Abstract
Soybean mosaic virus (SMV) is one of the most destructive viral diseases in soybeans (Glycine max). In this study, an interaction between the SMV P3 protein and cytochrome b5 was detected by yeast two-hybrid assay, and bimolecular fluorescence complementation assay showed that the interaction took place at the cell periphery. Further, the interaction was confirmed by co-immunoprecipitation analysis. Quantitative real-time polymerase chain reaction analysis revealed that GmCYB5 gene was differentially expressed in resistant and susceptible soybean plants after inoculation with SMV-SC15 strain. To test the involvement of this gene in SMV resistance, the GmCYB5 was silenced using a bean pod mottle virus (BPMV)-based vector construct. Results showed that GmCYB5-1 was 83% and 99% downregulated in susceptible (NN1138-2) and resistant (RN-9) cultivars, respectively, compared to the empty vector-treated plants. Silencing of GmCYB5 gene promotes SMV replication in soybean plants. Our results suggest that during SMV infection, the host CYB5 protein targets P3 protein to inhibit its proliferation. Taken together, these results suggest that CYB5 is an important factor in SMV infection and replication in soybeans, which could help soybean breeders develop SMV resistant soybean cultivars.
1. Introduction
Soybean (Glycine max (L.) Merr.) is an important protein and oil crop. Soybean mosaic virus (SMV), a member of Potyvirus, is seed-borne and aphid-transmitted. SMV causes severe reduction of soybean yield and destroys the seed quality [1,2]. In the United States, 98 isolates of SMV were divided into seven strains, namely G1 to G7, according to differential phenotypic reactions to five resistant and two susceptible varieties of soybeans [3,4]. Resistance loci containing dominant resistance (R) genes named Rsv1, Rsv3, and Rsv4 were mapped to chromosomes 13, 14, and 2, respectively [5,6,7,8,9]. Rsv1 was resistant to G1–G6 [10] while Rsv3 was resistant to strains G5–G7 [5]. Rsv4 was initially thought to provide resistance against all North American strains of SMV but later was shown to exhibit a late susceptible phenotype to strains G1 and G2 [11,12,13,14,15]. Based on the reaction to specific soybean cultivars, the SMV isolates were classified into 21 strains in China and were named SC1 to SC21 [16,17,18]. Resistance derived from the resistant to SC (Rsc) loci to the strains from China have been mapped to chromosome 2 in Kefeng 1, chromosome 13 in Qihuang and chromosome 14 in Dabaima, respectively [19,20,21].
SMV has a single-stranded positive sense RNA genome of ~9.6 kb in length, which is similar to other Potyvirus, in processing to eleven mature multi-functional proteins. Of these, the P3 protein is reported to participate in viral replication, movement, virulence, pathogenesis and in one case avirulence [22,23,24,25,26,27,28,29,30,31]. However, the precise biochemical function of SMV P3 protein, and the potyviral P3 protein in general, is largely uncharacterized due to the lack of structural or functional motifs in its sequence. Recent researches revealed that the P3 cistron plays a crucial role in SMV virulence on Rsv1 and Rsv4 soybean genotypes [32,33]. The P3 protein has also been reported to interact with ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO), thus affecting the systematic development of host plants [34]. The endoplasmic reticulum (ER)-localized SMV P3 targets eukaryotic elongation factor 1 alpha (eEF1A) to facilitate its survival in the soybean plant [35]. SMV P3 was also used to generate RNAi-mediated silencing plants. Transgenic lines exhibited stable and enhanced resistance to other Potyvirus [36].
Cytochrome B5 (CYB5) is a class of heme proteins associated with endoplasmic reticulum in plants, animals, and fungi. As a ubiquitous intercellular electron transporter, CYB5 participates in various redox reactions in cells thereby regulating the balance of reactive oxygen species (ROS) in plants. In plants, amino acid sequences of CYB5 have been identified in cauliflower [37], tobacco [38], and rice [39]. Sequence analysis showed that these proteins shared common characteristics of carboxyl terminal polar parts rich in positively-charged amino acids [40]. Previous studies have focused on the structural aspects of the CYB5 interaction with CYP450 monooxygenases [41] and the biochemical and kinetic aspects of CYB5 involved in the CYP450 monooxygenase reaction [42]. However, the function of CYB5 enzymes in virus infections, especially Potyvirus infections are unknown.
Based on the SMV P3 interaction network, a protein named GmCYB5 encoded by the Glyma18G154900 gene was selected for further characterization. Here, we studied the role of GmCYB5 in the process of SMV infection. We showed that GmCYB5 inhibited SMV proliferation by targeting the virus protein P3.
2. Materials and Methods
2.1. Plant Growth and Virus Strains
Soybean (Glycine max (L.)) cultivars NN1138-2 and RN-9, which are susceptible and resistant to SMV SC15 strain, respectively, were grown in an aphid-free greenhouse with day and night temperatures of 25 °C and 20 °C, in 65% relative humidity and during a 14 h photoperiod. We used SMV-SC15 strain in this study, which is one of the most virulent strains in China [18]. Fully expanded unifoliate leaves were mechanically inoculated by SMV-SC15. NMY51 strain of yeast was used in yeast two-hybrid analysis (Dualsystems Biotech, Schlieren, Switzerland), which is an ideal reporter strain for DUAL membrane screening systems which can be used to find novel interaction partners of a protein of interest by screening cDNA libraries, and compatible with most LexA based yeast two-hybrid systems. All the materials were provided by the National Center for Soybean Improvement, Nanjing Agricultural University, China.
2.2. Yeast Two-Hybrid Assay
A soybean cDNA library (~0.68 × 107 clones) from SMV-SC15 infected soybean (cv. NN1138-2) was cloned into the modified vector pPR3-N using Gateway technology. The P3 gene of SMV-SC15 was cloned in pBT3 and used as a bait to screen the library (3× clones) by co-transformation in yeast (NMY51). Yeast transformants expressing P3-interacting proteins were selected on synthetic dropout medium lacking tryptophan (Trp), leucine (Leu), histidine (His), and adenine (Ade). Yeast strains expressing P3 interactors were further assessed for β–galactosidase activity.
