Genetic Variability of Chrysodeixis Includens Nucleopolyhedrovirus (ChinNPV) and the Insecticidal Characteristics of Selected Genotypic Variants
Abstract
1. Introduction
2. Materials and Methods
2.1. Insects, Cells, and Viruses
2.2. Viral DNA Extraction and Restriction Endonuclease Analysis
2.3. Construction of the ChinNPV-Mex1 Mixture and Its Biological Characterization
2.4. Genotypic Characterization and Mortality Response
2.5. Genotype Selection and Characterization of Median Time to Death and OB Production
3. Results
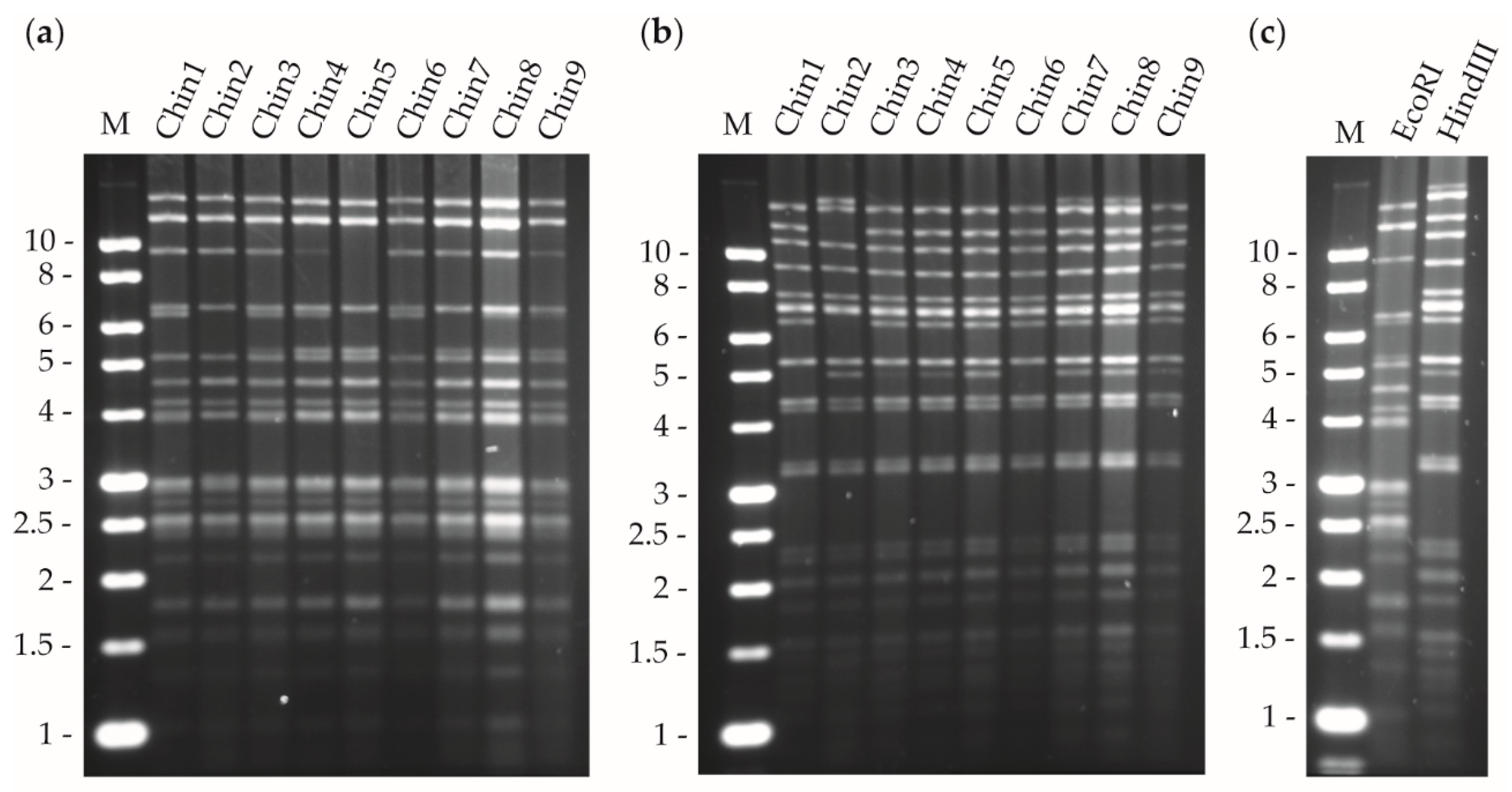
3.1. Identification by REN of Field ChinNPV Isolates
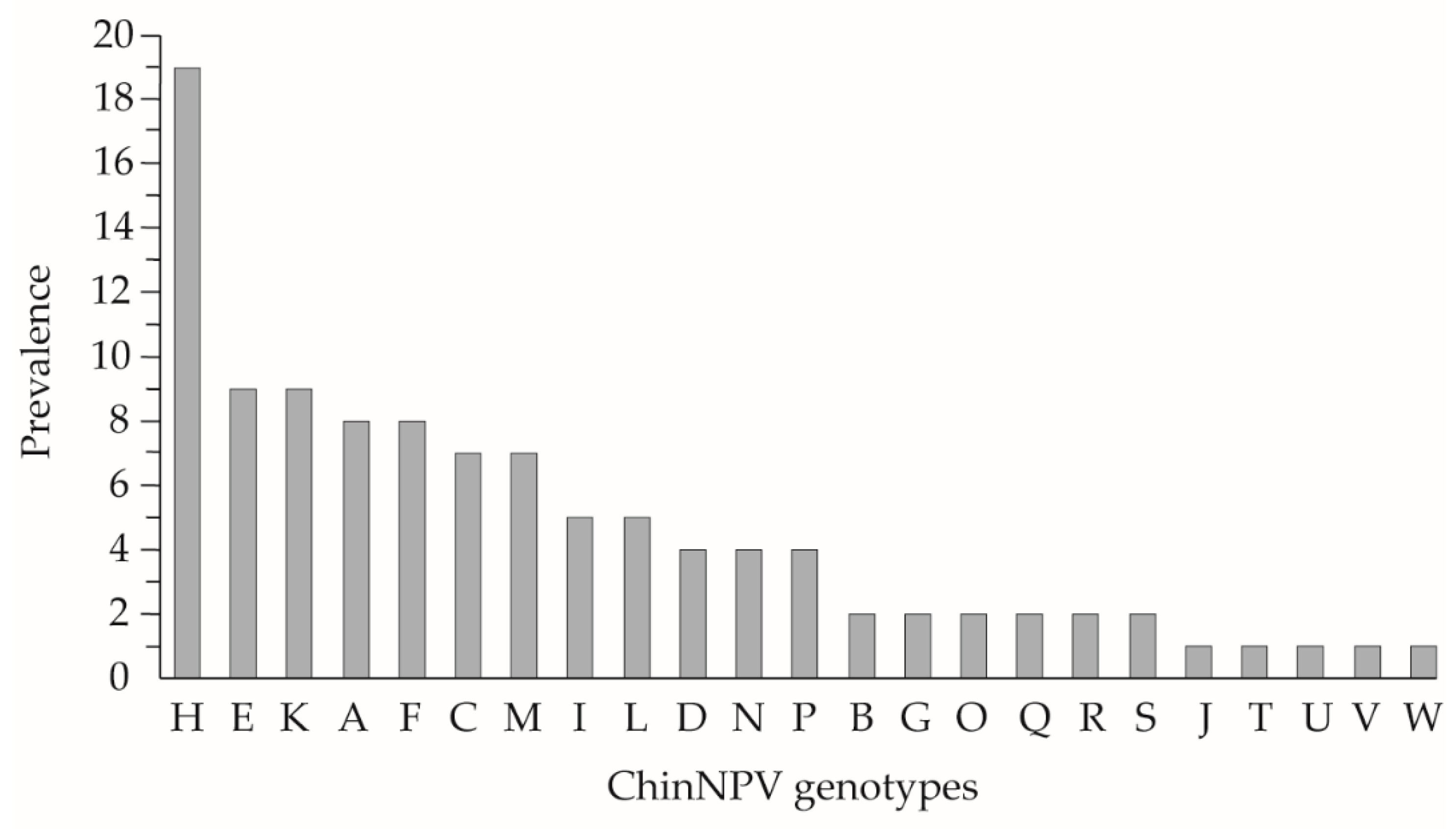
3.2. Biological and Genotypic Characterization of ChinNPV-Mex1
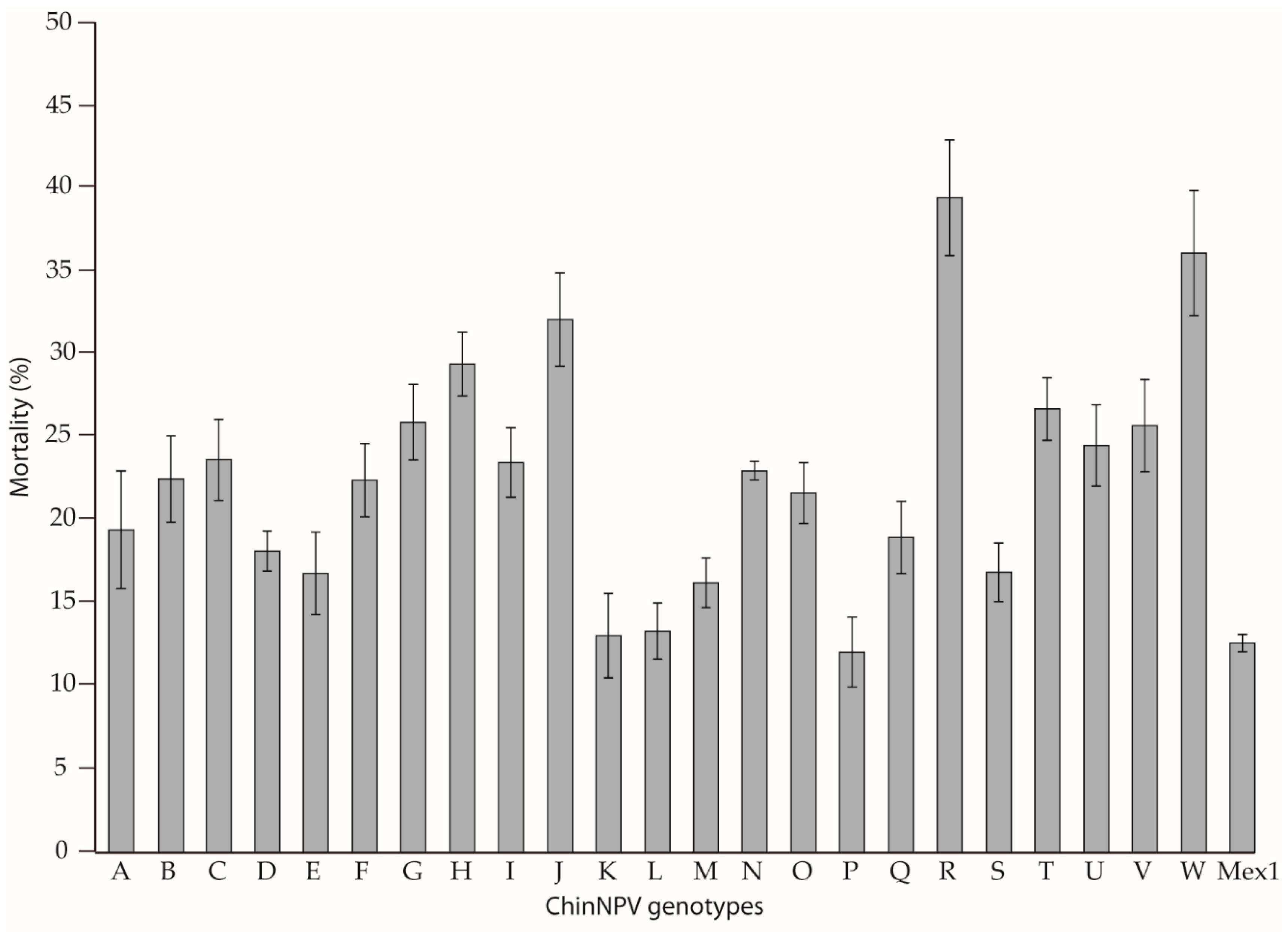
3.3. Biological Characterization of ChinNPV Genotypes
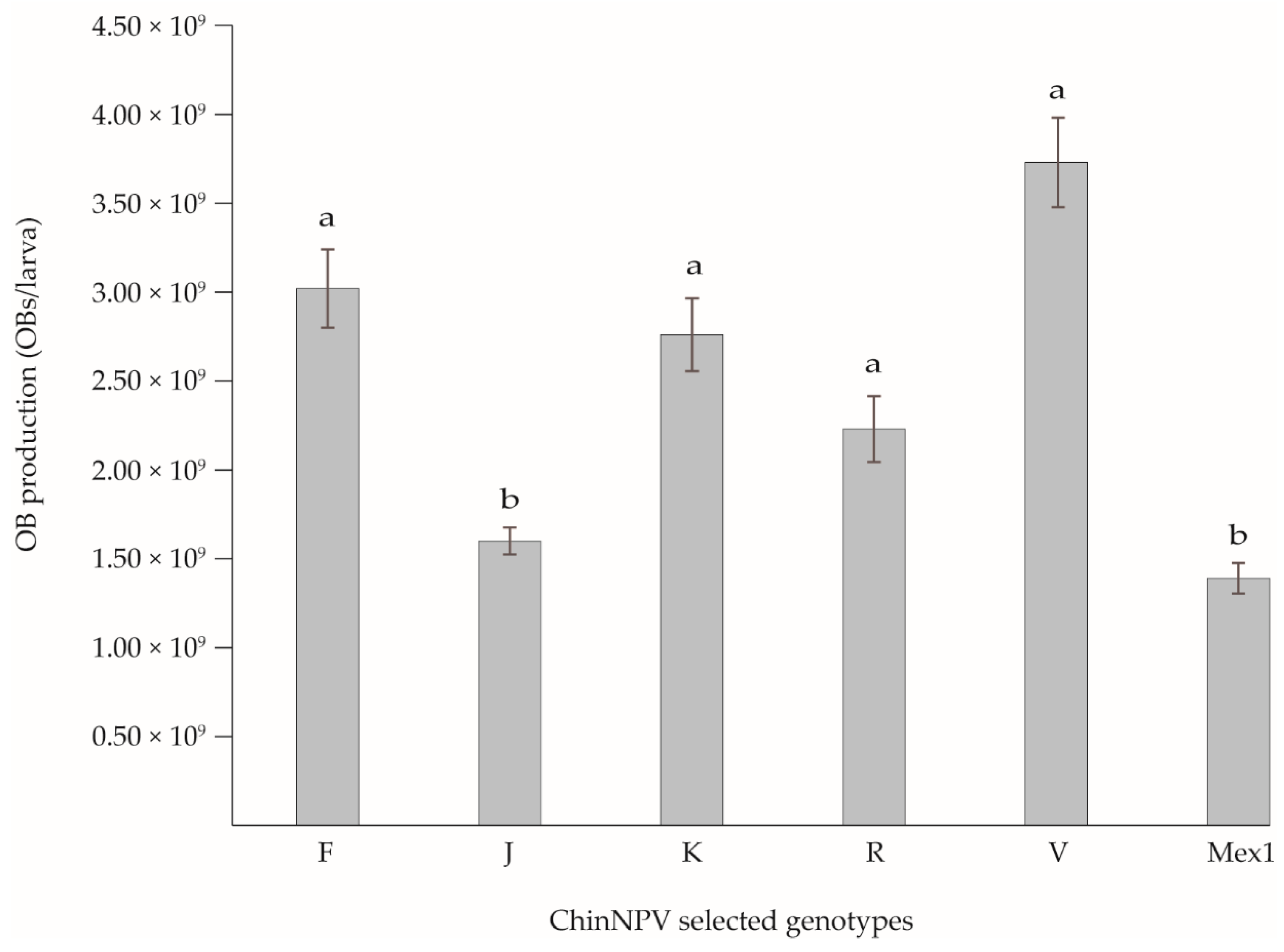
3.4. Virulence and OB Production of the Selected Genotypes
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Variants | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| P | 290.080 | ||||||||||
| A | 546.503 | ||||||||||
| L | 679.357 | 679.357 | |||||||||
| K | 811.045 | 811.045 | |||||||||
| M | 821.033 | 821.033 | |||||||||
| E | 948.565 | 948.565 | |||||||||
| S | 983.034 | 983.034 | |||||||||
| D | 983.703 | 983.703 | |||||||||
| Q | 1035.547 | ||||||||||
| V | 1267.990 | ||||||||||
| O | 1282.400 | 1282.400 | |||||||||
| B | 1304.449 | 1304.449 | 1304.449 | ||||||||
| I | 1392.007 | 1392.007 | 1392.007 | ||||||||
| N | 1413.618 | 1413.618 | 1413.618 | ||||||||
| U | 1449.109 | 1449.109 | 1449.109 | 1449.109 | |||||||
| C | 1451.831 | 1451.831 | 1451.831 | ||||||||
| F | 1467.757 | 1467.757 | 1467.757 | ||||||||
| G | 1664.041 | 1664.041 | |||||||||
| T | 1669.211 | ||||||||||
| H | 1670.921 | ||||||||||
| J | 2008.000 | ||||||||||
| W | 2181.689 | ||||||||||
| R | 2409.836 | ||||||||||
