Abstract
The baculovirus nucleocapsid is formed through a rod-like capsid encapsulating a genomic DNA molecule of 80~180 kbp. The viral capsid is a large oligomer composed of many copies of various protein subunits. The assembly of viral capsids is a complex oligomerization process. The timing of expression of nucleocapsid-related proteins, transport pathways, and their interactions can affect the assembly process of preformed capsids. In addition, the selection of viral DNA and the injection of the viral genome into empty capsids are the critical steps in nucleocapsid assembly. This paper reviews the replication and recombination of baculovirus DNA, expression and transport of capsid proteins, formation of preformed capsids, DNA encapsulation, and nucleocapsid formation. This review will provide a basis for further study of the nucleocapsid assembly mechanism of baculovirus.
1. Introduction
Baculoviridae consists of a large family of circular dsDNA viruses with DNA molecules sized from 80 to 180 kbp that are encapsulated in enveloped rod-like capsids [1,2]. Baculoviruses are insect-specific pathogens that have been extensively applied for insect control [3]. A baculovirus generates budded virions (BVs) and occlusion-derived virions (ODVs), which are two phenotypes of virions in its life cycle. Although BVs and ODVs contain the same nucleocapsid, they perform different functions in virus infection. Nucleocapsids bud from the plasma membrane to obtain the envelopes with fusion proteins F or GP64 to generate BVs. BVs are able to launch successful systematic infection through entering into many cells and tissues mediated by the fusion proteins within a host larva [4,5]. Nucleocapsids are coated by their envelopes containing a series of per os infectivity factors from the host cell nuclear membrane to form ODVs that are further embedded in paracrystalline polyhedrin (Polh) structures to form occlusion bodies (OBs). Liquefied host insects release OBs into the environment to cause epidemics among host larvae [6]. Since the nucleocapsids are identical between BVs and ODVs [6], the proper assembly of the nucleocapsid is essential for the production of BVs and ODVs, and also for the infectivity of the baculovirus. With all of this in mind, revealing the mechanism of nucleocapsid assembly in baculoviruses is important in order to help scientists to understand baculovirus transmission.
2. Assembly of Nucleocapsids in Viruses
Virus nucleocapsid assembly is generally divided into two steps: production of capsid proteins and genome package. The assembly mechanism of viral nucleocapsids is very complex and varies between viruses, but most viruses generally adopt two main strategies. For most DNA and RNA viruses with genome sizes less than 20 kb, such as simian virus 40 (a small DNA virus) [7] and tobacco mosaic virus (a famous RNA virus) [8], the energy-independent system is used, where the nucleocapsid is assembled around the genome by free capsid proteins or subunits. On the contrary, large viruses tend to use the energy-dependent system, where the empty capsids are assembled, and then the genomic nucleic acids are recognized and pumped into the preformed capsids by ATP-driven motors. This process has been well studied in bacteriophage lambda [9] and herpes viruses [10]. Baculoviruses appear to assemble their nucleocapsids using the latter strategy.
3. Replication and Processing of Baculovirus Genomes
Baculovirus DNA replication is carried out in the virogenic stroma (VS), a specific region in the host cell nucleus. A baculovirus replicates its genome by rolling circle replication, which is a strand displacement replication occurring on circular dsDNA [11,12]. This type of replication can produce numerous copies of the genome in a short time. Additionally, baculovirus replication leads to a high recombination status [13,14]. Recombination-dependent replication plays an essential role in the replication cycle of many dsDNA viruses, especially DNA viruses with large genomes. Baculoviruses produce high molecular weight DNA with an apparently branched structure [15]. A sequence alignment revealed that all baculoviruses encode a gene (alkaline nuclease) homologous to the lambda red α exonuclease in bacteriophages, which is a typical exonuclease in the Red recombination system [16]. Alkaline nuclease interacts with a virus-encoded single-strand DNA binding protein LEF-3 to form a complex that is involved in DNA recombination [17,18].
The intermediates in recombination-dependent replication are complex branched structures that should be processed into unit-length cyclic dsDNA genomes before nucleocapsid assembly. VLF-1, a member of the lambda integrase family [19], may play a role in the genome processing. VLF-1 exhibits a structure-dependent binding activity to DNA molecules, with the highest binding activity to the recombination intermediate with a cruciform DNA structure [20]. However, the processing mechanism is unclear. In addition to VLF-1, the host recombination and repair system is likely involved in viral DNA processing, which has been identified in a lambda phage [21].