2.3. Interaction of GmCYB5-1 and P3 Using Y2H
The full length of GmCYB5-1 was cloned into prey vector pPR3 by the gateway system. The NMY51 yeast cells harboring the pBT3-P3 reporter plasmid was transformed with pPR3-GmCYB5-1 plasmid. The transformants were placed on agar plates with synthetic media containing dextrose lacking Leu, Trp, His, and Ade (SD-Leu-Trp-His-Ade), and incubated for two days at 30 °C. The surviving transformants were re-cultured until OD600 0.8 and dropped at concentrations ranging from 1:10, 1:100, 1:1000 to 1:10,000 on SD-Leu-Trp-His-Ade medium with α-x-gal at final concentration of 4 mg/mL to determine the interaction affinity.
2.4. Bimolecular Fluorescence Complementation (BiFC) Assays
BiFC assays were carried out as described before [43]. The interaction proteins were cloned to pSITE-n/cEYFP vectors [44] and transformed into Agrobacterium tumefaciens strain LBA4404. Positive agrobacteria which fused with reciprocal halves of EYFP were co-infiltrated into transgenic N. benthamiana plants expressing nuclear localized H2B protein with a CFP tag [45]. Leaf tissues were immersed in water after 2 days and checked by confocal microscopy using PLAPO60XWLSM (NA 1.0) objective. The interaction was confirmed using both combinations of reciprocal nEYFP/cEYFP fusion proteins in two separate experiments (three replicates per experiment).
2.5. Sequence Analysis of GmCYB5
To amplify GmCYB5-1, primers were designed by Primer Premier5.0 software (Premier Biosoft, Palo Alto, CA, USA) (Supplemental Table S1). Based on the Williams82 soybean reference genome annotation Glyma v1.1, the coding sequence (CDS) region of the GmCYB5-1 gene was cloned by polymerase chain reaction (PCR) using soybean cDNA as templates. The sequence alignment and phylogenetic analysis were performed by DNASTAR package [46].
2.6. Expression Analysis of GmCYB5
When the first pair of true leaves had developed, RN-9 and NN1138-2 plants were rub-inoculated by SMV-SC15. Leaf samples were collected from infected plants at 0, 2, 8, 12, and 24 h post-inoculation. In addition, root, stem, leaf, flower, and pod were sampled from NN1138-2 plants without virus inoculation. All samples were in triplicates and flash frozen in liquid nitrogen immediately after collection and subsequently stored at −80 °C until use. Total RNA was extracted by TRIZOL using a total RNA isolation kit (Tiangen, Beijing, China). cDNA was synthesized using HiScript ® II QRT SuperMix according to the manufacturer’s instructions (Vazyme Biotech, Nanjing, China). The expression levels of target genes were analyzed by qRT-PCR using a Light-Cycler 480 (Roche Diagnostics, Indianapolis, IN, USA). Three technical replicates were performed for each biological replicate. The target gene sequences used in this study were obtained from Soybean Genome Sequences. Primers were designed to amplify gene specific PCR products of <200 bp in size. Primer sequences are listed in Supplemental Table S1. B-Tubulin was used as an internal control to normalize the cDNA concentrations. qRT-PCR was carried out in a 96-well plate using SYBR Green Mix, with cycling program as 95 °C for 1 min followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s. Gene expression was quantified using the relative quantification (2−ΔΔCt) method. Each sample or treatment was tested in at least three biological replicates and the same experiment was performed twice.
2.7. Construction of Recombinant Plasmids, in Vitro Transcription, and Plant Inoculation
The BPMV vectors, pGG7R1 and pGG7R2, were provided by the National Center for Soybean Improvement, Nanjing Agricultural University, China. To construct the VIGS vector, a 126 bp (E3-D44) cDNA fragment conserved in GmCYB5 isoforms was amplified from NN1138-2 with the primers listed in Supplemental Table S1. The PCR product was inserted into the pGG7R2 vector. After transformation into DH5α competent cells, plasmids were extracted and then sequenced by Invitrogen Biotechnology Co. Ltd (Shanghai, China). Resulting sequences were analyzed by BLAST to confirm that the target fragment was inserted into the vector.
FpGG7R2-GmCYB5 construct, carrying the fragment to silence the GmCYB5, and pGG7R1 vector were transcribed after linearizing with Sal I and Sal I/Not I, respectively. Briefly, capped RNA transcripts were synthesized by incubating 30 μL of linearized plasmids in 50 μL of reaction mixture containing 10× Buffer, T7 RNA polymerase, rGTP, rATP, rCTP, rUTP, m7G5’cap, RNase inhibitor, and DEPC H2O at 37 °C for 2 h. In vitro transcription products of pGG7R1 and pGG7R2-CYB5 were mixed in a ratio of 1:1 to produce an infectious transcript, and then immediately inoculated on the fully expanded primary leaves of 7-day-old NN 1138-2 to generate the CYB5-silenced plant (SCYB5). For silencing CYB5 gene in RN-9 plant, the desiccated leaf tissues from of NN1138-2-SCYB5 which contain the infectious transcript (pGG7R2-CYB5 and pGG7R1) were mixed with inoculation buffer and then rub-inoculated on RN-9 primary leaves. Empty-vector (V) inoculated plants served as mock-inoculated controls.
2.8. Silencing Efficiency of GmCYB5
To confirm target gene silencing efficiency, treatment groups were inoculated with SCYB5, and control group were inoculated with V. When plants mosaic symptoms showed up, the infected leaves were collected and the RNA expression levels of GmCYB5 were analyzed by qRT-PCR.
2.9. Resistance Analysis of SCYB5
When the first pair of trifoliolate leaves developed, resistant (RN-9) and susceptible (NN1138-2) soybean cultivars were inoculated with tissue from CYB5 silencing plants, and V as the control. When the second pair of trifoliolate leaves developed, all the plants were inoculated with SMV-SC15. SMV-SC15-infected leaves from RN-9 and NN1138-2 were collected at 0, 4, 7, 10, and 14 dpi in triplicates, flash frozen in liquid nitrogen immediately, and stored at −80 °C.
The RNA accumulation of SMV was analyzed by qRT-PCR using SMV-CP (coat protein) gene primers (Supplemental Table S1), and SMV-CP protein concentration was analyzed by double-antibody sandwich-ELISA.
