosin. Adv. Exp. Med. Biol. 1989, 255, 299–304. [Google Scholar]
- Shutova, M.S.; Spessott, W.A.; Giraudo, C.G.; Svitkina, T. Endogenous species of mammalian nonmuscle myosin IIA and IIB include activated monomers and heteropolymers. Curr. Biol. 2014, 24, 1958–1968. [Google Scholar] [CrossRef]
- Cureton, D.K.; Massol, R.H.; Saffarian, S.; Kirchhausen, T.L.; Whelan, S.P. Vesicular stomatitis virus enters cells through vesicles incompletely coated with clathrin that depend upon actin for internalization. PLoS Pathog. 2009, 5, e1000394. [Google Scholar] [CrossRef]
- Cureton, D.K.; Massol, R.H.; Whelan, S.P.; Kirchhausen, T. The length of vesicular stomatitis virus particles dictates a need for actin assembly during clathrin-dependent endocytosis. PLoS Pathog. 2010, 6, e1001127. [Google Scholar] [CrossRef]
- Piccinotti, S.; Kirchhausen, T.; Whelan, S.P. Uptake of rabies virus into epithelial cells by clathrin-mediated endocytosis depends upon actin. J. Virol. 2013, 87, 11637–11647. [Google Scholar] [CrossRef]
- Gao, J.; Xiao, S.; Xiao, Y.; Wang, X.; Zhang, C.; Zhao, Q.; Nan, Y.; Huang, B.; Liu, H.; Liu, N.; et al. MYH9 is an Essential Factor for Porcine Reproductive and Respiratory Syndrome Virus Infection. Sci. Rep. 2016, 6, 25120. [Google Scholar] [CrossRef] [PubMed]
- Zhou, E.M.; Xiao, Y.; Shi, Y.; Li, X.; Ma, L.; Jing, S.; Peng, J. Generation of internal image monoclonal anti-idiotypic antibodies against idiotypic antibodies to GP5 antigen of porcine reproductive and respiratory syndrome virus. J. Virol. Methods 2008, 149, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Jerne, N.K. Towards a network theory of the immune system. Ann. Immunol. 1974, 125, 373–389. [Google Scholar]
- Gaulton, G.N.; Sharpe, A.H.; Chang, D.W.; Fields, B.N.; Greene, M.I. Syngeneic monoclonal internal image anti-idiotopes as prophylactic vaccines. J. Immunol. 1986, 137, 2930–2936. [Google Scholar]
- Durrant, L.G.; Parsons, T.; Moss, R.; Spendlove, I.; Carter, G.; Carr, F. Human anti-idiotypic antibodies can be good immunogens as they target FC receptors on antigen-presenting cells allowing efficient stimulation of both helper and cytotoxic T-cell responses. Int. J. Cancer 2001, 92, 414–420. [Google Scholar] [CrossRef]
- Herlyn, D.; Somasundaram, R.; Li, W.; Maruyama, H. Anti-idiotype cancer vaccines: Past and future. Cancer Immunol. Immunother. 1996, 43, 65–76. [Google Scholar] [CrossRef]
- Uytdehaag, F.G.; Osterhaus, A.D. Induction of neutralizing antibody in mice against poliovirus type II with monoclonal anti-idiotypic antibody. J. Immunol. 1985, 134, 1225–1229. [Google Scholar]
- Co, M.S.; Gaulton, G.N.; Fields, B.N.; Greene, M.I. Isolation and biochemical characterization of the mammalian reovirus type 3 cell-surface receptor. Proc. Natl. Acad. Sci. USA 1985, 82, 1494–1498. [Google Scholar] [CrossRef]
- Fingeroth, J.D.; Weis, J.J.; Tedder, T.F.; Strominger, J.L.; Biro, P.A.; Fearon, D.T. Epstein-Barr virus receptor of human B lymphocytes is the C3d receptor CR2. Proc. Natl. Acad. Sci. USA 1984, 81, 4510–4514. [Google Scholar] [CrossRef]