4. Expression and Transport of Structural Proteins of Baculovirus Nucleocapsid
VP39 (the major capsid protein) [22,23] and P6.9 [24], the DNA binding protein that condenses the viral genome to form a nucleocapsid core, were identified as components of baculovirus nucleocapsid in the 1980s. With the development of technology, especially proteomics [25,26], an increasing number of structural proteins have been identified, including 38K [25,26,27], VP1054 [26,28], Ac53 [29], VP91 [30], VLF-1 [26], BV/ODV-C42 [25,26], P78/83 [25,26], BV/ODV-EC27 [26,31], 49K [26,31], VP80 [25,26,32], and so on. Almost all of the genes encoding these structural proteins are conserved in all baculoviruses and are essential to virus proliferation. The most probable reason is that without any of these structural proteins, the nucleocapsid will be damaged and the virus will lose its infectivity.
Interestingly, all structural genes appear to be late transcribed by a virus-encoded RNA polymerase that is resistant to α-amanitin [6]. The separation between early and late transcription is the onset of DNA replication. The mechanism of the coordination and consistency between structural protein expression and viral genome replication is unclear. This may be related to the balance between genome replication and virus nucleocapsid formation. If structural genes are early genes, they will be expressed before the replication of the viral genome and then viral genomic DNAs are packaged into viral nucleocapsids, which leads to few genomic DNAs being used as templates for DNA replication. In this case, the replication of genomic DNA is disrupted, which is not conducive to the reproduction of viruses [6]. It is thought that two major features of newly synthesized DNA are beneficial to late transcription. The first feature is that newly synthesized (naked) DNA may promote the activation of late promoters until p6.9 or other DNA-binding proteins accumulate to the level at which viral DNA is condensed and transcription is suppressed. Another major feature is that in the process of lagging strand synthesis, the synthesis of Okazaki fragments produces a large number of nicks and RNA-DNA junctions. It is suggested that the unligated junctions of Okazaki fragments may be used as short-term enhancers of late transcription. Once DNA replication is completed, the loading sites of late gene activators can be eliminated by removing RNA primers and ligating lagging stands, thus terminating late transcription [6]. Therefore, the coordinated relationship between structural gene expression and genome replication may be as follows.
(1) When viral genomic DNA begins to be replicated, structural proteins begin to be expressed. At first, the number of genomic DNAs and structural proteins are small, and only small quantities of nucleocapsids are formed in the VS. In this state, the nucleocapsids are more easily transported outside the nucleus and budded from the cell membrane to produce BVs.
(2) When the number of viral genomic DNAs is increasing and the expression of structural proteins is increasing, more and more nucleocapsids are assembled, and nucleocapsids begin to arrange in bundles. In this case, it becomes more difficult for the nucleocapsid to be transported outside the nucleus. Then the nuclear membrane invades and envelopes the nucleocapsids to form ODVs with single or multiple particle(s).
(3) When the viral genome stops replicating, and the nucleocapsid assembly is finished, the unpackaged genome DNA is coated by P6.9 and the expression of structural proteins is shut off. In this case, the very late genes are easily overexpressed in large quantities. A large number of P10 and polyhedrin proteins are produced, and finally ODVs are embedded to form polyhedra.
Since nucleocapsid assembly is performed at the VS in the host cell nucleus, all structural proteins of nucleocapsids need to be transported to the nucleus, and then aggregate at the VS. The exact mechanism of nucleocapsid protein transport is unclear. It is suggested that VP1054 is responsible for the correct localization of capsid proteins (including VP39, P78/83, and BV/ODV-C42) in the nucleocapsid assembly site by regulating the delivery of nucleocapsid proteins [28]. Additionally, BV/ODV-C42 is capable of transporting P78/83 to cell nuclei. It is hypothesized that BV/ODV-C42 interacts with P78/83 to form a complex and facilitates transportation from the cytoplasm to the nucleus in insect cell infected with the virus [33].
5. Formation of the Preformed Capsid
Baculovirus nucleocapsid has a cylindrical sheath with an apical cap and a basal structure. When capsid proteins are transported to the VS, empty capsids will be assembled. The preformed capsids of nearly full length appear to begin with the assembly of the capsid sheath on a basal structure in the pockets of the VS [34]. The main component of the preformed capsid is VP39, which is the richest protein in viral particles [26]. Interestingly, a lot of long empty tubular structures were detected within the matrix of the VS or accumulated at the edge of the nucleus in the insect cells when a variety of different individual viral genes were deleted from the viral genome, including ac53 [29], 38k [27], pk1 [1,35], vp1054 [36], bv/odv-c42 [31], p49 [31], bv/odv-ec27 [31], vlf-1 [37], and p6.9 [26], most of which are viral genes involved in nucleocapsid formation. It is unclear why the preformed capsid in cells transfected with these mutant viruses is not an empty capsid of suitable length, but an aberrant long tubular structure. The reasons may be as follows. (1) The normal empty capsids of nearly full length containing the apical cap, the cylindrical sheath, and the basal structure are able to be assembled in cells infected with replicative viable baculovirus (Figure 1A). (2) The normal empty capsids tend to grow longer and longer when they are not packaged into viral genomes in time (Figure 1B). (3) If the basal structure fails to be formed, and excess VP39 tends to self-assemble the long tubular structure (Figure 1C).