3. Results
3.1. P3 Protein Interact with GmCYB5-1
SMV P3 protein is a virulence determining factor. During SMV infection, there might be some host proteins promoting its infection and survival in the host plant. Therefore, we conducted a yeast two-hybrid (Y2H) screen to identify soybean proteins that interacted with the SMV P3 protein. Among the several identified P3-interacting proteins, GmCYB5-1 (Glyma18g154900) was selected for further characterization. The full length GmCYB5-1 was amplified from NN1138-2 and constructed into a yeast vector. The cells which contain both GmCYB5-1 and P3 were grown on synthetic dropout SD-Leu-Trp-His-Ade plates with and without X-α-gal at the dilutions of (1:10, 1:100, 1:1000, and 1:10,000). The results showed strong binding affinity between GmCYB5 and P3 proteins at 1:10 and 1:100 dilutions and weak binding affinity at 1:10,000 dilution (Figure 1A).
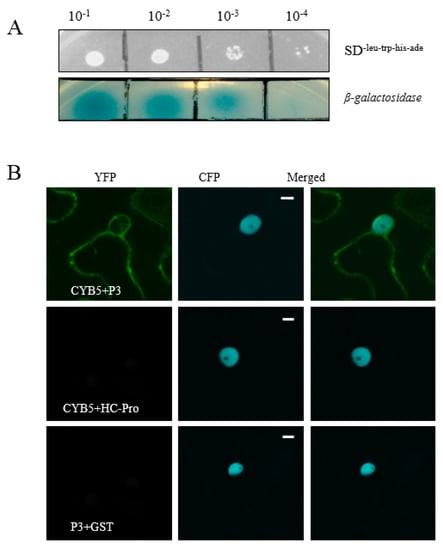
Figure 1.
GmCYB5-1 interacts with the P3 protein of soybean mosaic virus (SMV). (A) Interaction of SMV-P3 and GmCYB5 in yeast growing on selective medium SD-leu-trp-his-ade with X-α-gal at different concentrations. Images are representative two independent experiments for each interaction. (B) Bimolecular florescence complementation (BiFC) micrographs of 40× magnification at 48 h post-infiltration from plants co-expressing nYFP-GmCYB5 and cYFP-P3, with cYFP-HC-Pro and cYFP-GST are negative controls. Transgenic Nicotiana benthamiana, expressing CFP-H2B (nuclear localized histone 2B), were used for the BiFC assays. Images are representative of three separate infiltrations from two independent experiments for each interaction using both combinations of n/cYFP fused proteins. Scale bars: 10 µm.
Interaction between P3 and GmCYB5-1 proteins was also confirmed by bimolecular florescence complementation (BiFC) assays. In BiFC assays the reciprocal N- or C-terminal halves of enhanced yellow fluorescent proteins (nEYFP and cEYFP) were used to label the proteins, then labeled proteins were co-expressed in tobacco transiently. The results revealed that P3 interacted with GmCYB5-1 in plants; as YFP florescence was detected in plants co-infiltrated with P3 and GmCYB5-1. In contrast, P3 did not interact with GST (glutathione-S-transferase) and GmCYB5-1 did not interact with SMV helper component-proteinase (HC-Pro) (Figure 1B). Together, these results suggested that P3 could interact with GmCYB5-1.
3.2. Sequence Analysis of GmCYB5
The full length of GmCYB5-1 was amplified and cloned from both susceptible (NN1138-2) and resistant (RN-9) soybean cultivars. By sequence alignment two single nucleotide polymorphisms (SNPs) were identified in GmCYB5-1 of the two cultivars (Figure 2A). The Glycine. max genome contains three GmCYB5-like sequences including Glyma18g154900 (GmCYB5-1), Glyma03g080100 (GmCYB5-2), and Glyma08g354100 (GmCYB5-3); their predicted protein sequences shared a minimum of 70% and maximum of 98% identity with one another (Figure 2B). In order to determine the expression levels of GmCYB5 isoforms in different tissues, a quantitative (q)RT-PCR analysis was carried out and the results showed that transcript levels of GmCYB5 isoforms were higher in leaf tissue comparing to other analyzed tissues (cotyledon, stem, flower, and roots) (Figure 2C). To determine if GmCYB5 was involved in SMV infection, the expression pattern analysis was carried out in both susceptible (NN1138-2) and resistant (RN-9) cultivars with or without SMV infection using qRT-PCR. The three isoforms of GmCYB5 were all induced after infection with strain SC15 of SMV at the indicated time point in both susceptible and resistant plants (Figure 3A,B). Furthermore, the expression level of GmCYB5-1 was much higher than other isoforms after SMV inoculation. Taken together, these results indicated that GmCYB5 might be involved in SMV infection.
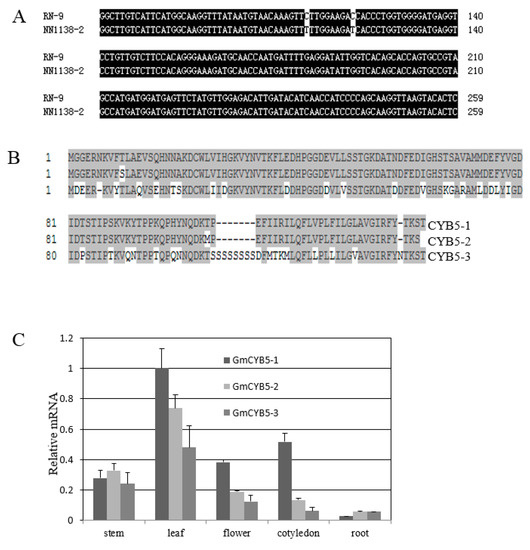
Figure 2.
Sequence analysis of GmCYB5. (A) The full cDNA sequence alignment of GmCYB5-1 from resistant (RN-9) and susceptible (NN1138-2) cultivars. (B) Amino acid sequence alignment of GmCYB5 isoforms was carried out using ClustalW in the Megalign program of the DNASTAR package, with identical residues shaded in gray. Numbers indicate the position of amino acid residues. Results are representative of two to three independent experiments. (C) Relative mRNA levels of GmCYB5 isoforms in different tissues of soybean plants (cv. NN1138-2) as determined by quantitative RT-PCR analysis. Error bars indicate SD (n = 3).
