Conserved Residue Asn-145 in the C-Terminal Heptad Repeat Region of HIV-1 gp41 is Critical for Viral Fusion and Regulates the Antiviral Activity of Fusion Inhibitors
Abstract
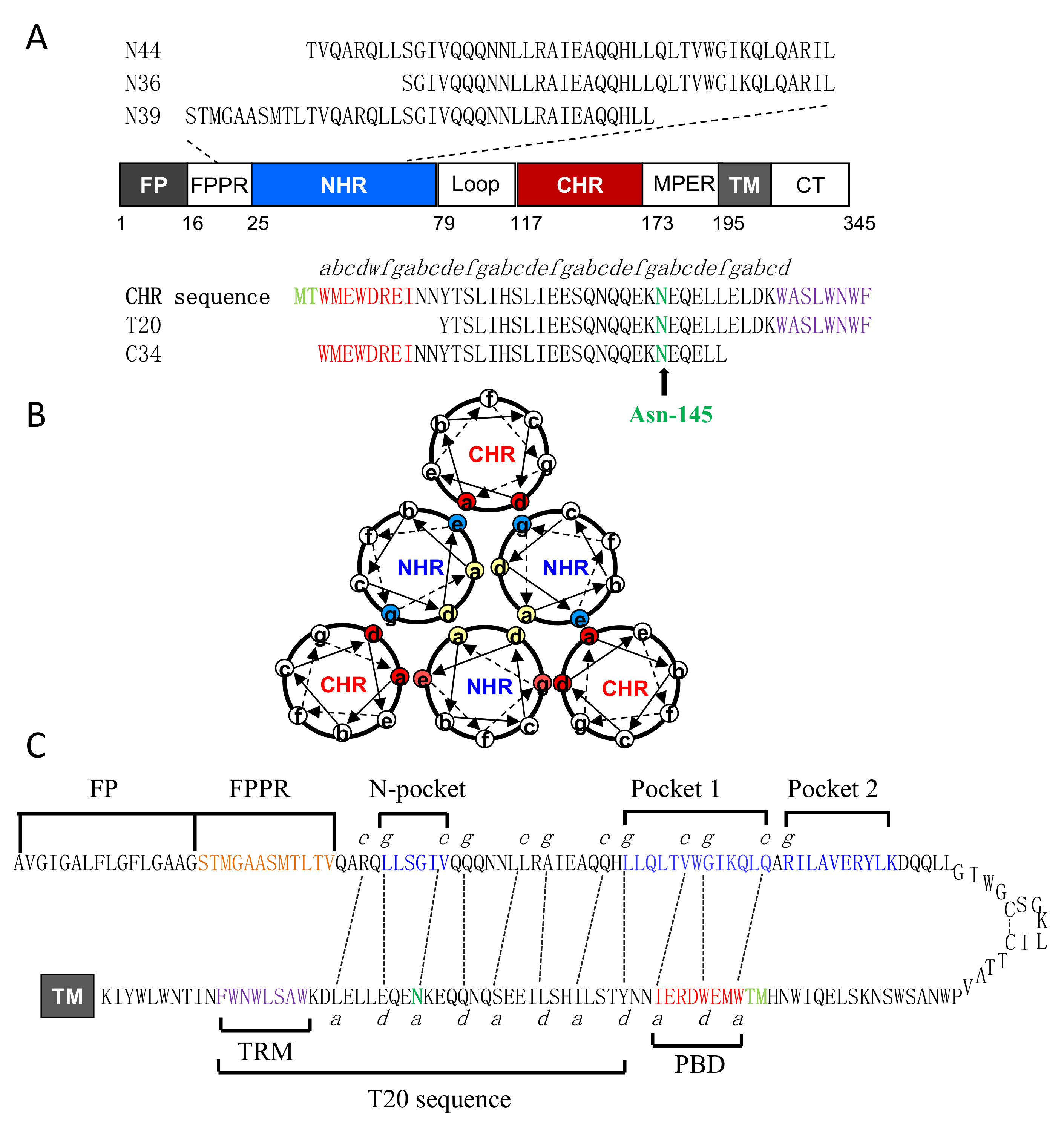
1. Introduction
2. Materials and Methods
2.1. Plasmids and Cell Lines
2.2. Peptide Synthesis
2.3. Site-Directed Mutagenesis
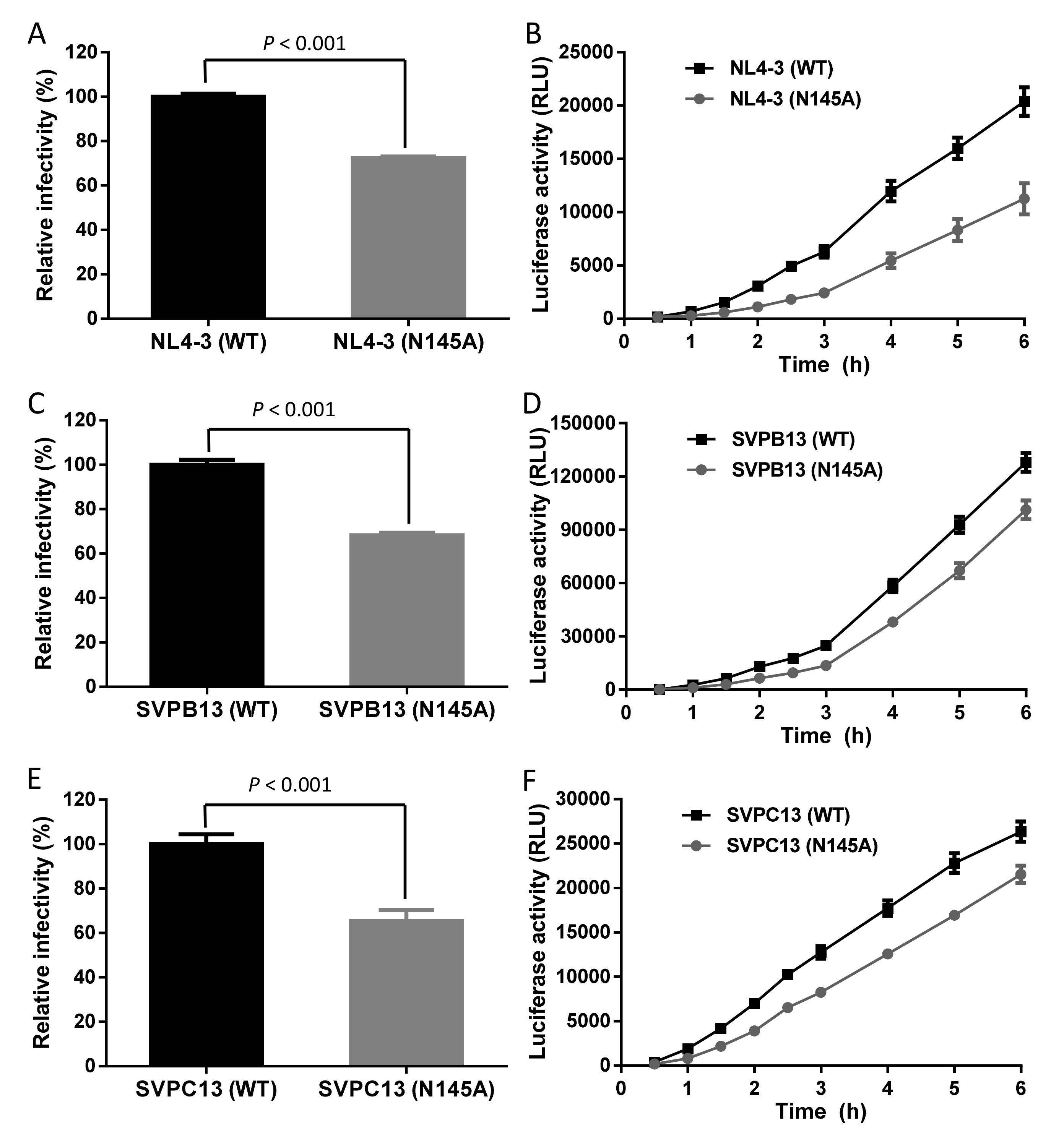
2.4. Single-Cycle Infection Assay
2.5. Cell-Cell Fusion Assay
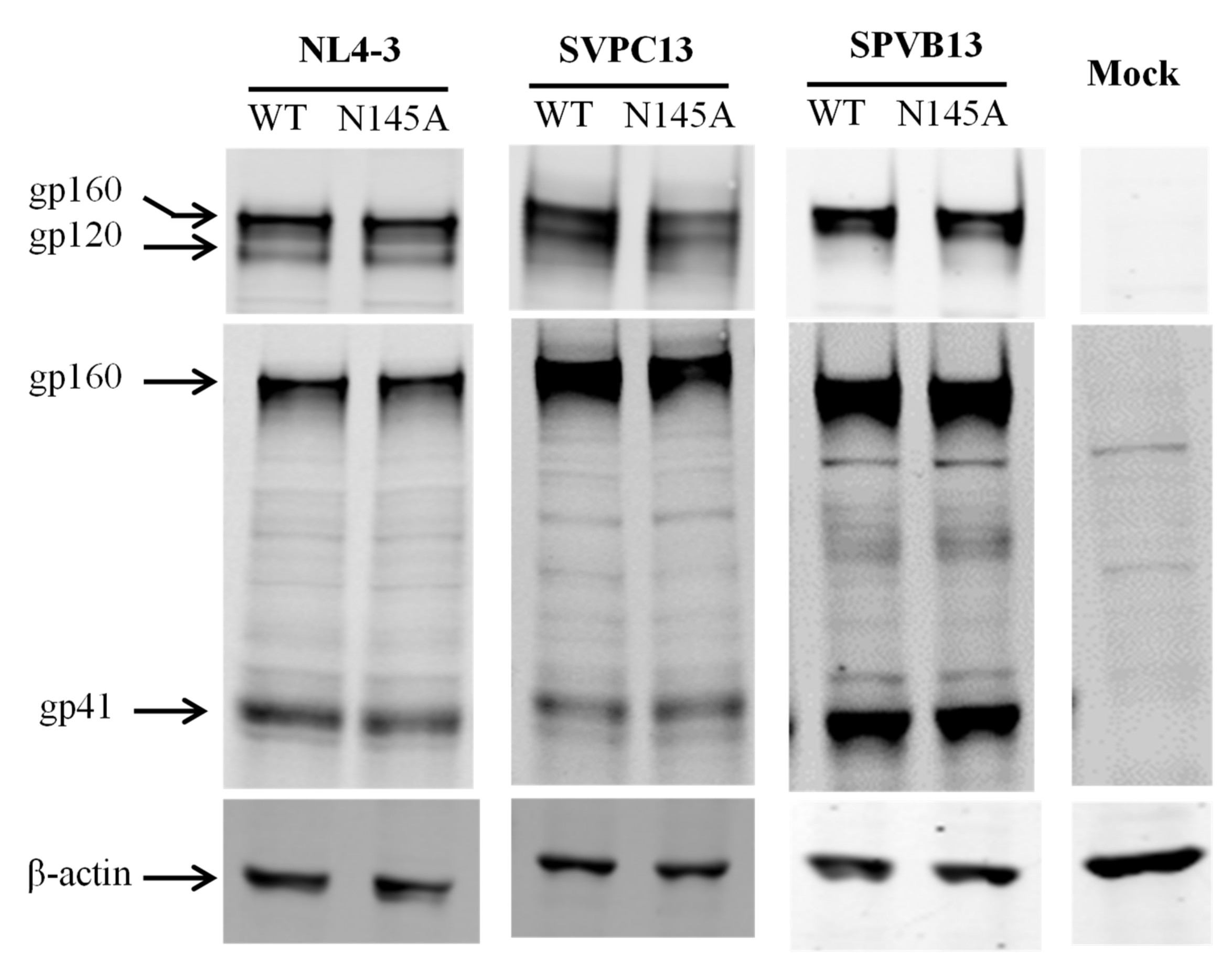
2.6. Western Blotting Assay
2.7. Circular Dichroism (CD) Spectroscopy
2.8. Isothermal Titration Calorimetry (ITC) Experiment
2.9. Crystallization and Structure Determination
3. Results
3.1. Asn-145 is Critical for HIV-1 Env-Mediated Cell Fusion
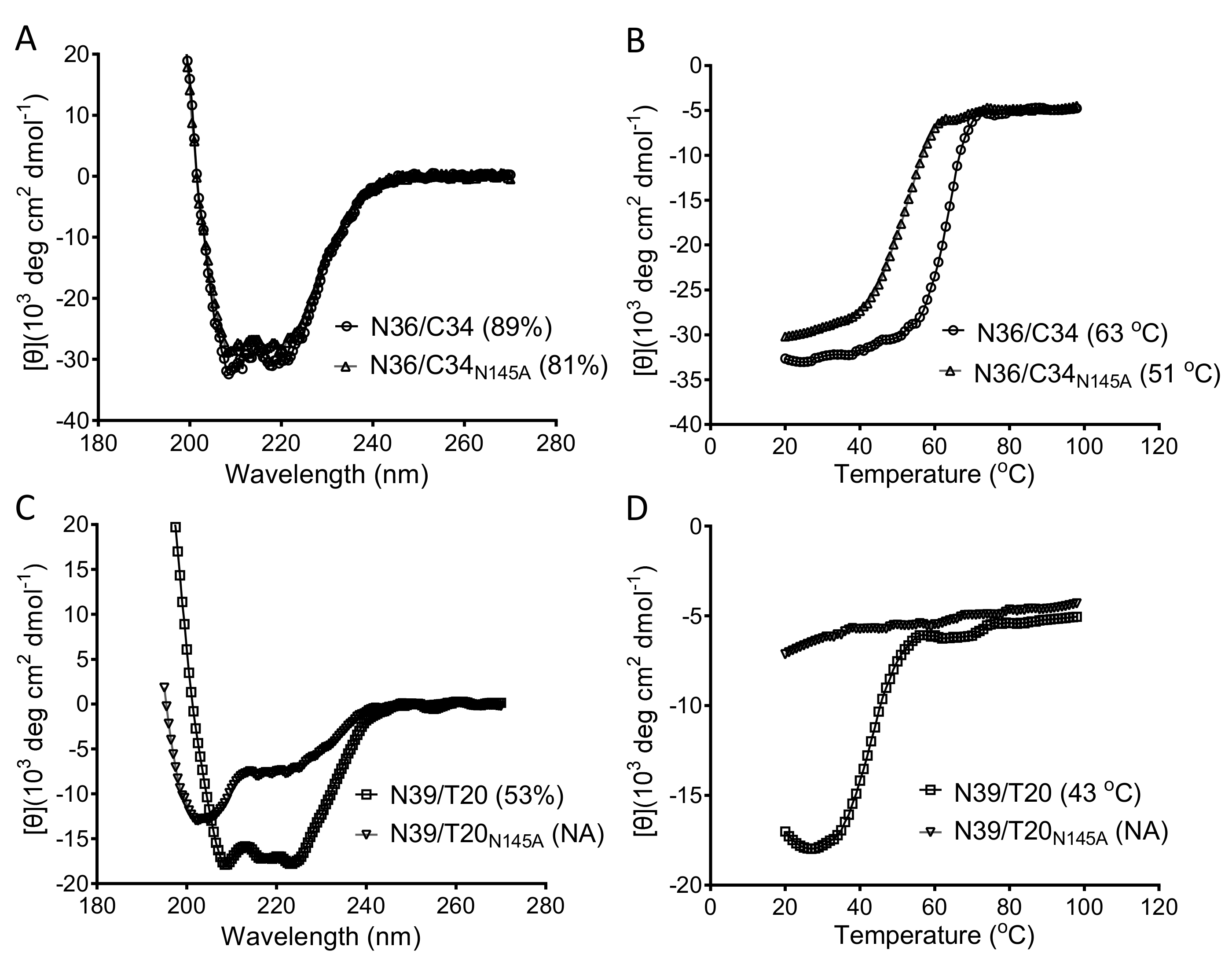
3.2. Asn-145 is Critical for the Interactions of the N- and C- Terminal Heptad Repeats (NHR and CHR) Helices
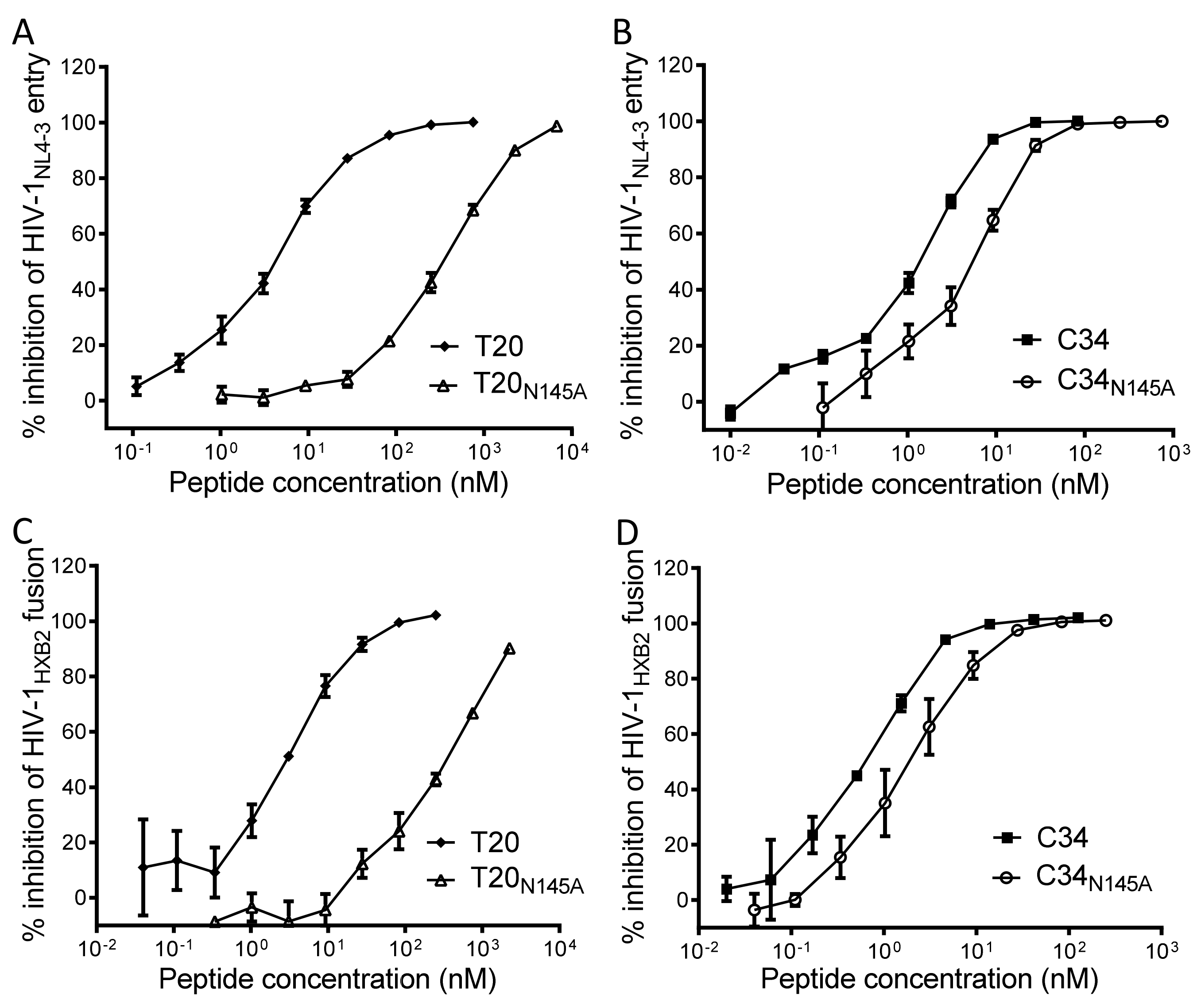
3.3. Asn-145 Determines the Antiviral Activity of Fusion Inhibitors
3.4. Asn-145 Serves as a Turning-Point for the Anti-HIV Activity of Helical Peptide Inhibitors
3.5. Asn-145 Regulates the Antiviral Function of the M-T Hook Structure
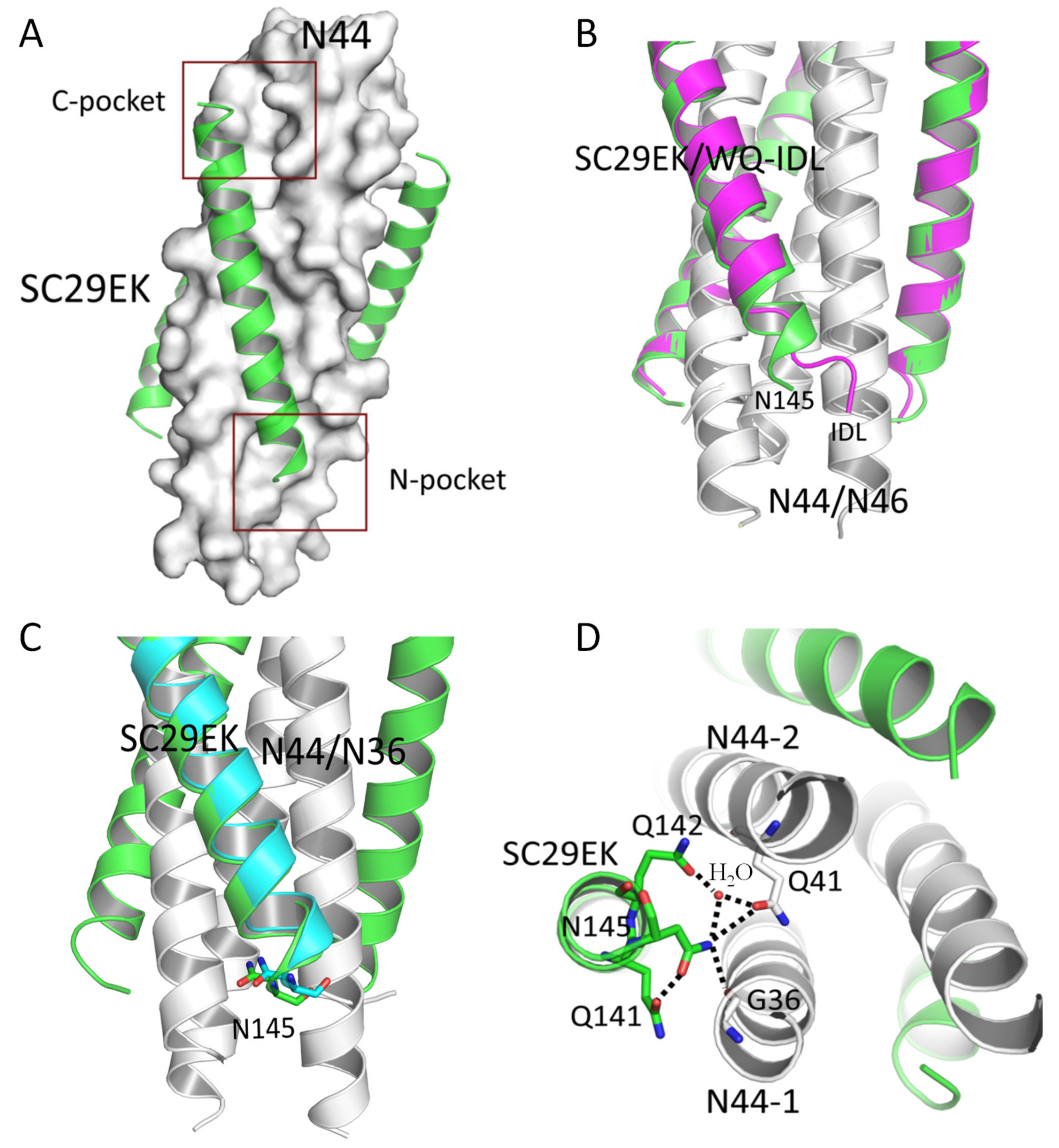
3.6. Substitution of an IDL Anchor for Asn-145 Slightly Improves the Binding But Not the Inhibitory Activities
3.7. Structural Basis for the Functionality of Asn-145 in SC29EK
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Eckert, D.M.; Kim, P.S. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 2001, 70, 777–810. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.C.; Kim, P.S. HIV entry and its inhibition. Cell 1998, 93, 681–684. [Google Scholar] [CrossRef]
- Weissenhorn, W.; Dessen, A.; Harrison, S.C.; Skehel, J.J.; Wiley, D.C. Atomic structure of the ectodomain from HIV-1 gp41. Nature 1997, 387, 426–430. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.C.; Fass, D.; Berger, J.M.; Kim, P.S. Core structure of gp41 from the HIV envelope glycoprotein. Cell 1997, 89, 263–273. [Google Scholar] [CrossRef]
- He, Y. Synthesized peptide inhibitors of HIV-1 gp41-dependent membrane fusion. Curr. Pharm. Des. 2013, 19, 1800–1809. [Google Scholar] [CrossRef] [PubMed]
- Matthews, T.; Salgo, M.; Greenberg, M.; Chung, J.; DeMasi, R.; Bolognesi, D. Enfuvirtide: The first therapy to inhibit the entry of HIV-1 into host cd4 lymphocytes. Nat. Rev. Drug Discov. 2004, 3, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Lalezari, J.P.; Henry, K.; O’Hearn, M.; Montaner, J.S.; Piliero, P.J.; Trottier, B.; Walmsley, S.; Cohen, C.; Kuritzkes, D.R.; Eron, J.J., Jr.; et al. Enfuvirtide, an HIV-1 fusion inhibitor, for drug-resistant HIV infection in north and south america. N. Engl. J. Med. 2003, 348, 2175–2185. [Google Scholar] [CrossRef] [PubMed]