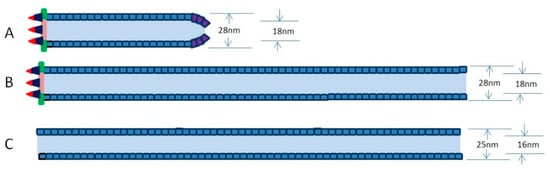
Figure 1.
Simplified schemes of preformed capsid and tubular structure in cells infected with viable viruses (A) or inviable viruses (B,C). (A) A preformed capsid of nearly full length containing an apical cap, a cylindrical sheath, and a basal structure. (B) A preformed capsid grows longer and longer as a long tube when they are not packaged into viral genomes in time. (C) Excess VP39 proteins tend to self-assemble into the long tubular structure. In (A,B), the tubes are similar to W-tube (outer diameter of 28 nm and inner diameter of 18 nm), and the tube in (C) is similar to an N-tube (outer diameter of 25 nm and inner diameter of 16 nm).
The basal structure may consist of VLF-1 [37,38,39], BV/ODV-C42 [40], P78/83 [40], BV/ODV-EC27 [41], and other proteins. VLF-1 is a structural protein that is detected in two types of virions (BV and ODV) [42] and localizes to the ends of nucleocapsid [37]. P78/83 is a Wiskott-Aldrich syndrome protein (WASP)-like protein that activates Arp2/3 to drive the polymerization of G-actin into F-actin. The actin polymerization is the main driving force of nucleocapsid movement in cells [43,44]. However, P78/83 is not able to enter into the cell nucleus without the aid of BV/ODV-C42, which contains a nuclear localization signal [33,45]. The interaction of BV/ODV-EC27 with BV/ODV-C42 is detected in yeast two-hybrid assay, so BV/ODV-EC27 may be one of components of the basal structure [41]. VP39 is the only detectable component of the cylindrical sheath [31]. 38K, VP1054, Ac53, VP91, 49K, and VP80 are also classified as minor capsid proteins, but the localizations of these proteins in nucleocapsids are unclear. In cells transfected with baculovirus genome with 49k gene deletion, some nucleocapsids appeared to be fully formed, but were unenveloped in the nucleus [46]. As such, the 49K may localize to the cap of nucleocapsids, because association of nucleocapsids with de novo envelopes is apparently mediated through the cap structure [34].
It has been suggested that VP39 self-assembles to form preformed capsid in host cells infected with baculovirus [31]. Recent report showed that Helicoverpa armigera nucleopolyhedrovirus (an Alphabaculovirus) VP39 expressed in Escherichia. coli self-assembled into long tubular structures in vitro. These long tubular structures contained two tube types, one was an N-tube (25 nm outer diameter and 16 nm inner diameter), and the other was a W-tube (28 nm outer diameter and 18 nm inner diameter) [47]. In Spodoptera litura granulosis virus (SpliGV, a Betabaculovirus), the cylindrical sheath of the empty nucleocapsid was detected to be similar to a W-tube [48]. The empty nucleocapsids of SpliGV extracted from granuloses are very similar to the normal nucleocapsid and contain the apical cap, the cylindrical sheath, and the basal structure [48]. Interestingly, the N-tubes are about 90% of the in vitro synthesis system, and the W-tubes are about 10% in vitro synthesis system, indicating that N-tubes are more easily formed in vitro [47]. As such, the assembly of the capsid sheath may need the help of other proteins in vivo. In our opinion, the capsid sheath containing the basal structure may be assembled according to the W-tube style (Figure 1B), while the capsid sheath lacking the basal structure may tend to be assembled according to the N-tube style in vivo (Figure 1C). Further investigation is required for a definitive confirmation.
6. DNA Packaging and Nucleocapsid Formation
Once viral DNA begins to replicate and its concentration becomes higher and higher, it triggers the viral DNA to be packaged into preformed capsid. DNA packaging may be coordinated with genome production. It has been suggested that the proteins that influence the replication of genome and DNA condensation may also influence the assembly of nucleocapsids. The knockout of DBP [49] or P6.9 [50] causes the empty tubular structure to be produced. P6.9 encoded by virus is a basic DNA-binding protein with only 50 aa residues [51] and is abundant in nucleocapsids [26]. Small basic proteins are able to condense genomic DNA and aid the packaging of DNA into capsids in DNA viruses. Host histones are frequently used for DNA packaging in some DNA viruses [52,53,54]. Interestingly, the phosphorylation status of P6.9 is very important to nucleocapsid assembly and viral gene transcription [55,56,57]. It has been reported that newly synthesized P6.9 is transiently phosphorylated [55], and hyperphosphorylation of P6.9 has been reported to be involved in viral very late gene hyperexpression [57]. However, the dephosphorylation of P6.9 plays an important role in viral genome packaging [24,58,59]. A recent report showed that 38K mediates the dephosphorylation of P6.9, which is essential to nucleocapsid assembly [60], suggesting that genomic DNA coated with dephosphorylated P6.9 is allowed to be condensed and packaged into the nucleocapsid.