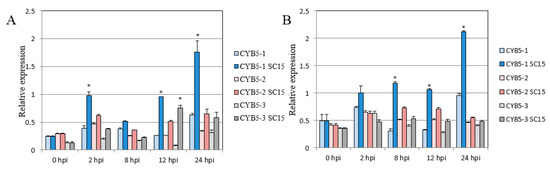
Figure 3.
Analysis of mRNA levels of CYB5 isoforms in soybean. Relative mRNA levels of GmCYB5 isoforms in (A) susceptible plants (cv. NN1138-2) or (B) resistant plants (cv. RN-9) with or without SMV (SC15) infection. Samples are collected at 0, 2, 8, 12, and 24 h post infection (hpi) and quantified by qRT-PCR. Error bars indicate SD (n = 3). Asterisks denote the significant difference between pathogen-treated and control plants for each respective isoform, t-test, p < 0.001. Results are representative of two–three independent experiments.
3.3. Silencing of GmCYB5
We next tested the role of GmCYB5 in resistance of soybean to SMV. We knocked down the GmCYB5 genes in soybean using the bean pod mottle virus (BPMV)-based VIGS (virus-induced gene silencing) vector [47,48]. A 126-nt fragment of GmCYB5-1 from NN1138-2 cultivar which showed high similarity among the three GmCYB5 isoforms, was selected and cloned (Figure 4A). The cloned fragment was verified by sequencing and subsequently inserted into the modified pGG7R2 vector. The pGG7R2-GmCYB5-1 construct was expected to knock down the expression of all isoforms simultaneously because the various isoforms share >82% nucleotide identity in the 126 nt fragment. Inoculated plants (cv. NN1138-2 and RN-9) with the construct were tested for GmCYB5-1 transcript level using qRT-PCR. Notably, mRNA level of GmCYB5-1 was reduced by 83% in the susceptible plant (NN1138-2) and 99% in the resistant plant (RN-9) in the GmCYB5 knock down plants (SCYB5) compared to control plants (inoculated with a BPMV control vector; containing a non-specific sequence, V) (Figure 4B). Silencing of GmCYB5 did not change the morphology of soybean plants.
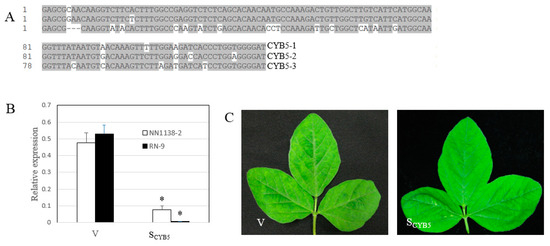
Figure 4.
Silencing GmCYB5 in soybean. (A) Sequence of GmCYB5 isoforms used for silencing. Sequence alignment was carried out using ClustalW in the Megalign program of the DNASTAR package, with identical residues shaded in gray. (B) Relative mRNA levels of GmCYB5-1 isoform in soybean plants (cv. NN1138-2 and RN-9) infected with empty vector (V), or vector targeting GmCYB5-1 (SCYB5) as determined by qRT-PCR analysis. Error bars indicate SD (n = 3). Asterisks denote significant difference in expression of the respective transcripts when compared to V plants, t-test, p < 0.001. Results are representative of two independent experiments. (C) Comparing symptoms of NN1138-2 plants with GmCYB5 mRNA expression (SCYB5) knocked down and that infected with the control bean pod mottle virus (BPMV) vector (V).
3.4. Knock Down of GmCYB5 Expression Promote SMV Accumulation in Soybean Plants
The V and SCYB5 plants (cvs. NN1138-2 and RN-9) were infected with the SC15 strain (virulent on NN1138-2, avirulent on RN-9) of SMV. The NN1128-2-SCYB5 plants developed more severe SMV-related symptoms compared to V plants (Figure 5A upper panel). In contrast, there was no visible SMV symptom of SCYB5 and V on RN-9 plants (Figure 5A lower panel). This correlated with the enzyme-linked immunosorbent assay (ELISA) analysis results of total protein extracts, which showed that there was no SMV accumulation in either V or SCYB5 RN-9 plants. Furthermore, SMV infection resulted in much higher induction of SMV coat protein (CP) expression in the SCYB5 plants than in V plants of NN1138-2 (Figure 5B). The expression level of SMV CP transcript was measured in RN-9 plants (SCYB5 and V) using qRT-PCR. SMV CP mRNA was detected at a low level at 4 dpi and increased in response to SMV infection in the SCYB5 plants (Figure 5C). The CP protein of SMV was detected by ELISA in SCYB5 and V plants of NN1138-2 cultivar. Based on the ELISA result there was more CP in SCYB5 plants than V plants in both inoculated (I) and systemic (S) leaves (Figure 5D). Together, these results indicated that knock down of GmCYB5 expression reduced the plant resistance to SMV.
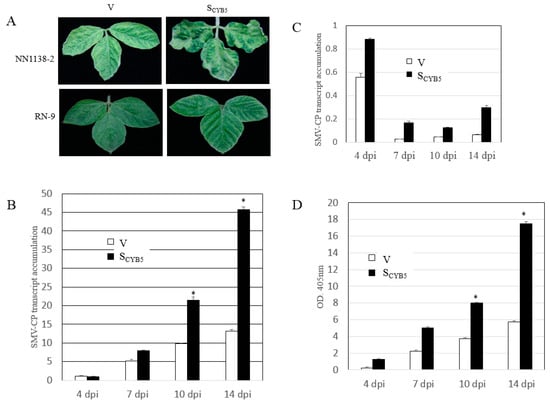
Figure 5.