| Test statistic | 0.0 1 | 3.078 | 2.236 | 7.309 | 5.246 | 7.014 | 10.204 | 11.173 | 10.290 | 1.068 | 0.0 1 |
| Sig.(2 sided test) | 0.0 | 0.215 | 0.327 | 0.063 | 0.155 | 0.220 | 0.116 | 0.083 | 0.067 | 0.301 | 0.0 |
| Adjusted Sig. (2 sided test) | 0.0 | 0.843 | 0.952 | 0.311 | 0.619 | 0.613 | 0.334 | 0.248 | 0.235 | 0.984 | 0.0 |
References
- Cory, J.S.; Myers, J.H. The ecology and evolution of insect baculoviruses. Annu. Rev. Ecol. Evol. Syst. 2003, 34, 239–272. [Google Scholar] [CrossRef]
- Hitchman, R.B.; Hodgson, D.J.; King, L.A.; Hails, R.S.; Cory, J.S.; Possee, R.D. Host mediated selection of pathogen genotypes as a mechanism for the maintenance of baculovirus diversity in the field. J. Invertebr. Pathol. 2007, 94, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, M.; Hamajima, R.; Kobayashi, M. Baculoviruses: Diversity, evolution and manipulation of insects. Entomol. Sci. 2015, 18, 1–20. [Google Scholar] [CrossRef]
- Williams, T. Viruses. In Ecology of Invertebrate Diseases; Hajek, A.E., Shapiro-Ilan, D.I., Eds.; John Wiley Ltd.: Chichester, UK, 2018; pp. 213–285. ISBN 9781119256106. [Google Scholar]
- Stiles, B.; Himmerich, S. Autographa californica NPV isolates: Restriction endonuclease analysis and comparative biological activity. J. Invertebr. Pathol. 1998, 72, 174–177. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, K.; Zhu, J.; Murata, M.; Laviña-Caoili, B.A.; Ikeda, M.; Kobayashi, M.; Kawamura, S. Cloning and comparative characterization of three distinct nucleopolyhedroviruses isolated from the common cutworm, Spodoptera litura (Lepidoptera: Noctuidae) in Japan. Biol. Control 2004, 31, 38–48. [Google Scholar] [CrossRef]
- Cory, J.S.; Green, B.M.; Paul, R.K.; Hunter-Fujita, F. Genotypic and phenotypic diversity of a baculovirus population within an individual insect host. J. Invertebr. Pathol. 2005, 89, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Harrison, R.L.; Puttler, B.; Popham, H.J.R. Genomic sequence analysis of a fast-killing isolate of Spodoptera frugiperda multiple nucleopolyhedrovirus. J. Gen. Virol. 2008, 89, 775–790. [Google Scholar] [CrossRef]