- Colman, P.M.; Lawrence, M.C. The structural biology of type i viral membrane fusion. Nat. Rev. Mol. Cell Biol. 2003, 4, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Caffrey, M.; Cai, M.; Kaufman, J.; Stahl, S.J.; Wingfield, P.T.; Covell, D.G.; Gronenborn, A.M.; Clore, G.M. Three-dimensional solution structure of the 44 kda ectodomain of siv gp41. EMBO J. 1998, 17, 4572–4584. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.; Liu, J.; Wang, J.; Shen, S.; Lu, M. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl. Acad. Sci. USA 1997, 94, 12303–12308. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Kim, P.S. A trimeric structural subdomain of the HIV-1 transmembrane glycoprotein. J. Biomol. Struct. Dyn. 1997, 15, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Sivaraman, V.; Zhang, L.; Meissner, E.G.; Jeffrey, J.L.; Su, L. The heptad repeat 2 domain is a major determinant for enhanced human immunodeficiency virus type 1 (HIV-1) fusion and pathogenicity of a highly pathogenic HIV-1 env. J. Virol. 2009, 83, 11715–11725. [Google Scholar] [CrossRef] [PubMed]
- Ray, N.; Blackburn, L.A.; Doms, R.W. Hr-2 mutations in human immunodeficiency virus type 1 gp41 restore fusion kinetics delayed by hr-1 mutations that cause clinical resistance to enfuvirtide. J. Virol. 2009, 83, 2989–2995. [Google Scholar] [CrossRef] [PubMed]
- Buzon, V.; Natrajan, G.; Schibli, D.; Campelo, F.; Kozlov, M.M.; Weissenhorn, W. Crystal structure of HIV-1 gp41 including both fusion peptide and membrane proximal external regions. PLoS Pathog. 2010, 6, e1000880. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.C.; Chutkowski, C.T.; Kim, P.S. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc. Natl. Acad. Sci. USA 1998, 95, 15613–15617. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Liu, S.; Li, J.; Lu, H.; Qi, Z.; Liu, Z.; Debnath, A.K.; Jiang, S. Conserved salt bridge between the n- and c-terminal heptad repeat regions of the human immunodeficiency virus type 1 gp41 core structure is critical for virus entry and inhibition. J. Virol. 2008, 82, 11129–11139. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Liu, S.; Jing, W.; Lu, H.; Cai, D.; Chin, D.J.; Debnath, A.K.; Kirchhoff, F.; Jiang, S. Conserved residue lys574 in the cavity of HIV-1 gp41 coiled-coil domain is critical for six-helix bundle stability and virus entry. J. Biol. Chem. 2007, 282, 25631–25639. [Google Scholar] [CrossRef]
- Chong, H.; Yao, X.; Sun, J.; Qiu, Z.; Zhang, M.; Waltersperger, S.; Wang, M.; Cui, S.; He, Y. The m-t hook structure is critical for design of HIV-1 fusion inhibitors. J. Biol. Chem. 2012, 287, 34558–34568. [Google Scholar] [CrossRef]