Silencing GmCYB5 alters soybean response to SMV. (A) Visual symptoms of SMV-SC15 on GmCYB5-silenced plant (SCYB5) (right panels), control plant inoculated with BPMV vector (V) (left panels), and susceptible plant (NN1138-2) (upper panels) and resistant plants (RN-9) (lower panels). (B) qRT-PCR analysis of SMV coat protein (CP) level at indicated dpi in V and SCYB5 plants of susceptible cultivar (NN1138-2) and (C) resistant cultivar (RN-9). Error bars indicate SD (n = 3). Asterisks denote significant differences from V for each time point, t-test, p < 0.001. Results are representative of two to three independent experiments. (D) ELISA results of SMV CP level in V and SCYB5 plants (cv. NN1138-2) inoculated with SMV-SC15 at indicated dpi (4, 7 dpi from I, 10 and 14 dpi from S) with SMV SC15. Error bars indicate SD (n = 3). Asterisks denote significant difference from V, t-test, p < 0.001. Results are representative of three independent experiments.
4. Discussion
Protein–protein interactions are fundamental to the development and survival of viruses in host plants and can serve as excellent targets for identifying interacting factors related to host-pathogen interactions. A widely used method to characterize the function of known proteins is to identify their interacting factors and infer their roles based on the functions of known interacting factors. Here, we identified a protein of soybean plant (CYB5-1) that interacted with SMV P3 protein using yeast two-hybrid screening. However, as this screening uses DUAL membrane system which can result in a certain number of false positives [49], BiFC and co-immunoprecipitation were used to confirm the reliability of interaction. In the BiFC assay, the fluorescence was detected at cell periplasm. Characterizing soybean gene function by gene transformation has limited value due to low efficiency and high cost. However, the BPMV-VIGS system offers a convenient way to downregulate gene expression and was successfully introduced to analyze the candidate gene function in soybean [50]. In this research, the CYB5 function during SMV infection was characterized by BPMV-VIGS system. Since three isoforms of GmCYB5 share high sequence similarity, a conserved region was selected to generate the BPMV-VIGS construct to silence the genes to the maximum extent possible. By qRT-PCR, a 83% reduction of GmCYB5 transcript accumulation in the susceptible cultivar (NN 1138-2) and 99% reduction in the resistant cultivar (RN-9) were observed, as compared with the vector control plants (inoculated with an empty vector). The VIGS systems do not completely silence the expression of target genes. The silencing efficiency can be affected by inserted regions [51], cultivars [52], and environment conditions even for the same plant [53].
We next investigated whether silencing of GmCYB5 gene altered the soybean resistance to SMV. Severe SMV symptoms were observed on the SCYB5 plants compared to V plants. These results were consistent with mRNA accumulation of SMV CP that was measured by qRT-PCR in susceptible cultivar (NN1138-2) and resistant cultivar (RN-9), as well as the virion concentration that is represented by the level of SMV CP in susceptible cultivar (NN1138-2) in the ELISA result.
Previous research showed that the difference in resistance to SMV SC15 between RN-9 and NN1138-2 was mainly attributed to the GmPEX14 gene [54]. In this study, sequence analysis showed that there are two single nucleotide polymorphisms (SNPs) in GmCYB5-1 coding region between RN-9 and NN1138-2. After SMV inoculation, the resistant RN-9 plants showed extreme resistance, and no SMV symptom at all. However, a small amount of SMV was detected by qRT-PCR in inoculated leaves. A possible biochemical mechanism of extreme resistance could be the direct antiviral effect of reactive oxygen species (ROS) accumulation at infection sites [55]. The resistant RN-9 plants have accumulated a high level of ROS early after SMV inoculation [54]. The accumulation of ROS is usually linked to the host plant defense responses during the plant–virus interactions. A slower host response allows a certain degree of virus replication and movement resulting in oxidative stress including the ROS and programmed death of affected plant cells before conferring pathogen arrest (hypersensitive response, HR) [56].
CYB5 is known to be an electron transfer component in a number of oxidative reactions in biological tissues. Since CYB5 has relatively high mid-point redox potential, it can deliver only the second electron to oxyferrous CYP450 [57]. In fact, most studies suggest that with certain P450 forms and certain substrates, CYB5 supplies electrons to the intermediate oxygenated-ferrous-P450-substrate complex more rapidly than does NADPH-P450 oxidoreductase, which is accompanied by a decrease in H2O2 formation by the enzyme [58]. The symptom development associated with compatible cucumber mosaic virus- and zucchini yellow mosaic virus-infected plants showed an enhanced lipid peroxidation indicating an advanced disintegration of membranes [59]. The increased H2O2 accumulation in sunflower prevented an early increase in antioxidant activities after the Sunflower chlorotic mottle virus (SuCMoV) infection which thus can facilitate virus systemization in compatible interactions [60]. The strong systemic burst of H2O2 in Plum pox virus (PPV)-infected plants that exhibited severe symptoms might be attributed, at least in part, to a delayed and failed attempt by the host to elicit resistance to the virus in systemic tissues [61,62,63]. This agrees with other findings that demonstrate the pivotal role of timely ROS accumulation in virus resistance [64,65]. Similarly, as GmCYB5 was silenced in the susceptible cultivar, the host plants showed more susceptibility to SMV. This observation suggests that GmCYB5 is required for cellular ROS accumulation which is important in resistance to SMV by host plants.
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4915/11/6/546/s1, Table S1: Primer sequence.
Author Contributions
Funding acquisition, H.Z.; investigation, H.L. and H.N.; project administration, H.Z.; resources, J.L.; writing—original draft, H.L.
Funding
This work was supported by the Fund of Transgenic Breeding for Soybean Resistance to Soybean mosaic virus (2016ZX08004-004), the Fundamental Research Funds for the Central Universities (KYT201801), Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT_17R55), the National Natural Science Foundation of China (31571690), the National Soybean Industrial Technology System of China (CARS-004), Jiangsu Collaborative Innovation Center for Modern Crop Production(JCIC-MCP), and the National Key R&D Program of China (2017YFD0101501).