- Cabodevilla, O.; Ibañez, I.; Simón, O.; Murillo, R.; Caballero, P.; Williams, T. Occlusion body pathogenicity, virulence and productivity traits vary with transmission strategy in a nucleopolyhedrovirus. Biol. Control 2011, 56, 184–192. [Google Scholar] [CrossRef]
- Williams, H.L.; Monge-Monge, K.S.; Otvos, I.S.; Reardon, R.; Ragenovich, I. Genotypic variation among Douglas-fir tussock moth nucleopolyhedrovirus (OpNPV) isolates in the western United States. J. Invertebr. Pathol. 2011, 108, 13–21. [Google Scholar] [CrossRef]
- Redman, E.M.; Wilson, K.; Grzywacz, D.; Cory, J.S. High levels of genetic diversity in Spodoptera exempta NPV from Tanzania. J. Invertebr. Pathol. 2010, 105, 190–193. [Google Scholar] [CrossRef]
- Rowley, D.L.; Popham, H.J.R.; Harrison, R.L. Genetic variation and virulence of nucleopolyhedroviruses isolated worldwide from the heliothine pests Helicoverpa armigera, Helicoverpa zea, and Heliothis virescens. J. Invertebr. Pathol. 2011, 107, 112–126. [Google Scholar] [CrossRef] [PubMed]
- Laviña, B.A.; Padua, L.E.; Wu, F.Q.; Shirata, N.; Ikeda, M.; Kobayashi, M. Biological characterization of a nucleopolyhedrovirus of Spodoptera litura (Lepidoptera: Noctuidae) isolated from the Philippines. Biol. Control 2001, 20, 39–47. [Google Scholar] [CrossRef]
- Harrison, R.L. Structural divergence among genomes of closely related baculoviruses and its implications for baculovirus evolution. J. Invertebr. Pathol. 2009, 101, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.H.; Miller, L.K. Isolation of genotypic variants of Autographa californica nuclear polyhedrosis virus. J. Virol. 1978, 27, 754–767. [Google Scholar] [PubMed]