- Chong, H.; Yao, X.; Qiu, Z.; Qin, B.; Han, R.; Waltersperger, S.; Wang, M.; Cui, S.; He, Y. Discovery of critical residues for viral entry and inhibition through structural insight of HIV-1 fusion inhibitor cp621-652. J. Biol. Chem. 2012, 287, 20281–20289. [Google Scholar] [CrossRef]
- He, Y.; Cheng, J.; Li, J.; Qi, Z.; Lu, H.; Dong, M.; Jiang, S.; Dai, Q. Identification of a critical motif for the human immunodeficiency virus type 1 (HIV-1) gp41 core structure: Implications for designing novel anti-HIV fusion inhibitors. J. Virol. 2008, 82, 6349–6358. [Google Scholar] [CrossRef]
- Qiu, Z.; Chong, H.; Yao, X.; Su, Y.; Cui, S.; He, Y. Identification and characterization of a subpocket on the n-trimer of HIV-1 gp41: Implication for viral entry and drug target. Aids 2015, 29, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ding, X.; Zhu, Y.; Chong, H.; Cui, S.; He, J.; Wang, X.; He, Y. Structural and functional characterization of HIV-1 cell fusion inhibitor t20. Aids 2019, 33, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Pancera, M.; Zhou, T.; Druz, A.; Georgiev, I.S.; Soto, C.; Gorman, J.; Huang, J.; Acharya, P.; Chuang, G.Y.; Ofek, G.; et al. Structure and immune recognition of trimeric pre-fusion HIV-1 env. Nature 2014, 514, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Lyumkis, D.; Julien, J.P.; de Val, N.; Cupo, A.; Potter, C.S.; Klasse, P.J.; Burton, D.R.; Sanders, R.W.; Moore, J.P.; Carragher, B.; et al. Cryo-em structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 2013, 342, 1484–1490. [Google Scholar] [CrossRef] [PubMed]
- Julien, J.P.; Cupo, A.; Sok, D.; Stanfield, R.L.; Lyumkis, D.; Deller, M.C.; Klasse, P.J.; Burton, D.R.; Sanders, R.W.; Moore, J.P.; et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 2013, 342, 1477–1483. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Zhu, Y.; Ye, S.; Qi, Q.; Xia, S.; Ma, Z.; Yu, F.; Wang, Q.; Zhang, R.; Jiang, S.; et al. Creating an artificial tail anchor as a novel strategy to enhance the potency of peptide-based HIV fusion inhibitors. J. Virol. 2017, 91, e01416–e01445. [Google Scholar] [CrossRef]
- Su, S.; Ma, Z.; Hua, C.; Li, W.; Lu, L.; Jiang, S. Adding an artificial tail-anchor to a peptide-based HIV-1 fusion inhibitor for improvement of its potency and resistance profile. Molecules 2017, 22. [Google Scholar] [CrossRef]
- Zhu, Y.; Su, S.; Qin, L.; Wang, Q.; Shi, L.; Ma, Z.; Tang, J.; Jiang, S.; Lu, L.; Ye, S.; et al. Rational improvement of gp41-targeting HIV-1 fusion inhibitors: An innovatively designed ile-asp-leu tail with alternative conformations. Sci. Rep. 2016, 6, 31983. [Google Scholar] [CrossRef]
- Chong, H.; Qiu, Z.; Su, Y.; He, Y. The n-terminal t-t motif of a third-generation HIV-1 fusion inhibitor is not required for binding affinity and antiviral activity. J. Med. Chem. 2015, 58, 6378–6388. [Google Scholar] [CrossRef]