Acknowledgments
We thank Qingming Gao and Keshun Yu for critical review of the manuscript and valuable suggestions, and Aardra Kachroo for providing materials and space to carry out the BIFC assay in her lab at the University of Kentucky.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Adams, M.J.; Antoniw, J.F.; Beaudoin, F. Overview and analysis of the polyprotein cleavage sites in the family Potyviridae. Mol. Plant Pathol. 2005, 6, 471–487. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.H.; Bailey, T.B.; Benner, H.I.; Tachibana, H.; Durand, D.P. Soybean mosaic virus: Effects of primary disease incidence on yield and seed quality. Plant Dis. 1987, 71, 237–239. [Google Scholar] [CrossRef]
- Cho, E.K.; Goodman, R.M. Strains of soybean mosaic virus: Classification based on virulence in resistant soybean cultivars. Phytopathology 1979, 69, 467–470. [Google Scholar] [CrossRef]
- Cho, E.K.; Goodman, R.M. Evaluation of resistance in soybeans to soybean mosaic virus strains 1. Crop Sci. 1982, 22, 1133–1136. [Google Scholar] [CrossRef]
- Hayes, A.J.; Ma, G.; Buss, G.R.; Maroof, M.A. Molecular marker mapping of Rsv 4, a gene conferring resistance to all known strains of soybean mosaic virus. Crop Sci. 2000, 40, 1434–1437. [Google Scholar] [CrossRef]
- Hayes, A.J.; Jeong, S.C.; Gore, M.A.; Yu, Y.G.; Buss, G.R.; Tolin, S.A.; Maroof, M.S. Recombination within a nucleotide-binding-site/leucine-rich-repeat gene cluster produces new variants conditioning resistance to soybean mosaic virus in soybeans. Genetics 2004, 166, 493–503. [Google Scholar] [CrossRef] [PubMed]
- Gore, M.A.; Hayes, A.J.; Jeong, S.C.; Yue, Y.G.; Buss, G.R.; Maroof, M.S. Mapping tightly linked genes controlling potyvirus infection at the Rsv1 and Rpv1 region in soybean. Genome 2002, 45, 592–599. [Google Scholar] [CrossRef]
- Jeong, S.C.; Saghai Maroof, M.A. Detection and genotyping of SNPs tightly linked to two disease resistance loci, Rsv1 and Rsv3, of soybean. Plant Breed. 2008, 123, 305–310. [Google Scholar] [CrossRef]
- Maroof, M.A.; Tucker, D.M.; Skoneczka, J.A.; Bowman, B.C.; Tripathy, S.; Tolin, S.A. Fine mapping and candidate gene discovery of the soybean mosaic virus resistance gene, Rsv4. Plant Genome 2010, 3, 14–22. [Google Scholar] [CrossRef]
- Gunduz, I.; Buss, G.R.; Chen, P.; Tolin, S.A. Characterization of SMV resistance genes in Tousan 140 and Hourei soybean. Crop Sci. 2002, 42, 90–95. [Google Scholar] [CrossRef]
- Chen, P.; Buss, G.R.; Tolin, S.A. Resistance to soybean mosaic virus conferred by two independent dominant genes in PI 486355. J. Hered. 1993, 84, 25–28. [Google Scholar] [CrossRef]
- Ma, G.; Chen, P.; Buss, G.R.; Tolin, S.A. Genetic characteristics of two genes for resistance to soybean mosaic virus in PI486355 soybean. Theor. Appl. Genet. 1995, 91, 907–914. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Chen, P.; Buss, G.R.; Tolin, S.A. Complementary action of two independent dominant genes in Columbia soybean for resistance to soybean mosaic virus. J. Hered. 2002, 93, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Gunduz, I.; Buss, G.R.; Chen, P.; Tolin, S.A. Genetic and phenotypic analysis of soybean mosaic virus resistance in PI 88788 soybean. Phytopathology 2004, 94, 687–692. [Google Scholar] [CrossRef] [PubMed]
- Maroof, S.; Jeong, S.C.; Gunduz, I.; Tucker, D.M.; Buss, G.R.; Tolin, S.A. Pyramiding of soybean mosaic virus resistance genes by marker-assisted selection. Crop Sci. 2008, 48, 517–526. [Google Scholar] [CrossRef]
- Wang, X.Q.; Gai, J.Y.; Pu, Z.Q. Classification and distribution of strains of soybean mosaic virus in middle and lower Huang-Huai and Changjiang Valleys. Soybean Sci. 2003, 22, 102–107. [Google Scholar]
- Guo, D.Q.; Zhi, H.J.; Yahwei, W. Identification and distribution of soybean mosaic virus strains in Middle and Northern Huang Huai Region of China. Chin. J. Oil Crop Sci. 2005, 27, 64–68. [Google Scholar]
- Li, K.; Yang, Q.H.; Zhi, H.J.; Gai, J.Y. Identification and distribution of soybean mosaic virus strains in southern China. Plant Dis. 2010, 94, 351–357. [Google Scholar] [CrossRef]
- Wang, D.; Ma, Y.; Yang, Y.; Liu, N.; Li, C.; Song, Y.; Zhi, H. Fine mapping and analyses of R SC8 resistance candidate genes to soybean mosaic virus in soybean. Theor. Appl. Genet. 2011, 122, 555–565. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, D.G.; Li, H.C.; Zheng, G.J.; Yang, Y.Q.; Li, H.W.; Zhi, H.J. Fine mapping of the RSC14Q locus for resistance to soybean mosaic virus in soybean. Euphytica 2011, 181, 127–135. [Google Scholar] [CrossRef]
- Zheng, G.J.; Yang, Y.Q.; Ying, M.A.; Yang, X.F.; Chen, S.Y.; Ren, R.; Wang, D.G.; Yang, Z.L.; Zhi, H.J. Fine mapping and candidate gene analysis of resistance gene RSC3Q to soybean mosaic virus in Qihuang 1. J. Integr. Agric. 2014, 13, 2608–2615. [Google Scholar] [CrossRef]
- Rodriguez-Cerezo, E.; Ammar, E.D.; Pirone, T.P.; Shaw, J.G. Association of the non-structural P3 viral protein with cylindrical inclusions in potyvirus-infected cells. J. Gen. Virol. 1993, 74, 1945–1949. [Google Scholar] [CrossRef] [PubMed]