In phages and some large viruses, genomic DNA is packaged into a preformed capsid by an ATP-driven motor that includes a portal protein and terminase-type enzyme(s) or other translocases. The portal protein acts as a channel for the injection of DNA, and the translocases pump the DNA into the preformed capsid [61]. This mechanism of packaging genomic DNA may also be used in baculoviruses. Combining the early microscopy observation of Autographa californica multiple nucleopolyhedrovirus nucleocapsid morphogenesis [34] with other knowledge of nucleocapsid assembly [6,61], we propose a model of baculovirus DNA packaging. In this model, DNA packaging may be generally divided into three steps. (1) The viral genome is docked onto the portal situated at the cap of a preformed capsid by packaging proteins (Figure 2A,B). (2) The genome is pumped into the preformed capsid through the portal by a packaging motor with the help of ATP hydrolysis (Figure 2C,D). (3) Packaging proteins are released from the nucleocapsid when the genome is fully packaged. The portal of the capsid is blocked with structure protein(s) to form mature nucleocapsid (Figure 2E,F).
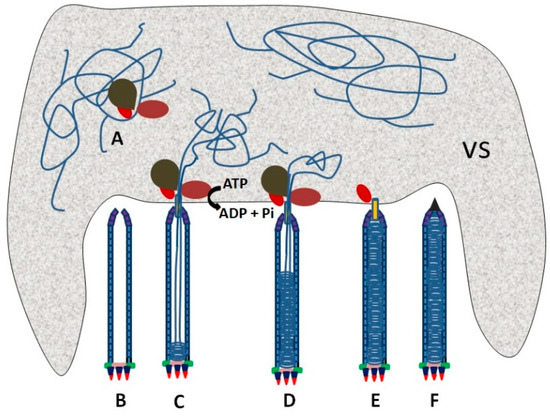
Figure 2.
Theoretical schematic of the genome packaging in a baculovirus. (A) A cis-acting element is recognized by a packaging protein complex. (B) A preformed capsid is assembled and waits for docking of the viral genome onto it though the packaging complex. (C,D) The viral genome is packaged through a portal at the apical cap of the preformed capsid with the aid of ATP hydrolysis. (E) The packaging complex is released after a genome DNA with one unit length is packaged. (F) The portal of a capsid is blocked with structure protein(s) to form mature nucleocapsid. The shaded area is the virogenic stroma (VS).
In order to ensure that only the viral genome is packaged, recognition of a specific sequence in the viral genome is essential for nucleocapsid assembly (Figure 2A). It is suggested that VP1054 may be involved in DNA recognition. VP1054 is similar to PURα family proteins and is able to bind to GGN-rich sequences [36]. A GGN-rich region in p78/83 was detected to interact with VP1054 [36]. Moreover, VP1054 was able to interact with the capsid protein 38K that catalyzed the dephosphorylation of P6.9 in genome packaging [62]. Interestingly, a cis-acting element named NAE containing eight conserved A/T-rich regions was identified in the ac83 coding region and is essential to DNA packaging [2]. Homologs of NAE are detected only in alphabaculoviruses. Another essential cis-acting element conserved in alphabaculoviruses is found in the ac152 coding region [63]. However, it is unknown which protein(s) can recognize those cis-acting elements.
To date, the composition of the baculovirus motor complex is unclear. Ac66 may act as a candidate for a motor protein. Ac66 is related to some motor proteins [6]. However, the function of Ac66 in nucleocapsid assembly still needs to be explored. Additionally, actin plays an important role in DNA packaging. Structure components of nucleocapsid are able to interact with actin in the infected cell nucleus, indicating that actin is transported from the cytoplasm into the nucleus [64]. Empty tubular structures were detected in cells when actin polymerization was blocked [65], suggesting that actin is involved in genome packaging into preformed capsids.