- Greve, J.M.; Davis, G.; Meyer, A.M.; Forte, C.P.; Yost, S.C.; Marlor, C.W.; Kamarck, M.E.; McClelland, A. The major human rhinovirus receptor is ICAM-1. Cell 1989, 56, 839–847. [Google Scholar] [CrossRef]
- Arii, J.; Goto, H.; Suenaga, T.; Oyama, M.; Kozuka-Hata, H.; Imai, T.; Minowa, A.; Akashi, H.; Arase, H.; Kawaoka, Y.; et al. Non-muscle myosin IIA is a functional entry receptor for herpes simplex virus-1. Nature 2010, 467, 859–862. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Qi, Y.; Liu, C.; Gao, W.; Chen, P.; Fu, L.; Peng, B.; Wang, H.; Jing, Z.; Zhong, G.; et al. Nonmuscle myosin heavy chain IIA is a critical factor contributing to the efficiency of early infection of severe fever with thrombocytopenia syndrome virus. J. Virol. 2014, 88, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Xiong, D.; Du, Y.; Wang, H.B.; Zhao, B.; Zhang, H.; Li, Y.; Hu, L.J.; Cao, J.Y.; Zhong, Q.; Liu, W.L.; et al. Nonmuscle myosin heavy chain IIA mediates Epstein-Barr virus infection of nasopharyngeal epithelial cells. Proc. Natl. Acad. Sci. USA 2015, 112, 11036–11041. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Zhang, A.; Zhang, C.; Ni, H.; Gao, J.; Wang, C.; Zhao, Q.; Wang, X.; Ma, C.; Liu, H.; et al. Heme oxygenase-1 acts as an antiviral factor for porcine reproductive and respiratory syndrome virus infection and over-expression inhibits virus replication in vitro. Antivir. Res. 2014, 110, 60–69. [Google Scholar] [CrossRef]
- Li, L.; Xue, B.; Sun, W.; Gu, G.; Hou, G.; Zhang, L.; Wu, C.; Zhao, Q.; Zhang, Y.; Zhang, G.; et al. Recombinant MYH9 protein C-terminal domain blocks porcine reproductive and respiratory syndrome virus internalization by direct interaction with viral glycoprotein 5. Antivir. Res. 2018, 156, 10–20. [Google Scholar] [CrossRef]
- Zhang, A.; Zhao, L.; Li, N.; Duan, H.; Liu, H.; Pu, F.; Zhang, G.; Zhou, E.M.; Xiao, S. Carbon Monoxide Inhibits Porcine Reproductive and Respiratory Syndrome Virus Replication by the Cyclic GMP/Protein Kinase G and NF-kappaB Signaling Pathway. J. Virol. 2017, 91, e01866–16. [Google Scholar] [CrossRef]
- Mu, Y.; Li, L.; Zhang, B.; Huang, B.; Gao, J.; Wang, X.; Wang, C.; Xiao, S.; Zhao, Q.; Sun, Y.; et al. Glycoprotein 5 of porcine reproductive and respiratory syndrome virus strain SD16 inhibits viral replication and causes G2/M cell cycle arrest, but does not induce cellular apoptosis in Marc-145 cells. Virology 2015, 484, 136–145. [Google Scholar] [CrossRef]
- Wu, X.; Wu, D.; Lu, Z.; Chen, W.; Hu, X.; Ding, Y. A novel method for high-level production of TEV protease by superfolder GFP tag. J. Biomed. Biotechnol. 2009, 2009, 591923. [Google Scholar] [CrossRef]
- Wang, X.; Wei, R.; Li, Q.; Liu, H.; Huang, B.; Gao, J.; Mu, Y.; Wang, C.; Hsu, W.H.; Hiscox, J.A.; et al. PK-15 cells transfected with porcine CD163 by PiggyBac transposon system are susceptible to porcine reproductive and respiratory syndrome virus. J. Virol Methods 2013, 193, 383–390. [Google Scholar] [CrossRef]
- Fang, Z.; Takizawa, N.; Wilson, K.A.; Smith, T.C.; Delprato, A.; Davidson, M.W.; Lambright, D.G.; Luna, E.J. The membrane-associated protein, supervillin, accelerates F-actin-dependent rapid integrin recycling and cell motility. Traffic 2010, 11, 782–799. [Google Scholar] [CrossRef]