- Weitzman, M.D.; Possee, R.D.; King, L.A. Characterization of two variants of Panolis flammea multiple nucleocapsid nuclear polyhedrosis virus. J. Gen. Virol. 1992, 73, 1881–1886. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, D.; Murillo, R.; Krell, P.J.; Vlak, J.M.; Caballero, P. Four genotypic variants of a Spodoptera exigua nucleopolyhedrovirus (Se-SP2) are distinguishable by a hypervariable genomic region. Virus Res. 1999, 59, 61–74. [Google Scholar] [CrossRef]
- Simón, O.; Williams, T.; López-Ferber, M.; Caballero, P. Genetic structure of a Spodoptera frugiperda nucleopolyhedrovirus population: High prevalence of deletion genotypes. Appl. Environ. Microbiol. 2004, 70, 5579–5588. [Google Scholar] [CrossRef] [PubMed]
- Bernal, A.; Simón, O.; Williams, T.; Muñoz, D.; Caballero, P. A Chrysodeixis chalcites single-nucleocapsid nucleopolyhedrovirus population from the Canary Islands is genotypically structured to maximize survival. Appl. Environ. Microbiol. 2013, 79, 7709–7718. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Caballero, P.; Murillo, R.; Muñoz, D.; Williams, T. El nucleopoliedrovirus de Spodoptera exigua (Lepidoptera: Noctuidae) como bioplaguicida: Análisis de avances recientes en España. Rev. Colomb. Entomol. 2009, 35, 105–115. [Google Scholar]
- Caballero, P.; Bernal, A.; Simón, O.; Carnero, A.; Hernández-Suárez, E.; Williams, T. Nuevos genotipos del NPV simple de Chrysodeixis chalcites (ChchSNPV): Procedimiento para su producción y uso como agente de control biológico. Patent ES2504866, 8 October 2014. [Google Scholar]
- Caballero, P.; Arrizubieta, M.; Simón, O.; Williams, T. Nuevos genotipos del nucleopoliedrovirus simple de Helicoverpa armigera (HearSNPV), procedimiento para su producción y uso como agente de control biológico. Patent ES2555165, 29 December 2015. [Google Scholar]