- Sáenz, P.; Cervera, M.T.; Dallot, S.; Quiot, L.; Quiot, J.B.; Riechmann, J.L.; García, J.A. Identification of a pathogenicity determinant of Plum pox virus in the sequence encoding the C-terminal region of protein P3+6K1. J. Gen. Virol. 2000, 81, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Desbiez, C.; Gal-On, A.; Girard, M.; Wipf-Scheibel, C.; Lecoq, H. Increase in zucchini yellow mosaic virus symptom severity in tolerant zucchini cultivars is related to a point mutation in P3 protein and is associated with a loss of relative fitness on susceptible plants. Phytopathology 2003, 93, 1478–1484. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jenner, C.E.; Wang, X.; Tomimura, K.; Ohshima, K.; Ponz, F.; Walsh, J.A. The dual role of the potyvirus P3 protein of turnip mosaic virus as a symptom and avirulence determinant in brassicas. Mol. Plant-Microbe Interact. 2003, 16, 777–784. [Google Scholar] [CrossRef] [PubMed]
- Suehiro, N.; Natsuaki, T.; Watanabe, T.; Okuda, S. An important determinant of the ability of turnip mosaic virus to infect Brassica spp. and/or Raphanus sativus is in its P3 protein. J. Gen. Virol. 2004, 85, 2087–2098. [Google Scholar] [CrossRef] [PubMed]
- Choi, I.R.; Horken, K.M.; Stenger, D.C.; French, R. An internal RNA element in the P3 cistron of Wheat streak mosaic virus revealed by synonymous mutations that affect both movement and replication. J. Gen. Virol. 2005, 86, 2605–2614. [Google Scholar] [CrossRef]
- Hajimorad, M.R.; Eggenberger, A.L.; Hill, J.H. Loss and gain of elicitor function of soybean mosaic virus G7 provoking Rsv1-mediated lethal systemic hypersensitive response maps to P3. J. Virol. 2005, 79, 1215–1222. [Google Scholar] [CrossRef]
- Hajimorad, M.R.; Eggenberger, A.L.; Hill, J.H. Strain-specific P3 of soybean mosaic virus elicits Rsv1-mediated extreme resistance, but absence of P3 elicitor function alone is insufficient for virulence on Rsv1-genotype soybean. Virology 2006, 345, 156–166. [Google Scholar] [CrossRef][Green Version]
- Hjulsager, C.K.; Olsen, B.S.; Jensen, D.M.K.; Cordea, M.I.; Krath, B.N.; Johansen, I.E.; Lund, O.S. Multiple determinants in the coding region of pea seed-borne mosaic virus P3 are involved in virulence against sbm-2 resistance. Virology 2006, 355, 52–61. [Google Scholar] [CrossRef]
- Eggenberger, A.L.; Hajimorad, M.R.; Hill, J.H. Gain of virulence on Rsv1-genotype soybean by an avirulent soybean mosaic virus requires concurrent mutations in both P3 and HC-Pro. Mol. Plant-Microbe Interact. 2008, 21, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Wen, R.H.; Maroof, M.A.; Hajimorad, M.R. Amino acid changes in P3, and not the overlapping pipo-encoded protein, determine virulence of soybean mosaic virus on functionally immune Rsv1-genotype soybean. Mol. Plant Pathol. 2011, 12, 799–807. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Khatabi, B.; Hajimorad, M.R. Amino acid substitution in p3 of soybean mosaic virus to convert avirulence to virulence on Rsv4-genotype soybean is influenced by the genetic composition of P3. Mol. Plant Pathol. 2015, 16, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Luo, Z.; Yan, F.; Lu, Y.; Zheng, H.; Chen, J. Interaction between potyvirus P3 and ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) of host plants. Virus Genes 2011, 43, 90–92. [Google Scholar] [CrossRef] [PubMed]
- Luan, H.; Shine, M.B.; Cui, X.; Chen, X.; Ma, N.; Kachroo, P.; Zhi, H.; Kachroo, A. The potyviral P3 protein targets eukaryotic elongation factor 1A to promote the unfolded protein response and viral pathogenesis. Plant Physiol. 2016, 172, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Niu, L.; Zhang, W.; Yang, J.; Xing, G.; He, H.; Guo, D.; Du, Q.; Qian, X.; Yao, Y.; et al. RNAi-mediated SMV P3 cistron silencing confers significantly enhanced resistance to multiple Potyvirus strains and isolates in transgenic soybean. Plant Cell Rep. 2018, 37, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Kearns, E.V.; Keck, P.; Somerville, C.R. Primary structure of cytochrome b5 from cauliflower (Brassica oleracea L.) deduced from peptide and cDNA sequences. Plant Physiol. 1992, 99, 1254–1257. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.A.; Stobart, A.K.; Shewry, P.R.; Napier, J.A. Tobacco cytochromeb5: cDNA isolation, expression analysis and in vitro protein targeting. Plant Mol. Biol. 1994, 25, 527–537. [Google Scholar] [CrossRef]
- Saito, T.; Morio, T.; Ochiai, H. A second functional Δ5 fatty acid desaturase in the cellular slime mould Dictyostelium discoideum. Eur. J. Biochem. 2000, 267, 1813–1818. [Google Scholar] [CrossRef]
- Zhao, J.; Onduka, T.; Kinoshita, J.Y.; Honsho, M.; Kinoshita, T.; Shimazaki, K.I.; Ito, A. Dual subcellular distribution of cytochrome b5 in plant, cauliflower, cells. J. Biochem. 2003, 133, 115–121. [Google Scholar] [CrossRef]
- Hlavica, P.; Lewis, D.F. Allosteric phenomena in cytochrome P450-catalyzed monooxygenations. Eur. J. Biochem. 2001, 268, 4817–4832. [Google Scholar] [CrossRef] [PubMed]
- Schenkman, J.B.; Jansson, I. The many roles of cytochrome b5. Pharmacol. Ther. 2003, 97, 139–152. [Google Scholar] [CrossRef]
- Selote, D.; Kachroo, A. RPG1-B-derived resistance to avrb-expressing Pseudomonas syringae requires RIN4-like proteins in soybean. Plant Physiol. 2010, 153, 1199–1211. [Google Scholar] [CrossRef]