- Su, Y.; Chong, H.; Qiu, Z.; Xiong, S.; He, Y. Mechanism of HIV-1 resistance to short-peptide fusion inhibitors targeting the gp41 pocket. J. Virol. 2015, 89, 5801–5811. [Google Scholar] [CrossRef]
- Chong, H.; Qiu, Z.; Su, Y.; Yang, L.; He, Y. Design of a highly potent HIV-1 fusion inhibitor targeting the gp41 pocket. Aids 2015, 29, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Ding, X.; Liu, Z.; Wu, X.; Zhu, Y.; Wei, H.; Chong, H.; Cui, S.; He, Y. Molecular mechanism of HIV-1 resistance to sifuvirtide, a clinical trial-approved membrane fusion inhibitor. J. Biol. Chem. 2018, 293, 12703–12718. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. Xds. Acta Crystallogr. Sect. D Biol. Cryst. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Chen, S.; Yu, J.; Behrens, R.; Wiggins, J.; Sherer, N.; Liu, S.L.; Xiong, Y.; Xiang, S.H.; Wu, L. The polar region of HIV-1 envelope protein determines viral fusion and infectivity by stabilizing gp120-gp41 association. J. Virol. 2019, 93, e02118–e02188. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Cheng, J.; Lu, H.; Li, J.; Hu, J.; Qi, Z.; Liu, Z.; Jiang, S.; Dai, Q. Potent HIV fusion inhibitors against enfuvirtide-resistant HIV-1 strains. Proc. Natl. Acad. Sci. USA 2008, 105, 16332–16337. [Google Scholar] [CrossRef] [PubMed]
- Xiong, S.; Borrego, P.; Ding, X.; Zhu, Y.; Martins, A.; Chong, H.; Taveira, N.; He, Y. A helical short-peptide fusion inhibitor with highly potent activity against human immunodeficiency virus type 1 (HIV-1), HIV-2, and simian immunodeficiency virus. J. Virol. 2017, 91, e01816–e01839. [Google Scholar] [CrossRef]
- Chong, H.; Yao, X.; Qiu, Z.; Sun, J.; Zhang, M.; Waltersperger, S.; Wang, M.; Liu, S.L.; Cui, S.; He, Y. Short-peptide fusion inhibitors with high potency against wild-type and enfuvirtide-resistant HIV-1. Faseb J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2013, 27, 1203–1213. [Google Scholar] [CrossRef]
- Wu, X.; Liu, Z.; Ding, X.; Yu, D.; Wei, H.; Qin, B.; Zhu, Y.; Chong, H.; Cui, S.; He, Y. Mechanism of HIV-1 resistance to an electronically constrained alpha-helical peptide membrane fusion inhibitor. J. Virol. 2018, 92, e02017–e02044. [Google Scholar] [CrossRef]
- He, Y.; Xiao, Y.; Song, H.; Liang, Q.; Ju, D.; Chen, X.; Lu, H.; Jing, W.; Jiang, S.; Zhang, L. Design and evaluation of sifuvirtide, a novel HIV-1 fusion inhibitor. J. Biol. Chem. 2008, 283, 11126–11134. [Google Scholar] [CrossRef]
- Chong, H.; Wu, X.; Su, Y.; He, Y. Development of potent and long-acting HIV-1 fusion inhibitors. Aids 2016, 30, 1187–1196. [Google Scholar] [CrossRef]