- Arrizubieta, M.; Simón, O.; Williams, T.; Caballero, P. A novel binary mixture of Helicoverpa armigera single nucleopolyhedrovirus genotypic variants has improved insecticidal characteristics for control of cotton bollworms. Appl. Environ. Microbiol. 2015, 81, 3984–3993. [Google Scholar] [CrossRef] [PubMed]
- Gebhardt, M.M.; Eberle, K.E.; Radtke, P.; Jehle, J.A. Baculovirus resistance in codling moth is virus isolate-dependent and the consequence of a mutation in viral gene pe38. Proc. Natl. Acad. Sci. USA 2014, 111, 15711–15716. [Google Scholar] [CrossRef] [PubMed]
- Jehle, J.A.; Schulze-Bopp, S.; Undorf-Spahn, K.; Fritsch, E. Evidence for a second type of resistance against Cydia pomonella granulovirus in field populations of codling moths. Appl. Environ. Microbiol. 2017, 83, e02330-16. [Google Scholar] [CrossRef] [PubMed]
- Eichlin, T.D.; Cunningham, H.B. The Plusiinae (Lepidoptera: Noctuidae) of America north of Mexico, emphasizing genitalic and larval morphology. Tech. Bull. 1567 1978, 14–15. [Google Scholar]
- Craveiro, S.R.; Cunha, F.; Fonseca, I.C.B.; Castro, M.E.B.; Alexandre, T.M.; Moscardi, F.; Ribeiro, Z.M.A. Evaluation of seven viral isolates as potential biocontrol agents against Pseudoplusia includens (Lepidoptera: Noctuidae) caterpillars. J. Invertebr. Pathol. 2010, 105, 98–104. [Google Scholar]
- Wennmann, J.T.; Keilwagen, J.; Jehle, J.A. Baculovirus Kimura two-parameter species demarcation criterion is confirmed by the distances of 38 core gene nucleotide sequences. J. Gen. Virol. 2018, 99, 1307–1320. [Google Scholar] [CrossRef]
- Thézé, J.; Lopez-Vaamonde, C.; Cory, J.S.; Herniou, E.A. Biodiversity, evolution and ecological specialization of baculoviruses: A treasure trove for future applied research. Viruses 2018, 10, 366. [Google Scholar] [CrossRef]
- Felland, C.M.; Pitre, H.N.; Luttrell, R.G.; Hamer, J.L. Resistance to pyrethroid insecticides in soybean looper (Lepidoptera: Noctuidae) in Mississippi. J. Econ. Entomol. 1990, 83, 35–40. [Google Scholar] [CrossRef]