7. Outstanding Question and Conclusions
An outstanding question concerns the parameters that regulate the length of the nucleocapsid to fit the size of the encapsulated DNA molecule. It has been reported that the length of capsids may be resilient to genome size [66]. It is unknown whether the lengths of preformed capsids could be changed according to the size of the genome. In addition, it is also possible that a preformed capsid of a fixed length could be elongated in DNA packaging according to the size of the genome. A lot of aberrant long electron-lucent tubular structures are frequently observed in cells infected with gene-deleted baculoviruses, suggesting that the nucleocapsid assembly of baculoviruses is very easy to disturb. Recently, an increasing number of nucleocapsid proteins and cis-acting elements were identified in order to expand our knowledge of baculovirus nucleocapsid assembly. Here, we made an attempt to review baculovirus nucleocapsid assembly. We believe that the mechanism of nucleocapsid assembly will aid us to better understand the life cycle of baculoviruses.
Author Contributions
All authors contributed sections for the completion of this manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (Grant No. 31600122).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Liang, C.; Li, M.; Dai, X.; Zhao, S.; Hou, Y.; Zhang, Y.; Lan, D.; Wang, Y.; Chen, X. Autographa californica multiple nucleopolyhedrovirus PK-1 is essential for nucleocapsid assembly. Virology 2013, 443, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Pan, M.; Zhu, S.; Zhang, H.; Wu, W.; Yuan, M.; Yang, K. The Autographa californica multiple nucleopolyhedrovirus ac83 gene contains a cis-acting element that is essential for nucleocapsid assembly. J. Virol. 2017, 91, e02110-16. [Google Scholar] [CrossRef] [PubMed]
- Qin, F.; Xu, C.; Lei, C.; Hu, J.; Sun, X. Autographa californica multiple nucleopolyhedrovirus enters host cells via clathrin-mediated endocytosis and direct fusion with the plasma membrane. Viruses 2018, 10, 632. [Google Scholar] [CrossRef] [PubMed]
- Kadlec, J.; Loureiro, S.; Abrescia, N.G.; Stuart, D.I.; Jones, I.M. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat. Struct. Mol. Biol. 2008, 15, 1024–1030. [Google Scholar] [CrossRef] [PubMed]
- Blissard, G.W.; Theilmann, D.A. Baculovirus entry and egress from insect cells. Annu. Rev. Virol. 2018, 5, 113–139. [Google Scholar] [CrossRef] [PubMed]
- Rohrmann, G.F. Baculovirus Molecular Biology, 3rd ed.; National Library of Medicine (US), National Center for Biotechnology Information: Bethesda, MD, USA, 2013. [Google Scholar]
- Ben-nun-Shaul, O.; Bronfeld, H.; Reshef, D.; Schueler-Furman, O.; Oppenheim, A. The SV40 capsid is stabilized by a conserved pentapeptide hinge of the major capsid protein VP1. J. Mol. Biol. 2009, 386, 1382–1391. [Google Scholar] [CrossRef] [PubMed]
- Chu, S.; Brown, A.D.; Culver, J.N.; Ghodssi, R. Tobacco mosaic virus as a versatile platform for molecular assembly and device fabrication. Biotechnol. J. 2018, 13, e1800147. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Maluf, N.K.; Catalano, C.E. Packaging of a unit-length viral genome: The role of nucleotides and the gpD decoration protein in stable nucleocapsid assembly in bacteriophage lambda. J. Mol. Biol. 2008, 383, 1037–1048. [Google Scholar] [CrossRef] [PubMed]
- Dohner, K.; Ramos-Nascimento, A.; Bialy, D.; Anderson, F.; Hickford-Martinez, A.; Rother, F.; Koithan, T.; Rudolph, K.; Buch, A.; Prank, U.; et al. Importin alpha1 is required for nuclear import of herpes simplex virus proteins and capsid assembly in fibroblasts and neurons. PLoS Pathog. 2018, 14, e1006823. [Google Scholar] [CrossRef] [PubMed]
- Leisy, D.J.; Rohrmann, G.F. Characterization of the replication of plasmids containing hr sequences in baculovirus-infected Spodoptera frugiperda cells. Virology 1993, 196, 722–730. [Google Scholar] [CrossRef] [PubMed]
- Oppenheimer, D.I.; Volkman, L.E. Evidence for rolling circle replication of Autographa californica M nucleopolyhedrovirus genomic DNA. Arch. Virol. 1997, 142, 2107–2113. [Google Scholar] [CrossRef] [PubMed]