- Chong, H.; Xue, J.; Xiong, S.; Cong, Z.; Ding, X.; Zhu, Y.; Liu, Z.; Chen, T.; Feng, Y.; He, L.; et al. A lipopeptide HIV-1/2 fusion inhibitor with highly potent in vitro, ex vivo, and in vivo antiviral activity. J. Virol. 2017, 91, e00217–e00288. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhang, X.; Ding, X.; Chong, H.; Cui, S.; He, J.; Wang, X.; He, Y. Exceptional potency and structural basis of a t1249-derived lipopeptide fusion inhibitor against HIV-1, HIV-2, and simian immunodeficiency virus. J. Biol. Chem. 2018, 293, 5323–5334. [Google Scholar] [CrossRef] [PubMed]
- Chong, H.; Xue, J.; Zhu, Y.; Cong, Z.; Chen, T.; Guo, Y.; Wei, Q.; Zhou, Y.; Qin, C.; He, Y. Design of novel HIV-1/2 fusion inhibitors with high therapeutic efficacy in rhesus monkey models. J. Virol. 2018, 92, e00718–e00775. [Google Scholar] [CrossRef] [PubMed]
- Chong, H.; Zhu, Y.; Yu, D.; He, Y. Structural and functional characterization of membrane fusion inhibitors with extremely potent activity against HIV-1, HIV-2, and simian immunodeficiency virus. J. Virol. 2018, 92, e01018–e01088. [Google Scholar] [CrossRef] [PubMed]
- Chong, H.; Xue, J.; Zhu, Y.; Cong, Z.; Chen, T.; Wei, Q.; Qin, C.; He, Y. Monotherapy with a low-dose lipopeptide HIV fusion inhibitor maintains long-term viral suppression in rhesus macaques. PLoS Pathog. 2019, 15, e1007552. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Chong, H.; Yu, D.; Guo, Y.; Zhou, Y.; He, Y. Design and characterization of cholesterylated peptide HIV-1/2 fusion inhibitors with extremely potent and long-lasting antiviral activity. J. Virol. 2019, 93, e02312–e02318. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Chong, H.; Xiong, S.; Qiao, Y.; Qiu, Z.; He, Y. Genetic pathway of HIV-1 resistance to novel fusion inhibitors targeting the gp41 pocket. J. Virol. 2015, 89, 12467–12479. [Google Scholar] [CrossRef]
- Eggink, D.; Bontjer, I.; Langedijk, J.P.; Berkhout, B.; Sanders, R.W. Resistance of human immunodeficiency virus type 1 to a third-generation fusion inhibitor requires multiple mutations in gp41 and is accompanied by a dramatic loss of gp41 function. J. Virol. 2011, 85, 10785–10797. [Google Scholar] [CrossRef]
- Yao, X.; Chong, H.; Zhang, C.; Qiu, Z.; Qin, B.; Han, R.; Waltersperger, S.; Wang, M.; He, Y.; Cui, S. Structural basis of potent and broad HIV-1 fusion inhibitor cp32m. J. Biol. Chem. 2012, 287, 26618–26629. [Google Scholar] [CrossRef]
- Yao, X.; Chong, H.; Zhang, C.; Waltersperger, S.; Wang, M.; Cui, S.; He, Y. Broad antiviral activity and crystal structure of HIV-1 fusion inhibitor sifuvirtide. J. Biol. Chem. 2012, 287, 6788–6796. [Google Scholar] [CrossRef]
- Chong, H.; Yao, X.; Qiu, Z.; Sun, J.; Qiao, Y.; Zhang, M.; Wang, M.; Cui, S.; He, Y. The m-t hook structure increases the potency of HIV-1 fusion inhibitor sifuvirtide and overcomes drug resistance. J. Antimicrob. Chemother. 2014, 69, 6759. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhu, Y.; Hu, H.; Zhang, S.; Wang, P.; Chong, H.; He, J.; Wang, X.; He, Y. Structural insights into the mechanisms of action of short-peptide HIV-1 fusion inhibitors targeting the gp41 pocket. Front. Cell. Infect. Microbiol. 2018, 8, 51. [Google Scholar] [CrossRef]
