Surveillance for Adenoviruses in Bats in Italy
Abstract
1. Introduction
2. Materials and Methods
2.1. Origin of Samples
2.2. DNA Extraction and PCR Amplification
2.3. Virus Isolation
2.4. Next-Generation Sequencing
2.5. Genome Annotation and Comparison
2.6. Sequence and Phylogenetic Analysis
3. Results
3.1. Molecular Screening
3.2. Virus Isolation
3.3. NGS Analysis and Genome Structure of the Adenovirus Strain ITA/2018/251170-16
3.4. Sequence and Phylogenetic Analysis
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Harrach, B.; Benkö, M.; Both, G.; Brown, M.; Davison, A.; Echavarría, M.; Hess, M.; Jones, M.; Kajon, A.; Lehmkuhl, H.; et al. Family Adenoviridae. In Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses; King, A., Adams, M., Carstens, E., Lefkowitz, E., Eds.; Elsevier: San Diego, CA, USA, 2011; pp. 125–141. [Google Scholar]
- Tan, B.; Yang, X.L.; Ge, X.Y.; Peng, C.; Liu, H.Z.; Zhang, Y.Z.; Zhang, L.B.; Shi, Z.L. Novel bat adenoviruses with low G+C content shed new light on the evolution of adenoviruses. J. Gen. Virol. 2017, 98, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Davison, A.J.; Benko, M.; Harrach, B. Genetic content and evolution of adenoviruses. J. Gen. Virol. 2003, 84, 2895–2908. [Google Scholar] [CrossRef] [PubMed]
- International Committee on Taxonomy of Viruses (ICTV) Virus Taxonomy: 2018b Release. Available online: http://talk.ictvonline.org/taxonomy/ (accessed on 2 April 2019).
- Pauly, M.; Hoppe, E.; Mugisha, L.; Petrzelkova, K.; Akoua-Koffi, C.; Couacy-Hymann, E.; Anoh, A.E.; Mossoun, A.; Schubert, G.; Wiersma, L.; et al. High prevalence and diversity of species D adenoviruses (HAdV-D) in human populations of four Sub-Saharan countries. Virol. J. 2014, 11, 25. [Google Scholar] [CrossRef] [PubMed]
- Kendall Scott, M.; Chommanard, C.; Lu, X.; Appelgate, D.; Grenz, L.; Schneider, E.; Gerber, S.I.; Erdman, D.D.; Thomas, A. Human adenovirus associated with severe respiratory infection, Oregon, USA, 2013-2014. Emerg. Infect. Dis. 2016, 22, 1044–1051. [Google Scholar] [CrossRef] [PubMed]
- Benkö, M.; Harrach, B. Molecular Evolution of Adenoviruses. Immunology 2003, 272, 3–35. [Google Scholar]
- Robinson, C.M.; Singh, G.; Lee, J.Y.; Dehghan, S.; Rajaiya, J.; Liu, E.B.; Yousuf, M.A.; Betensky, R.A.; Jones, M.S.; Dyer, D.W.; et al. Molecular evolution of human adenoviruses. Sci. Rep. 2013, 3, 1812. [Google Scholar] [CrossRef] [PubMed]
- Hoppe, E.; Pauly, M.; Gillespie, T.R.; Akoua-Koffi, C.; Hohmann, G.; Fruth, B.; Karhemere, S.; Madinda, N.F.; Mugisha, L.; Muyembe, J.J.; et al. Multiple cross-species transmission events of human adenoviruses (HAdV) during hominine evolution. Mol. Biol. Evol. 2015, 32, 2072–2084. [Google Scholar] [CrossRef] [PubMed]
- Calisher, C.H.; Childs, J.E.; Field, H.E.; Holmes, K.V.; Schountz, T. Bats: Important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006, 19, 531–545. [Google Scholar] [CrossRef]
- Smith, I.; Wang, L.F. Bats and their virome: An important source of emerging viruses capable of infecting humans. Curr. Opin. Virol. 2013, 3, 84–91. [Google Scholar] [CrossRef]
- Li, W.; Shi, Z.; Yu, M.; Ren, W.; Smith, C.; Epstein, J.H.; Wang, H.; Crameri, G.; Hu, Z.; Zhang, H.; et al. Bats are natural reservoirs of SARS-like coronaviruses. Science 2005, 310, 676–679. [Google Scholar] [CrossRef]
- Rahman, S.A.; Hassan, S.S.; Olival, K.J.; Mohamed, M.; Chang, L.Y.; Hassan, L.; Saad, N.M.; Shohaimi, S.A.; Mamat, Z.C.; Naim, M.S.; et al. Characterization of Nipah virus from naturally infected Pteropus vampyrus bats, Malaysia. Emerg. Infect. Dis. 2010, 16, 1990–1993. [Google Scholar] [CrossRef] [PubMed]
- Mcelhinney, L.M.; Marston, D.A.; Leech, S.; Freuling, C.M.; van der Poel, W.H.M.; Echevarria, J.; Vázquez-Moron, S.; Horton, D.L.; Müller, T.; Fooks, A.R. Molecular Epidemiology of Bat Lyssaviruses in Europe. Zoonoses Public Health 2013, 60, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Banyard, A.C.; Evans, J.S.; Luo, T.R.; Fooks, A.R. Lyssaviruses and bats: Emergence and zoonotic threat. Viruses 2014, 6, 2974–2990. [Google Scholar] [CrossRef] [PubMed]