- Leonard, R.B.; Boethel, D.J.; Sparks, A.N.; Layton, B.M.; Mink, J.S.; Pavloff, A.M.; Burris, E.; Graves, J.B. Variations in response of soybean looper (Lepidoptera: Noctuidae) to selected insecticides in Louisiana. J. Econ. Entomol. 2015, 83, 27–34. [Google Scholar] [CrossRef]
- Mink, J.S.; Boethel, D.J. Development of a diagnostic technique for monitoring permethrin resistance in soybean looper (Lepidoptera: Noctuidae) larvae. J. Econ. Entomol. 2015, 85, 1056–1062. [Google Scholar] [CrossRef]
- Mascarenhas, R.N.; Boethel, D.J. Responses of field-collected strains of soybean looper (Lepidoptera: Noctuidae) to selected insecticides using an artificial diet overlay bioassay. J. Econ. Entomol. 1997, 90, 1117–1124. [Google Scholar] [CrossRef]
- Craveiro, S.R.; Santos, L.A.V.M.; Togawa, R.C.; Inglis, P.W.; Grynberg, P.; Ribeiro, Z.M.A.; Ribeiro, B.M.; Castro, M.E.B. Complete genome sequences of six Chrysodeixis includens nucleopolyhedrovirus isolates from Brazil and Guatemala. Genome Announc. 2016, 4, 8–9. [Google Scholar] [CrossRef]
- Greene, G.L.; Leppla, N.C.; Dickerson, W.A. Velvetbean caterpillar: A rearing procedure and artificial medium. J. Econ. Entomol. 1976, 69, 487–488. [Google Scholar] [CrossRef]
- Del-Angel, C.; Lasa, R.; Rodríguez-del-Bosque, L.A.; Mercado, G.; Beperet, I.; Caballero, P.; Williams, T. Anticarsia gemmatalis nucleopolyhedrovirus from soybean crops in Tamaulipas, Mexico: Diversity and insecticidal characteristics of individual variants and their co-occluded mixtures. Fla. Entomol. 2018, 101, 404–410. [Google Scholar] [CrossRef]
- Hughes, P.R.; Wood, H.A. In vivo and in vitro bioassay methods for baculoviruses. In The Biology of Baculoviruses; Granados, R.R., Federici, B.A., Eds.; CRC Press: Boca Raton, FL, USA, 1987; Volume 2, pp. 1–30. [Google Scholar]
- Polo Plus, version 1.0; LeOra Software: Parma, MO, USA, 2003.