- Guo, R.; Katz, B.B.; Tomich, J.M.; Gallagher, T.; Fang, Y. Porcine Reproductive and Respiratory Syndrome Virus Utilizes Nanotubes for Intercellular Spread. J. Virol. 2016, 90, 5163–5175. [Google Scholar] [CrossRef] [PubMed]
- Vicente-Manzanares, M.; Ma, X.; Adelstein, R.S.; Horwitz, A.R. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. 2009, 10, 778–790. [Google Scholar] [CrossRef] [PubMed]
- Li, L. Northwest A&F University, Yangling, China. Inhibition assay for Mab2-5G2 against the PRRSV-1 GZ11 strain in PAMs. Unpublished work. 2018. [Google Scholar]
- Ma, H.; Jiang, L.; Qiao, S.; Zhi, Y.; Chen, X.X.; Yang, Y.; Huang, X.; Huang, M.; Li, R.; Zhang, G.P. The Crystal Structure of the Fifth Scavenger Receptor Cysteine-Rich Domain of Porcine CD163 Reveals an Important Residue Involved in Porcine Reproductive and Respiratory Syndrome Virus Infection. J. Virol. 2017, 91, e01897-16. [Google Scholar] [CrossRef] [PubMed]
- Whitworth, K.M.; Rowland, R.R.; Ewen, C.L.; Trible, B.R.; Kerrigan, M.A.; Cino-Ozuna, A.G.; Samuel, M.S.; Lightner, J.E.; McLaren, D.G.; Mileham, A.J.; et al. Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nat. Biotechnol. 2016, 34, 20–22. [Google Scholar] [CrossRef] [PubMed]
- Van Gorp, H.; Van Breedam, W.; Van Doorsselaere, J.; Delputte, P.L.; Nauwynck, H.J. Identification of the CD163 protein domains involved in infection of the porcine reproductive and respiratory syndrome virus. J. Virol. 2010, 84, 3101–3105. [Google Scholar] [CrossRef]
- Burkard, C.; Lillico, S.G.; Reid, E.; Jackson, B.; Mileham, A.J.; Ait-Ali, T.; Whitelaw, C.B.; Archibald, A.L. Precision engineering for PRRSV resistance in pigs: Macrophages from genome edited pigs lacking CD163 SRCR5 domain are fully resistant to both PRRSV genotypes while maintaining biological function. PLoS Pathog. 2017, 13, e1006206. [Google Scholar] [CrossRef] [PubMed]
- Wells, K.D.; Bardot, R.; Whitworth, K.M.; Trible, B.R.; Fang, Y.; Mileham, A.; Kerrigan, M.A.; Samuel, M.S.; Prather, R.S.; Rowland, R.R. Replacement of Porcine CD163 Scavenger Receptor Cysteine-Rich Domain 5 with a CD163-Like Homolog Confers Resistance of Pigs to Genotype 1 but Not Genotype 2 Porcine Reproductive and Respiratory Syndrome Virus. J. Virol. 2017, 91, e01521-16. [Google Scholar] [CrossRef]
- Zhang, Q.; Yoo, D. PRRS virus receptors and their role for pathogenesis. Vet. Microbiol. 2015, 177, 229–241. [Google Scholar] [CrossRef]
- Yoon, K.J.; Wu, L.L.; Zimmerman, J.J.; Hill, H.T.; Platt, K.B. Antibody-dependent enhancement (ADE) of porcine reproductive and respiratory syndrome virus (PRRSV) infection in pigs. Viral Immunol. 1996, 9, 51–63. [Google Scholar] [CrossRef]
- Yoon, K.J.; Wu, L.L.; Zimmerman, J.J.; Platt, K.B. Field isolates of porcine reproductive and respiratory syndrome virus (PRRSV) vary in their susceptibility to antibody dependent enhancement (ADE) of infection. Vet. Microbiol. 1997, 55, 277–287. [Google Scholar] [CrossRef]