- Fischer, K.; dos Reis, V.P.; Balkema-Buschmann, A. Bat astroviruses: Towards understanding the transmission dynamics of a neglected virus family. Viruses 2017, 9, 34. [Google Scholar] [CrossRef] [PubMed]
- Johnson, N.; Aréchiga-Ceballos, N.; Aguilar-Setien, A. Vampire bat rabies: Ecology, epidemiology and control. Viruses 2014, 6, 1911–1928. [Google Scholar] [CrossRef] [PubMed]
- Moratelli, R.; Calisher, C.H. Bats and zoonotic viruses: Can we confidently link bats with emerging deadly viruses? Mem. Inst. Oswaldo Cruz 2015, 110, 1–22. [Google Scholar] [CrossRef]
- Serra-Cobo, J.; López-Roig, M. Bats and emerging infections: An ecological and virological puzzle. Adv. Exp. Med. Biol. 2017, 972, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Maeda, K.; Hondo, E.; Terakawa, J.; Kiso, Y.; Nakaichi, N.; Endoh, D.; Sakai, K.; Morikawa, S.; Mizutani, T. Isolation of novel adenovirus from fruit bat (Pteropus dasymallus yayeyamae). Emerg. Infect. Dis. 2008, 14, 347–349. [Google Scholar] [CrossRef]
- Tan, B.; Yang, X.L.; Ge, X.Y.; Peng, C.; Zhang, Y.Z.; Zhang, L.B.; Shi, Z.L. Novel bat adenoviruses with an extremely large E3 gene. J. Gen. Virol. 2016, 97, 1625–1635. [Google Scholar] [CrossRef]
- Hackenbrack, N.; Rogers, M.B.; Ashley, R.E.; Keel, M.K.; Kubiski, S.V.; Bryan, J.A.; Ghedin, E.; Holmes, E.C.; Hafenstein, S.L.; Allison, A.B. Evolution and Cryo-electron Microscopy Capsid Structure of a North American Bat Adenovirus and Its Relationship to Other Mastadenoviruses. J. Virol. 2016, 91, e01504–e01516. [Google Scholar] [CrossRef]
- Ogawa, H.; Kajihara, M.; Nao, N.; Shigeno, A.; Fujikura, D.; Hang’Ombe, B.M.; Mweene, A.S.; Mutemwa, A.; Squarre, D.; Yamada, M.; et al. Characterization of a novel bat adenovirus isolated from straw-colored fruit bat (Eidolon helvum). Viruses 2017, 9, 371. [Google Scholar] [CrossRef] [PubMed]
- Sonntag, M.; Mühldorfer, K.; Speck, S.; Wibbelt, G.; Kurth, A. New adenovirus in bats, Germany. Emerg. Infect. Dis. 2009, 15, 2052–2055. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ge, X.; Zhang, H.; Zhou, P.; Zhu, Y.; Zhang, Y.; Yuan, J.; Wang, L.F.; Shi, Z. Host range, prevalence, and genetic diversity of adenoviruses in bats. J. Virol. 2010, 84, 3889–3897. [Google Scholar] [CrossRef] [PubMed]
- Kohl, C.; Vidovszky, M.Z.; Mühldorfer, K.; Dabrowski, P.W.; Radonić, A.; Nitsche, A.; Wibbelt, G.; Kurth, A.; Harrach, B. Genome analysis of bat adenovirus 2: Indications of interspecies transmission. J. Virol. 2012, 86, 1888–1892. [Google Scholar] [CrossRef] [PubMed]
- Balboni, A.; Verin, R.; Morandi, F.; Poli, A.; Prosperi, S.; Battilani, M. Molecular epidemiology of canine adenovirus type 1 and type 2 in free-ranging red foxes (Vulpes vulpes) in Italy. Vet. Microbiol. 2013, 162, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Cortés-Hinojosa, G.; Gulland, F.M.D.; Goldstein, T.; Venn-Watson, S.; Rivera, R.; Waltzek, T.B.; Salemi, M.; Wellehan, J.F.X. Phylogenomic characterization of California sea lion adenovirus-1. Infect. Genet. Evol. 2015, 31, 270–276. [Google Scholar] [CrossRef] [PubMed]
- Wellehan, J.F.X.; Johnson, A.J.; Harrach, B.; Benko, M.; Pessier, A.P.; Johnson, C.M.; Garner, M.M.; Childress, A.; Jacobson, E.R. Detection and Analysis of Six Lizard Adenoviruses by Consensus Primer PCR Provides Further Evidence of a Reptilian Origin for the Atadenoviruses. J. Virol. 2004, 78, 13366–13369. [Google Scholar] [CrossRef] [PubMed]
- Lavazza, A.; Pascucci, S.; Gelmetti, D. Rod-shaped virus-like particles in intestinal contents of three avian species. Vet. Rec. 1990, 126, 581. [Google Scholar]
- Andrews, S. FastQC—A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 1 May 2019).