- R Foundation for Statistical Computing. A Language and Environment for Statistical Computing, version 3.6.0; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- IBM SPSS Statistics. IBM SPSS Statistics for Windows, version 25.0; IBM Corp: Armonk, NY, USA, 2017. [Google Scholar]
- King, L.A.; Possee, R.D. The Baculovirus Expression System. A Laboratory Guide; Chapman & Hall: London, UK, 1992. [Google Scholar]
- Fuentes, E.G.; Hernández-Suárez, E.; Simón, O.; Williams, T.; Caballero, P. Chrysodeixis chalcites nucleopolyhedrovirus (ChchNPV): Natural occurrence and efficacy as a biological insecticide on young banana plants in greenhouse and open-field conditions on the Canary Islands. PLoS ONE 2017, 12, 1–19. [Google Scholar] [CrossRef]
- Erlandson, M. Genetic variation in field populations of baculoviruses: Mechanisms for generating variation and its potential role in baculovirus epizootiology. Virol. Sin. 2009, 24, 458–469. [Google Scholar] [CrossRef]
- Martignoni, M.E.; Iwai, P.J. Propagation of multicapsid nuclear polyhedrosis virus of Orgyia pseudotsugata in larvae of Trichoplusia ni. J. Invertebr. Pathol. 1986, 47, 32–41. [Google Scholar] [CrossRef]
- Kolodny-Hirsch, D.M.; Beek, N.A.M. Van Selection of a morphological variant of Autographa californica nuclear polyhedrosis virus with increased virulence following serial passage in Plutella xylostella. J. Invertebr. Pathol. 1997, 69, 205–211. [Google Scholar] [CrossRef]
- Harrison, R.L.; Hoover, K. Baculoviruses and other occluded insect viruses. In Insect Pathology; Vega, F.E., Kaya, H.K., Eds.; Academic Press: London, UK, 2012; pp. 73–131. [Google Scholar]
- Caballero, P.; Williams, T.; Muñoz, D.; Murillo, R.; Lasa, R. Nuevos genotipos del nucleopoliedrovirus de Spodoptera exigua y uso de los mismos en el control de las plagas producidas por este insecto. WO2007122278A2, 1 November 2017. Available online: https://patents.google.com/patent/WO2007122278A2/es (accessed on 17 Febrary 2019).