- Ding, X.; Zhang, X.; Chong, H.; Zhu, Y.; Wei, H.; Wu, X.; He, J.; Wang, X.; He, Y. Enfuvirtide (t20)-based lipopeptide is a potent HIV-1 cell fusion inhibitor: Implication for viral entry and inhibition. J. Virol. 2017, 91, e00817–e00831. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Bergeron, L.; Helseth, E.; Thali, M.; Repke, H.; Sodroski, J. Effects of amino acid changes in the extracellular domain of the human immunodeficiency virus type 1 gp41 envelope glycoprotein. J. Virol. 1993, 67, 2747–2755. [Google Scholar] [PubMed]
- Chen, S.S.; Lee, S.F.; Hao, H.J.; Chuang, C.K. Mutations in the leucine zipper-like heptad repeat sequence of human immunodeficiency virus type 1 gp41 dominantly interfere with wild-type virus infectivity. J. Virol. 1998, 72, 4765–4774. [Google Scholar]
- Chen, S.S.; Lee, C.N.; Lee, W.R.; McIntosh, K.; Lee, T.H. Mutational analysis of the leucine zipper-like motif of the human immunodeficiency virus type 1 envelope transmembrane glycoprotein. J. Virol. 1993, 67, 3615–3619. [Google Scholar]
- Dubay, J.W.; Roberts, S.J.; Brody, B.; Hunter, E. Mutations in the leucine zipper of the human immunodeficiency virus type 1 transmembrane glycoprotein affect fusion and infectivity. J. Virol. 1992, 66, 4748–4756. [Google Scholar] [PubMed]
- Follis, K.E.; Larson, S.J.; Lu, M.; Nunberg, J.H. Genetic evidence that interhelical packing interactions in the gp41 core are critical for transition of the human immunodeficiency virus type 1 envelope glycoprotein to the fusion-active state. J. Virol. 2002, 76, 7356–7362. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lu, M.; Stoller, M.O.; Wang, S.; Liu, J.; Fagan, M.B.; Nunberg, J.H. Structural and functional analysis of interhelical interactions in the human immunodeficiency virus type 1 gp41 envelope glycoprotein by alanine-scanning mutagenesis. J. Virol. 2001, 75, 11146–11156. [Google Scholar] [CrossRef]
- Salzwedel, K.; West, J.T.; Hunter, E. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for env-mediated fusion and virus infectivity. J. Virol. 1999, 73, 2469–2480. [Google Scholar]
- Weng, Y.; Yang, Z.; Weiss, C.D. Structure-function studies of the self-assembly domain of the human immunodeficiency virus type 1 transmembrane protein gp41. J. Virol. 2000, 74, 5368–5372. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Pozniak, A.; Wildfire, A.; Stanfield-Oakley, S.A.; Mosier, S.M.; Ratcliffe, D.; Workman, J.; Joall, A.; Myers, R.; Smit, E.; et al. Emergence and evolution of enfuvirtide resistance following long-term therapy involves heptad repeat 2 mutations within gp41. Antimicrob. Agents Chemother. 2005, 49, 1113–1119. [Google Scholar] [CrossRef] [PubMed]