- Wan, B.; Chen, X.; Li, Y.; Pang, M.; Chen, H.; Nie, X.; Pan, Y.; Qiao, S.; Bao, D. Porcine FcgammaRIIb mediated PRRSV ADE infection through inhibiting IFN-beta by cytoplasmic inhibitory signal transduction. Int. J. Biol. Macromol. 2019, 138, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhou, Y.; Yang, Q.; Mu, C.; Duan, E.; Chen, J.; Yang, M.; Xia, P.; Cui, B. Ligation of Fc gamma receptor IIB enhances levels of antiviral cytokine in response to PRRSV infection in vitro. Vet. Microbiol. 2012, 160, 473–480. [Google Scholar] [CrossRef] [PubMed]




| Genes Name | (Sense 5′–3′) |
|---|---|
| PRA-F | ACGCGTCGACATGCGGGAGCTGGAG |
| PRA-R | CGGGATCCTCATTGTACTTCAGAGT |
| PRA1651-1716F | ATGTCGACATGCGGGAGCTGGAGGACAC |
| PRA1651-1716-R | ATTCTAGATTTGCCGCTGCTGTTGGCAA |
| PRA1714-1960-F | CGGGATCCATGGGCAAAGGGGCG |
| PRA1714-1960-R | ATTCTAGATTATTCGGCAGGTTT |
| PRA1670-F | GCACAGGCCAAGGCAAACGAGAAGAAGCTGAAAAGCATGGAGGCC |
| PRA1670-R | CTTCTTCTCGTTTGCCTTGGCCTGTGCCAGGATCTCCTCGCGGGA |
| PRA1673-F | AAGGAGAACGAGGCAAAGCTGAAAAGCATGGAGGCCGAGATGATC |
| PRA1673-R | GCTTTTCAGCTTTGCCTCGTTCTCCTTGGCCTGTGCCAGGATCTC |
| PRA1679-F | CTGAAAAGCATGGCAGCCGAGATGATCCAGCTGCAGGAGGAGCTG |
| PRA1679-R | GATCATCTCGGCTGCCATGCTTTTCAGCTTCTTCTCGTTCTCCTT |
| PRA1683-F | GAGGCCGAGATGGCACAGCTGCAGGAGGAGCTGGCCGCCGCCGA |
| PRA1683-R | CTCCTGCAGCTGTGCCATCTCGGCCTCCATGCTTTTCAGCTTCTT |
| ORF-7-F | ATGCCAAATAACAACGGCAAGCAGC |
| ORF-7-R | TCATGCTGAGGGTGATGCTGTG |
| GAPDH-F | CCTTCCGTGTCCCTACTGCCAAC |
| GAPDH-R | GACGCCTGCTTCACCACCTTCT |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Zhang, L.; Hu, Q.; Zhao, L.; Nan, Y.; Hou, G.; Chen, Y.; Han, X.; Ren, X.; Zhao, Q.; et al. MYH9 Key Amino Acid Residues Identified by the Anti-Idiotypic Antibody to Porcine Reproductive and Respiratory Syndrome Virus Glycoprotein 5 Involve in the Virus Internalization by Porcine Alveolar Macrophages. Viruses 2020, 12, 40. https://doi.org/10.3390/v12010040
Li L, Zhang L, Hu Q, Zhao L, Nan Y, Hou G, Chen Y, Han X, Ren X, Zhao Q, et al. MYH9 Key Amino Acid Residues Identified by the Anti-Idiotypic Antibody to Porcine Reproductive and Respiratory Syndrome Virus Glycoprotein 5 Involve in the Virus Internalization by Porcine Alveolar Macrophages. Viruses. 2020; 12(1):40. https://doi.org/10.3390/v12010040
Chicago/Turabian StyleLi, Liangliang, Lu Zhang, Qifan Hu, Liang Zhao, Yuchen Nan, Gaopeng Hou, Yiyang Chen, Ximeng Han, Xiaolei Ren, Qin Zhao, and et al. 2020. "MYH9 Key Amino Acid Residues Identified by the Anti-Idiotypic Antibody to Porcine Reproductive and Respiratory Syndrome Virus Glycoprotein 5 Involve in the Virus Internalization by Porcine Alveolar Macrophages" Viruses 12, no. 1: 40. https://doi.org/10.3390/v12010040
APA StyleLi, L., Zhang, L., Hu, Q., Zhao, L., Nan, Y., Hou, G., Chen, Y., Han, X., Ren, X., Zhao, Q., Tao, H., Sun, Z., Zhang, G., Wu, C., Wang, J., & Zhou, E.-M. (2020). MYH9 Key Amino Acid Residues Identified by the Anti-Idiotypic Antibody to Porcine Reproductive and Respiratory Syndrome Virus Glycoprotein 5 Involve in the Virus Internalization by Porcine Alveolar Macrophages. Viruses, 12(1), 40. https://doi.org/10.3390/v12010040