- Godoy, D.N.; Popham, H.J.R.; Giacomelli, T.; Muraro, D.S.; Bernardi, O.; Marçon, P.; Stacke, R.F. Baseline susceptibility of Brazilian populations of Chrysodeixis includens (Lepidoptera: Noctuidae) to C. includens nucleopolyhedrovirus and diagnostic concentration for resistance monitoring. J. Econ. Entomol. 2018, 112, 349–354. [Google Scholar]
- Hodgson, D.J.; Vanbergen, A.J.; Watt, A.D.; Hails, R.S.; Cory, J.S. Phenotypic variation between naturally co-existing genotypes of a lepidopteran baculovirus. Evol. Ecol. Res. 2001, 3, 687–701. [Google Scholar]
- Corsaro, B.G.; Fraser, M.J. Characterization of genotypic and phenotypic variation in plaque-purified strains of HzSNPV elkar isolate. Intervirology 1987, 28, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Vanbergen, A.J.; Hodgson, D.J.; Hails, R.S.; Cory, J.S.; Hartley, S.E. Differential selection of baculovirus genotypes mediated by different species of host food plant. Ecol. Lett. 2002, 5, 512–518. [Google Scholar]
- Chateigner, A.; Bézier, A.; Labrousse, C.; Jiolle, D.; Barbe, V.; Herniou, E.A. Ultra deep sequencing of a baculovirus population reveals widespread genomic variations. Viruses 2015, 7, 3625–3646. [Google Scholar] [CrossRef] [PubMed]
- Baillie, V.L.; Bouwer, G. The effect of inoculum dose on the genetic diversity detected within Helicoverpa armigera nucleopolyhedrovirus populations. J. Gen. Virol. 2013, 94, 2524–2529. [Google Scholar] [CrossRef] [PubMed]
- Barrera, G.; Williams, T.; Villamizar, L.; Caballero, P.; Simón, O. Deletion genotypes reduce occlusion body potency but increase occlusion body production in a Colombian Spodoptera frugiperda nucleopolyhedrovirus population. PLoS ONE 2013, 8, e77271. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, D.; Castillejo, J.I.; Caballero, P. Naturally occurring deletion mutants are parasitic genotypes in a wild-type nucleopolyhedrovirus population of Spodoptera exigua. Appl. Environ. Microbiol. 1998, 64, 4372–4377. [Google Scholar] [PubMed]
| Species | 5 × 106 OBs/mL | 5 × 108 OBs/mL |
|---|---|---|
| Anticarsia gemmatalis | N.P. | N.P. |
| Chrysodeixis chalcites | ++ | +++ |
| Chrysodeixis includens | ++++ | ++++ |
| Helicoverpa armigera | N.P. | N.P. |
| Lobesia botrana | N.P. | N.P. |
| Mamestra brassicae | N.P. | N.P. |
| Spodoptera eridania | N.P. | N.P. |
| Spodoptera exigua | N.P. | N.P. |
| Spodoptera frugiperda | N.P. | N.P. |
| Spodoptera littoralis | N.P. | N.P. |
| Trichoplusia ni | + | ++++ |
| Virus | LC50 * (OBs/mL) | 95% Confidence Limits | MTD (h) | 95% Confidence Limits | OB Production (OBs/larva) | 95% Confidence Limits | |||
|---|---|---|---|---|---|---|---|---|---|
| Low | High | Low | High | Low | High | ||||
| ChinNPV-Mex1 | 6.5 × 104 | 2.4 × 104 | 2.6 × 105 | 125.0 | 121.8 | 128.2 | 2.2 × 109 | 1.2 × 109 | 3.1 × 109 |
| Variant | MTD (h) | 95% Confidence Limits | |
|---|---|---|---|
| Low | High | ||
| ChinNPV-F | 125 | 120.3 | 129.7 |
| ChinNPV-J | 125 | 120.4 | 129.6 |
| ChinNPV-K | 119 | 115.5 | 122.5 |
| ChinNPV-R | 125 | 120.4 | 129.6 |
| ChinNPV-V | 119 | 115.7 | 122.3 |
| ChinNPV-Mex1 | 125 | 121.8 | 128.2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aguirre, E.; Beperet, I.; Williams, T.; Caballero, P. Genetic Variability of Chrysodeixis Includens Nucleopolyhedrovirus (ChinNPV) and the Insecticidal Characteristics of Selected Genotypic Variants. Viruses 2019, 11, 581. https://doi.org/10.3390/v11070581
Aguirre E, Beperet I, Williams T, Caballero P. Genetic Variability of Chrysodeixis Includens Nucleopolyhedrovirus (ChinNPV) and the Insecticidal Characteristics of Selected Genotypic Variants. Viruses. 2019; 11(7):581. https://doi.org/10.3390/v11070581
Chicago/Turabian StyleAguirre, Eduardo, Inés Beperet, Trevor Williams, and Primitivo Caballero. 2019. "Genetic Variability of Chrysodeixis Includens Nucleopolyhedrovirus (ChinNPV) and the Insecticidal Characteristics of Selected Genotypic Variants" Viruses 11, no. 7: 581. https://doi.org/10.3390/v11070581
APA StyleAguirre, E., Beperet, I., Williams, T., & Caballero, P. (2019). Genetic Variability of Chrysodeixis Includens Nucleopolyhedrovirus (ChinNPV) and the Insecticidal Characteristics of Selected Genotypic Variants. Viruses, 11(7), 581. https://doi.org/10.3390/v11070581