- Eggink, D.; Berkhout, B.; Sanders, R.W. Inhibition of HIV-1 by fusion inhibitors. Curr. Pharm. Des. 2010, 16, 3716–3728. [Google Scholar] [CrossRef] [PubMed]
- Ashkenazi, A.; Wexler-Cohen, Y.; Shai, Y. Multifaceted action of fuzeon as virus-cell membrane fusion inhibitor. Biochim. Biophys. Acta 2011, 1808, 2352–2358. [Google Scholar] [CrossRef] [PubMed]
- Berkhout, B.; Eggink, D.; Sanders, R.W. Is there a future for antiviral fusion inhibitors? Curr. Opin. Virol. 2012, 2, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Dwyer, J.J.; Wilson, K.L.; Davison, D.K.; Freel, S.A.; Seedorff, J.E.; Wring, S.A.; Tvermoes, N.A.; Matthews, T.J.; Greenberg, M.L.; Delmedico, M.K. Design of helical, oligomeric HIV-1 fusion inhibitor peptides with potent activity against enfuvirtide-resistant virus. Proc. Natl. Acad. Sci. USA 2007, 104, 12772–12777. [Google Scholar] [CrossRef] [PubMed]
- Otaka, A.; Nakamura, M.; Nameki, D.; Kodama, E.; Uchiyama, S.; Nakamura, S.; Nakano, H.; Tamamura, H.; Kobayashi, Y.; Matsuoka, M.; et al. Remodeling of gp41-c34 peptide leads to highly effective inhibitors of the fusion of HIV-1 with target cells. Angew. Chem. Int. Engl. 2002, 41, 2937–2940. [Google Scholar] [CrossRef]
- Welch, B.D.; VanDemark, A.P.; Heroux, A.; Hill, C.P.; Kay, M.S. Potent d-peptide inhibitors of HIV-1 entry. Proc. Natl. Acad. Sci. USA 2007, 104, 16828–16833. [Google Scholar] [CrossRef] [PubMed]
- Stephens, O.M.; Kim, S.; Welch, B.D.; Hodsdon, M.E.; Kay, M.S.; Schepartz, A. Inhibiting HIV fusion with a beta-peptide foldamer. J. Am. Chem. Soc. 2005, 127, 13126–13127. [Google Scholar] [CrossRef]
- Ingallinella, P.; Bianchi, E.; Ladwa, N.A.; Wang, Y.J.; Hrin, R.; Veneziano, M.; Bonelli, F.; Ketas, T.J.; Moore, J.P.; Miller, M.D.; et al. Addition of a cholesterol group to an HIV-1 peptide fusion inhibitor dramatically increases its antiviral potency. Proc. Natl. Acad. Sci. USA 2009, 106, 5801–5806. [Google Scholar] [CrossRef]
- Redman, J.S.; Francis, J.N.; Marquardt, R.; Papac, D.; Mueller, A.L.; Eckert, D.M.; Welch, B.D.; Kay, M.S. Pharmacokinetic and chemical synthesis optimization of a potent d-peptide HIV entry inhibitor suitable for extended-release delivery. Mol. Pharm. 2018, 15, 1169–1179. [Google Scholar] [CrossRef] [PubMed]
- Chong, H.; Qiu, Z.; Sun, J.; Qiao, Y.; Li, X.; He, Y. Two m-t hook residues greatly improve the antiviral activity and resistance profile of the HIV-1 fusion inhibitor sc29ek. Retrovirology 2014, 11, 40. [Google Scholar] [CrossRef] [PubMed]
- Naito, T.; Izumi, K.; Kodama, E.; Sakagami, Y.; Kajiwara, K.; Nishikawa, H.; Watanabe, K.; Sarafianos, S.G.; Oishi, S.; Fujii, N.; et al. Sc29ek, a peptide fusion inhibitor with enhanced alpha-helicity, inhibits replication of human immunodeficiency virus type 1 mutants resistant to enfuvirtide. Antimicrob. Agents Chemother. 2009, 53, 1013–1018. [Google Scholar] [CrossRef] [PubMed]