- Kamita, S.G.; Maeda, S.; Hammock, B.D. High-frequency homologous recombination between baculoviruses involves DNA replication. J. Virol. 2003, 77, 13053–13061. [Google Scholar] [CrossRef] [PubMed]
- Crouch, E.A.; Passarelli, A.L. Genetic requirements for homologous recombination in Autographa californica nucleopolyhedrovirus. J. Virol. 2002, 76, 9323–9334. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Martin, D.W.; Weber, P.C. Replication of simian virus 40 origin-containing DNA during infection with a recombinant Autographa californica multiple nuclear polyhedrosis virus expressing large T antigen. J. Virol. 1997, 71, 501–506. [Google Scholar] [PubMed]
- Maresca, M.; Erler, A.; Fu, J.; Friedrich, A.; Zhang, Y.; Stewart, A.F. Single-stranded heteroduplex intermediates in lambda Red homologous recombination. BMC Mol. Biol. 2010, 11, 54. [Google Scholar] [CrossRef] [PubMed]
- Mikhailov, V.S.; Okano, K.; Rohrmann, G.F. Baculovirus alkaline nuclease possesses a 5′-->3′ exonuclease activity and associates with the DNA-binding protein LEF-3. J. Virol. 2003, 77, 2436–2444. [Google Scholar] [CrossRef] [PubMed]
- Mikhailov, V.S.; Okano, K.; Rohrmann, G.F. Specificity of the endonuclease activity of the baculovirus alkaline nuclease for single-stranded DNA. J. Biol. Chem. 2004, 279, 14734–14745. [Google Scholar] [CrossRef] [PubMed]
- McLachlin, J.R.; Miller, L.K. Identification and characterization of vlf-1, a baculovirus gene involved in very late gene expression. J. Virol. 1994, 68, 7746–7756. [Google Scholar] [PubMed]
- Mikhailov, V.S.; Rohrmann, G.F. Binding of the baculovirus very late expression factor 1 (VLF-1) to different DNA structures. BMC Mol. Biol. 2002, 3, 14. [Google Scholar] [CrossRef]
- Poteete, A.R. What makes the bacteriophage lambda Red system useful for genetic engineering: Molecular mechanism and biological function. FEMS Microbiol. Lett. 2001, 201, 9–14. [Google Scholar] [PubMed]
- Zhang, J.; Feng, M.; Fan, Y.; Xu, W.; Zheng, Q.; Wu, X. Networks of protein-protein interactions among structural proteins of budded virus of Bombyx mori nucleopolyhedrovirus. Virology 2018, 518, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Katsuma, S.; Kokusho, R. A Conserved Glycine Residue Is Required for Proper Functioning of a Baculovirus VP39 Protein. J. Virol. 2017, 91, JVI-02253. [Google Scholar] [CrossRef] [PubMed]
- Singh, C.P.; Singh, J.; Nagaraju, J. bmnpv-miR-3 facilitates BmNPV infection by modulating the expression of viral P6.9 and other late genes in Bombyx mori. Insect Biochem. Mol. Biol. 2014, 49, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Hou, D.; Zhang, L.; Deng, F.; Fang, W.; Wang, R.; Liu, X.; Guo, L.; Rayner, S.; Chen, X.; Wang, H.; et al. Comparative proteomics reveal fundamental structural and functional differences between the two progeny phenotypes of a baculovirus. J. Virol. 2013, 87, 829–839. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Deng, F.; Hou, D.; Zhao, Y.; Guo, L.; Wang, H.; Hu, Z. Proteomics of the Autographa californica nucleopolyhedrovirus budded virions. J. Virol. 2010, 84, 7233–7242. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Lin, T.; Pan, L.; Yu, M.; Li, Z.; Pang, Y.; Yang, K. Autographa californica multiple nucleopolyhedrovirus nucleocapsid assembly is interrupted upon deletion of the 38K gene. J. Virol. 2006, 80, 11475–11485. [Google Scholar] [CrossRef] [PubMed]
- Guan, Z.; Zhong, L.; Li, C.; Wu, W.; Yuan, M.; Yang, K. The Autographa californica multiple nucleopolyhedrovirus ac54 gene is crucial for localization of the major capsid protein VP39 at the site of nucleocapsid assembly. J. Virol. 2016, 90, 4115–4126. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, Z.; Wu, W.; Li, L.; Yuan, M.; Pan, L.; Yang, K.; Pang, Y. Autographa californica multiple nucleopolyhedrovirus ac53 plays a role in nucleocapsid assembly. Virology 2008, 382, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Wang, W.; Wang, Y.; Yuan, M.; Yang, K. The baculovirus core gene ac83 is required for nucleocapsid assembly and per os infectivity of Autographa californica nucleopolyhedrovirus. J. Virol. 2013, 87, 10573–10586. [Google Scholar] [CrossRef] [PubMed]