- Martin, K.; Kopperud, K.; Chakrabarty, R.; Banerjee, R.; Brooks, R.; Goodin, M.M. Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J. 2009, 59, 150–162. [Google Scholar] [CrossRef] [PubMed]
- Goodin, M.M.; Chakrabarty, R.; Yelton, S.; Martin, K.; Clark, A.; Brooks, R. Membrane and protein dynamics in live plant nuclei infected with sonchus yellow net virus, a plant-adapted rhabdovirus. J. Gen. Virol. 2007, 88, 1810–1820. [Google Scholar] [CrossRef] [PubMed]
- Swofford, D.L. PAUP: Phylogenetic Analysis Using Parsimony and Other Methods; Software Version 4.0 b10; Sinaur Associates: Sunderland, MA, USA, 2000. [Google Scholar]
- Zhang, C.; Ghabrial, S.A. Development of bean pod mottle virus-based vectors for stable protein expression and sequence-specific virus-induced gene silencing in soybean. Virology. 2006, 344, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Kachroo, A.; Ghabrial, S. Virus-induced gene silencing in soybean. Methods Mol. Biol. 2012, 894, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Maple, J.; Møller, S.G. Yeast two-hybrid screening. In Circadian Rhythms: Methods and Protocols; Humana Press: Totowa, NJ, USA, 2007; pp. 207–224. [Google Scholar]
- Zhang, C.; Bradshaw, J.D.; Whitham, S.A.; Hill, J.H. The development of an efficient multipurpose bean pod mottle virus viral vector set for foreign gene expression and RNA silencing. Plant Physiol. 2010, 153, 52–65. [Google Scholar] [CrossRef]
- Zhang, C.; Grosic, S.; Whitham, S.A.; Hill, J.H. The requirement of multiple defense genes in soybean Rsv1-mediated extreme resistance to soybean mosaic virus. Mol. Plant-Microbe Interact. 2012, 25, 1307–1313. [Google Scholar] [CrossRef]
- Jiang, C.Z.; Lu, F.; Imsabai, W.; Meir, S.; Reid, M.S. Silencing polygalacturonase expression inhibits tomato petiole abscission. J. Exp. Bot. 2008, 59, 973–979. [Google Scholar] [CrossRef]
- Broderick, S.R.; Jones, M.L. An optimized protocol to increase virus-induced gene silencing efficiency and minimize viral symptoms in Petunia. Plant Mol. Biol. Report. 2014, 32, 219–233. [Google Scholar] [CrossRef] [PubMed]
- Rui, R.; Liu, S.; Karthikeyan, A.; Wang, T.; Niu, H.; Yin, J.; Yang, Y.; Wang, L.; Li, K. Fine-mapping and identification of a novel locus Rsc15 underlying soybean resistance to soybean mosaic virus. Theor. Appl. Genet. 2017, 130, 2395–2410. [Google Scholar] [CrossRef] [PubMed]
- Bendahmane, A.; Kanyuka, K.; Baulcombe, D.C. The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 1999, 11, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Hernández, J.A.; Gullner, G.; Clemente-Moreno, M.J.; Künstler, A.; Juhász, C.; Díaz-Vivancos, P.; Király, L. Oxidative stress and antioxidative responses in plant–virus interactions. Physiol. Mol. Plant Pathol. 2016, 94, 134–148. [Google Scholar] [CrossRef]
- Zhang, H.; Hamdane, D.; Im, S.C.; Waskell, L. Cytochrome b5 inhibits electron transfer from NADPH-cytochrome P450 reductase to ferric cytochrome P450 2B4. J. Biol. Chem. 2007, 283, 5217–5225. [Google Scholar] [CrossRef] [PubMed]
- Aoyama, T.; Nagata, K.; Yamazoe, Y.; Kato, R.; Matsunaga, E.; Gelboin, H.V.; Gonzalez, F.J. Cytochrome b5 potentiation of cytochrome P-450 catalytic activity demonstrated by a vaccinia virus-mediated in situ reconstitution system. Proc. Natl. Acad. Sci. USA 1990, 87, 5425–5429. [Google Scholar] [CrossRef] [PubMed]
- Riedle-Bauer, M. Role of reactive oxygen species and antioxidant enzymes in systemic virus infections of plants. J. Phytopathol. 2000, 148, 297–302. [Google Scholar] [CrossRef]
- Arias, M.C.; Luna, C.; Rodríguez, M.; Lenardon, S.; Taleisnik, E. Sunflower chlorotic mottle virus in compatible interactions with sunflower: ROS generation and antioxidant response. Eur. J. Plant Pathol. 2005, 113, 223–232. [Google Scholar] [CrossRef]
- Díaz-Vivancos, P.; Rubio, M.; Mesonero, V.; Periago, P.M.; Ros Barceló, A.; Martínez-Gómez, P.; Hernández, J.A. The apoplastic antioxidant system in Prunus: Response to long-term plum pox virus infection. J. Exp. Bot. 2006, 57, 3813–3824. [Google Scholar] [CrossRef]
- Díaz-Vivancos, P.; Clemente-Moreno, M.J.; Rubio, M.; Olmos, E.; García, J.A.; Martínez-Gómez, P.; Hernández, J.A. Alteration in the chloroplastic metabolism leads to ROS accumulation in pea plants in response to plum pox virus. J. Exp. Bot. 2008, 59, 2147–2160. [Google Scholar] [CrossRef]
- Hernández, J.A.; Díaz-Vivancos, P.; Rubio, M.; Olmos, E.; Ros-Barceló, A.; Martínez-Gómez, P. Long-term plum pox virus infection produces an oxidative stress in a susceptible apricot, Prunus armeniaca, cultivar but not in a resistant cultivar. Physiol. Plant. 2006, 126, 140–152. [Google Scholar] [CrossRef]
- Király, L.; Hafez, Y.M.; Fodor, J.; Király, Z. Suppression of tobacco mosaic virus-induced hypersensitive-type necrotization in tobacco at high temperature is associated with downregulation of NADPH oxidase and superoxide and stimulation of dehydroascorbate reductase. J. Gen. Virol. 2008, 89, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Mathioudakis, M.M.; Veiga, R.S.; Canto, T.; Medina, V.; Mossialos, D.; Makris, A.M.; Livieratos, I. Pepino mosaic virus triple gene block protein 1 (TGBp1) interacts with and increases tomato catalase 1 activity to enhance virus accumulation. Mol. Plant Pathol. 2013, 14, 589–601. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).