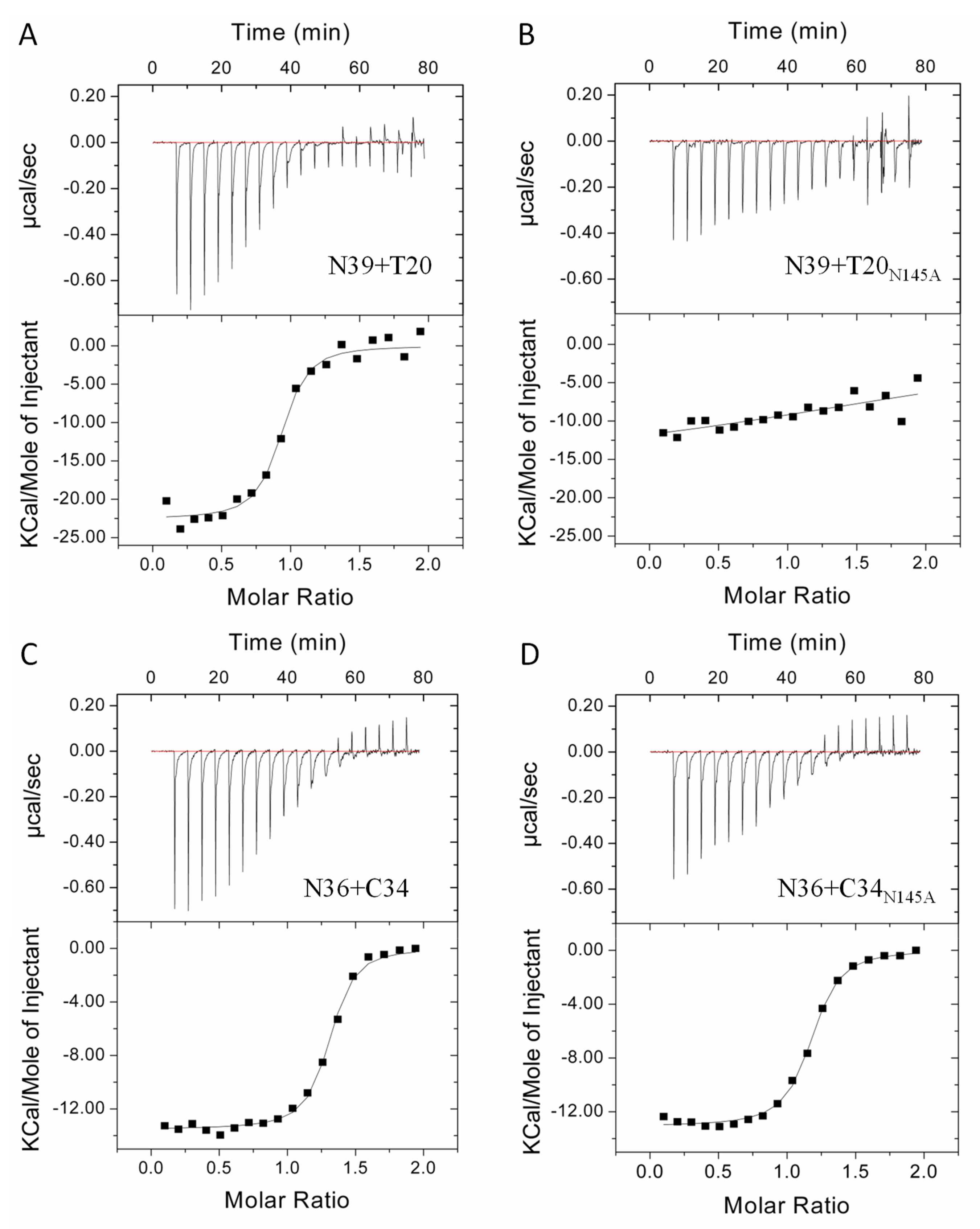


| Peptide Pair | N | K (M−1) | △H (Kcal/mol) | △S (cal/mol/deg) |
|---|---|---|---|---|
| N39/T20 | 0.9 | 3.2 × 106 | −22.6 | −45.9 |
| N39/T20N145A | 2.6 | 3.6 × 104 | −13.5 | −30.7 |
| N36/C34 | 1.3 | 3.3 × 106 | −15.4 | −15.5 |
| N36/C34N145A | 1.1 | 2.2 × 106 | −13.1 | −14.8 |
| N36/SC29EK | 1.1 | 2.6 × 106 | −15.8 | −23.6 |
| N36/SC28EK | 1.3 | 5.2 × 105 | −9.0 | −3.66 |
| Parameter | Value a |
|---|---|
| Data Collection | |
| Beamline | SSRF BL17U |
| Wavelength (Å) | 0.97915 |
| Resolution rage | 45.000–2.33 (2.47–2.33) |
| Space group | P212121 |
| Cell dimesions | |
| a, b, c (Å) | 36.500, 39.860, 171.503 |
| a, b, g (°) | 90.00, 90.00, 90.00 |
| Redundancy | 3.19 |
| Total no of reflections | 66,040 |
| No. of unique reflections | 20,721 |
| Rmergeb(%) | 8.3 (86.1) |
| I/SIGMA | 10.16 (1.53) |
| Completeness (%) | 98.0 (90.6) |
| Refinement | |
| Resolution (Å) | 38.825–2.330 (2.44–2.33) |
| No. of reflections | 11,172 |
| Rwork/Rfreec | 0.2330/0.2859 |
| No. of atoms | |
| Protein | 1762 |
| Water | 10 |
| B factors (Å2) | |
| Protein | 67.28 |
| Water | 60.77 |
| RMS deviations | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.491 |
| Ramachandran plot (%) | |
| Favored | 97.95% |
| Allowed | 1.54% |
| Disallowed | 0.51% |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geng, X.; Liu, Z.; Yu, D.; Qin, B.; Zhu, Y.; Cui, S.; Chong, H.; He, Y. Conserved Residue Asn-145 in the C-Terminal Heptad Repeat Region of HIV-1 gp41 is Critical for Viral Fusion and Regulates the Antiviral Activity of Fusion Inhibitors. Viruses 2019, 11, 609. https://doi.org/10.3390/v11070609
Geng X, Liu Z, Yu D, Qin B, Zhu Y, Cui S, Chong H, He Y. Conserved Residue Asn-145 in the C-Terminal Heptad Repeat Region of HIV-1 gp41 is Critical for Viral Fusion and Regulates the Antiviral Activity of Fusion Inhibitors. Viruses. 2019; 11(7):609. https://doi.org/10.3390/v11070609
Chicago/Turabian StyleGeng, Xiuzhu, Zixuan Liu, Danwei Yu, Bo Qin, Yuanmei Zhu, Sheng Cui, Huihui Chong, and Yuxian He. 2019. "Conserved Residue Asn-145 in the C-Terminal Heptad Repeat Region of HIV-1 gp41 is Critical for Viral Fusion and Regulates the Antiviral Activity of Fusion Inhibitors" Viruses 11, no. 7: 609. https://doi.org/10.3390/v11070609
APA StyleGeng, X., Liu, Z., Yu, D., Qin, B., Zhu, Y., Cui, S., Chong, H., & He, Y. (2019). Conserved Residue Asn-145 in the C-Terminal Heptad Repeat Region of HIV-1 gp41 is Critical for Viral Fusion and Regulates the Antiviral Activity of Fusion Inhibitors. Viruses, 11(7), 609. https://doi.org/10.3390/v11070609