- Vanarsdall, A.L.; Pearson, M.N.; Rohrmann, G.F. Characterization of baculovirus constructs lacking either the Ac 101, Ac 142, or the Ac 144 open reading frame. Virology 2007, 367, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.D.; Xu, Y.P.; Yu, L.L.; Lang, G.J.; Tian, C.H.; Zhao, J.F.; Zhang, C.X. Characterization of a Bombyx mori nucleopolyhedrovirus with Bmvp80 disruption. Virus Res. 2008, 138, 81–88. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Q.; Liang, C.; Song, J.; Li, N.; Shi, H.; Chen, X. Autographa californica multiple nucleopolyhedrovirus nucleocapsid protein BV/ODV-C42 mediates the nuclear entry of P78/83. J. Virol. 2008, 82, 4554–4561. [Google Scholar] [CrossRef]
- Fraser, M.J. Ultrastructural observations of virion maturation in Autographa californica nuclear polyhedrosis virus infected Spodoptera frugiperda cell cultures. J. Ultrastruct. Mol. Struct. Res. 1986, 95, 189–195. [Google Scholar] [CrossRef]
- Liang, C.; Su, X.; Xu, G.; Dai, X.; Zhao, S. Autographa californica multiple nucleopolyhedrovirus PK1 is a factor that regulates high-level expression of very late genes in viral infection. Virology 2017, 512, 56–65. [Google Scholar] [CrossRef]
- Marek, M.; Romier, C.; Galibert, L.; Merten, O.W.; van Oers, M.M. Baculovirus VP1054 is an acquired cellular PURalpha, a nucleic acid-binding protein specific for GGN repeats. J. Virol. 2013, 87, 8465–8480. [Google Scholar] [CrossRef]
- Vanarsdall, A.L.; Okano, K.; Rohrmann, G.F. Characterization of the role of very late expression factor 1 in baculovirus capsid structure and DNA processing. J. Virol. 2006, 80, 1724–1733. [Google Scholar] [CrossRef]
- Vanarsdall, A.L.; Okano, K.; Rohrmann, G.F. Characterization of a baculovirus with a deletion of vlf-1. Virology 2004, 326, 191–201. [Google Scholar] [CrossRef]
- Li, Y.; Wang, J.; Deng, R.; Zhang, Q.; Yang, K.; Wang, X. vlf-1 deletion brought AcMNPV to defect in nucleocapsid formation. Virus Genes 2005, 31, 275–284. [Google Scholar] [CrossRef]
- Russell, R.L.; Funk, C.J.; Rohrmann, G.F. Association of a baculovirus-encoded protein with the capsid basal region. Virology 1997, 227, 142–152. [Google Scholar] [CrossRef]
- Braunagel, S.C.; Guidry, P.A.; Rosas-Acosta, G.; Engelking, L.; Summers, M.D. Identification of BV/ODV-C42, an Autographa californica nucleopolyhedrovirus orf101-encoded structural protein detected in infected-cell complexes with ODV-EC27 and p78/83. J. Virol. 2001, 75, 12331–12338. [Google Scholar] [CrossRef]
- Yang, S.; Miller, L.K. Expression and mutational analysis of the baculovirus very late factor 1 (vlf-1) gene. Virology 1998, 245, 99–109. [Google Scholar] [CrossRef]
- Goley, E.D.; Ohkawa, T.; Mancuso, J.; Woodruff, J.B.; D’Alessio, J.A.; Cande, W.Z.; Volkman, L.E.; Welch, M.D. Dynamic nuclear actin assembly by Arp2/3 complex and a baculovirus WASP-like protein. Science 2006, 314, 464–467. [Google Scholar] [CrossRef]
- Ohkawa, T.; Volkman, L.E.; Welch, M.D. Actin-based motility drives baculovirus transit to the nucleus and cell surface. J. Cell Biol. 2010, 190, 187–195. [Google Scholar] [CrossRef]
- Li, K.; Wang, Y.; Bai, H.; Wang, Q.; Song, J.; Zhou, Y.; Wu, C.; Chen, X. The putative pocket protein binding site of Autographa californica nucleopolyhedrovirus BV/ODV-C42 is required for virus-induced nuclear actin polymerization. J. Virol. 2010, 84, 7857–7868. [Google Scholar] [CrossRef]
- McCarthy, C.B.; Dai, X.; Donly, C.; Theilmann, D.A. Autographa californica multiple nucleopolyhedrovirus ac142, a core gene that is essential for BV production and ODV envelopment. Virology 2008, 372, 325–339. [Google Scholar] [CrossRef][Green Version]
- Rao, G.; Fu, Y.; Li, N.; Yin, J.; Zhang, J.; Wang, M.; Hu, Z.; Cao, S. Controllable assembly of flexible protein nanotubes for loading multifunctional modules. ACS Appl. Mater. Interfaces 2018, 10, 25135–25145. [Google Scholar] [CrossRef]
- Burley, S.K.; Miller, A.; Harrap, K.A.; Kelly, D.C. Structure of the baculovirus nucleocapsid. Virology 1982, 120, 433–440. [Google Scholar] [CrossRef]
- Vanarsdall, A.L.; Mikhailov, V.S.; Rohrmann, G.F. Characterization of a baculovirus lacking the DBP (DNA-binding protein) gene. Virology 2007, 364, 475–485. [Google Scholar] [CrossRef]
- Wang, M.; Tuladhar, E.; Shen, S.; Wang, H.; van Oers, M.M.; Vlak, J.M.; Westenberg, M. Specificity of baculovirus P6.9 basic DNA-binding proteins and critical role of the C terminus in virion formation. J. Virol. 2010, 84, 8821–8828. [Google Scholar] [CrossRef]
- Wilson, M.E.; Mainprize, T.H.; Friesen, P.D.; Miller, L.K. Location, transcription, and sequence of a baculovirus gene encoding a small arginine-rich polypeptide. J. Virol. 1987, 61, 661–666. [Google Scholar]
- Fang, C.Y.; Shen, C.H.; Wang, M.; Chen, P.L.; Chan, M.W.; Hsu, P.H.; Chang, D. Global profiling of histone modifications in the polyomavirus BK virion minichromosome. Virology 2015, 483, 1–12. [Google Scholar] [CrossRef]
- Kumar, M.A.; Kasti, K.; Balakrishnan, L.; Milavetz, B. Directed nucleosome sliding during the formation of the simian virus 40 particle exposes DNA sequences required for early transcription. J. Virol. 2019, 93, e01678-18. [Google Scholar] [CrossRef]
- Gautam, D.; Johnson, B.A.; Mac, M.; Moody, C.A. SETD2-dependent H3K36me3 plays a critical role in epigenetic regulation of the HPV31 life cycle. PLoS Pathog. 2018, 14, e1007367. [Google Scholar] [CrossRef]
- Kelly, D.C.; Lescott, T. Baculovirus replication: Phosphorylation of polypeptides synthesized in Trichoplusia ni nuclear polyhedrosis virus-infected cells. J. Gen. Virol. 1984, 65 Pt 7, 1183–1191. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, H.; Fang, Z.; Yuan, M.; Yang, K.; Pang, Y. Distribution and phosphorylation of the basic protein P6.9 of Autographa californica nucleopolyhedrovirus. J. Virol. 2012, 86, 12217–12227. [Google Scholar] [CrossRef]
- Li, A.; Zhao, H.; Lai, Q.; Huang, Z.; Yuan, M.; Yang, K. Posttranslational modifications of baculovirus protamine-Like protein P6.9 and the significance of its hyperphosphorylation for viral very late gene hyperexpression. J. Virol. 2015, 89, 7646–7659. [Google Scholar] [CrossRef]
- Funk, C.J.; Consigli, R.A. Phosphate cycling on the basic protein of Plodia interpunctella granulosis virus. Virology 1993, 193, 396–402. [Google Scholar] [CrossRef]
- Oppenheimer, D.I.; Volkman, L.E. Proteolysis of p6.9 induced by cytochalasin D in Autographa californica M nuclear polyhedrosis virus-infected cells. Virology 1995, 207, 1–11. [Google Scholar] [CrossRef]
- Lai, Q.; Wu, W.; Li, A.; Wang, W.; Yuan, M.; Yang, K. The 38K-mediated specific dephosphorylation of the viral core protein P6.9 plays an important role in the nucleocapsid assembly of Autographa californica multiple nucleopolyhedrovirus. J. Virol. 2018, 92, JVI-01989. [Google Scholar] [CrossRef]
- Chelikani, V.; Ranjan, T.; Kondabagil, K. Revisiting the genome packaging in viruses with lessons from the “Giants”. Virology 2014, 466–467, 15–26. [Google Scholar] [CrossRef]
- Wu, W.; Liang, H.; Kan, J.; Liu, C.; Yuan, M.; Liang, C.; Yang, K.; Pang, Y. Autographa californica multiple nucleopolyhedrovirus 38K is a novel nucleocapsid protein that interacts with VP1054, VP39, VP80, and itself. J. Virol. 2008, 82, 12356–12364. [Google Scholar] [CrossRef]
- Kikhno, I. Identification of a conserved non-protein-coding genomic element that plays an essential role in Alphabaculovirus pathogenesis. PLoS ONE 2014, 9, e95322. [Google Scholar] [CrossRef]
- Lanier, L.M.; Volkman, L.E. Actin binding and nucleation by Autographa california M nucleopolyhedrovirus. Virology 1998, 243, 167–177. [Google Scholar] [CrossRef]
- Volkman, L.E. Autographa californica MNPV nucleocapsid assembly: Inhibition by cytochalasin D. Virology 1988, 163, 547–553. [Google Scholar] [CrossRef]
- Kool, M.; Voncken, J.W.; van Lier, F.L.; Tramper, J.; Vlak, J.M. Detection and analysis of Autographa californica nuclear polyhedrosis virus mutants with defective interfering properties. Virology 1991, 183, 739–746. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).