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Klempa, B.; Krüger, D.H.; Auste, B.; Stanko, M.; Krawczyk, A.; Nickel, K.F.; Uberla, K.; Stanga, A. A novel cardiotropic murine adenovirus representing a distinct species of mastadenoviruses. J. Virol. 2009, 83, 5749–5759. [Google Scholar] [CrossRef]
- Ursu, K.; Harrach, B.; Matiz, K.; Benko, M. Dna sequencing and analysis of the right-hand part of the genome of the unique bovine adenovirus type 10. J. Gen. Virol. 2004, 85, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Pearson, W.R. Searching Protein Sequence Libraries: Comparison of the Sensitivity and Selectivity of the Smith-Waterman and FASTA Algorithms. Genomics 1991, 3, 635–650. [Google Scholar] [CrossRef]
- Waruhiu, C.; Ommeh, S.; Obanda, V.; Agwanda, B.; Gakuya, F.; Ge, X.Y.; Yang, X.L.; Wu, L.J.; Zohaib, A.; Hu, B.; et al. Molecular detection of viruses in Kenyan bats and discovery of novel astroviruses, caliciviruses and rotaviruses. Virol. Sin. 2017, 32, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Iglesias-Caballero, M.; Juste, J.; Vázquez-Morón, S.; Falcon, A.; Aznar-Lopez, C.; Ibáñez, C.; Pozo, F.; Ruiz, G.; Berciano, J.; Garin, I.; et al. New Adenovirus Groups in Western Palaearctic Bats. Viruses 2018, 10, 443. [Google Scholar] [CrossRef] [PubMed]
- Cortez-Hinojosa, G.; Adkesson, M.J.; Cárdenas-Alayza, S.; Seguel, M.; Pavés, H.; Wellehan, J., Jr. Effect of environmental stress of an el niño event on adenoviral diversity in marine animal rookeries. In Proceedings of the AAZV Annual Conference, Virginia Beach, VA, USA, 21–26 May 2016. [Google Scholar]
- Harrach, B. Reptile Adenoviruses in Cattle? Acta Vet. Hung. 2000, 48, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Dowgier, G.; Lahoreau, J.; Lanave, G.; Losurdo, M.; Varello, K.; Lucente, M.S.; Ventriglia, G.; Bozzetta, E.; Martella, V.; Buonavoglia, C.; et al. Sequential circulation of canine adenoviruses 1 and 2 in captive wild carnivores, France. Vet. Microbiol. 2018, 221, 67–73. [Google Scholar] [CrossRef] [PubMed]
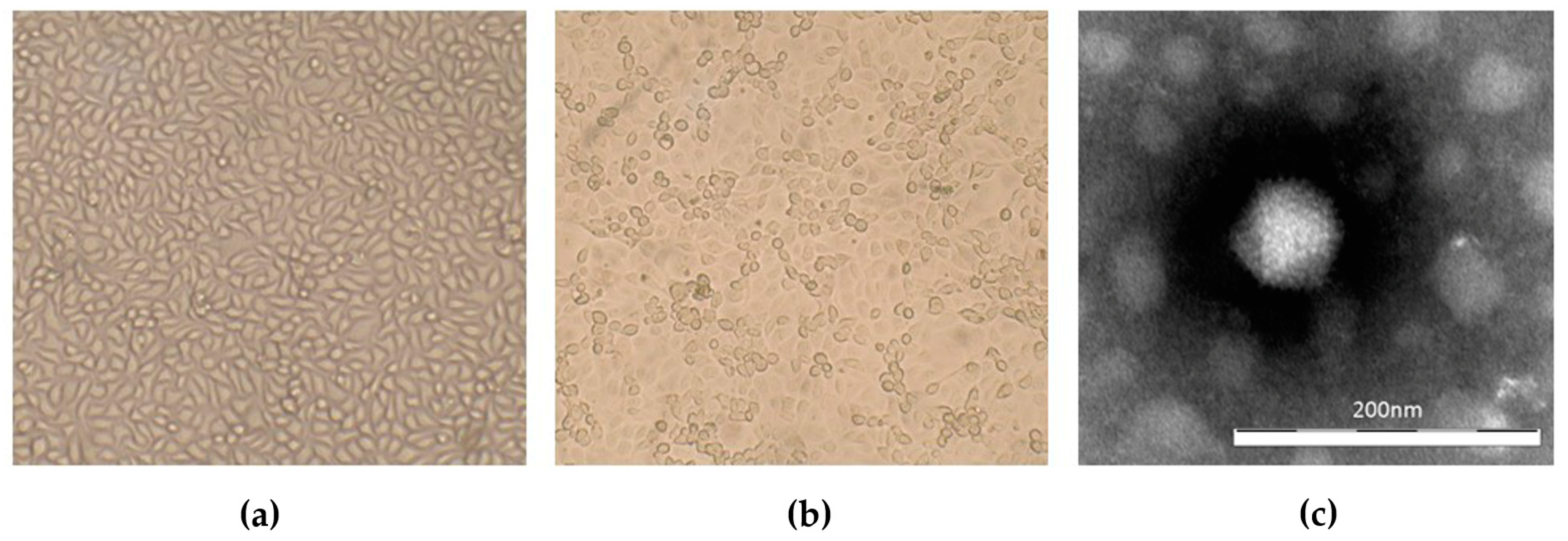
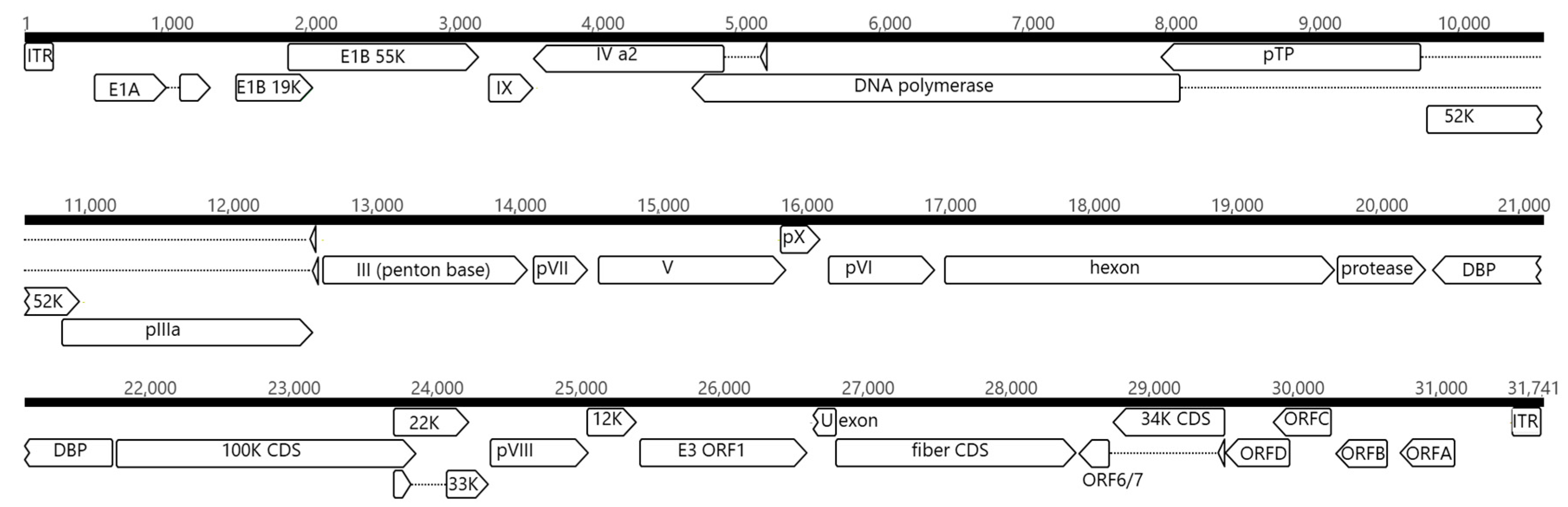
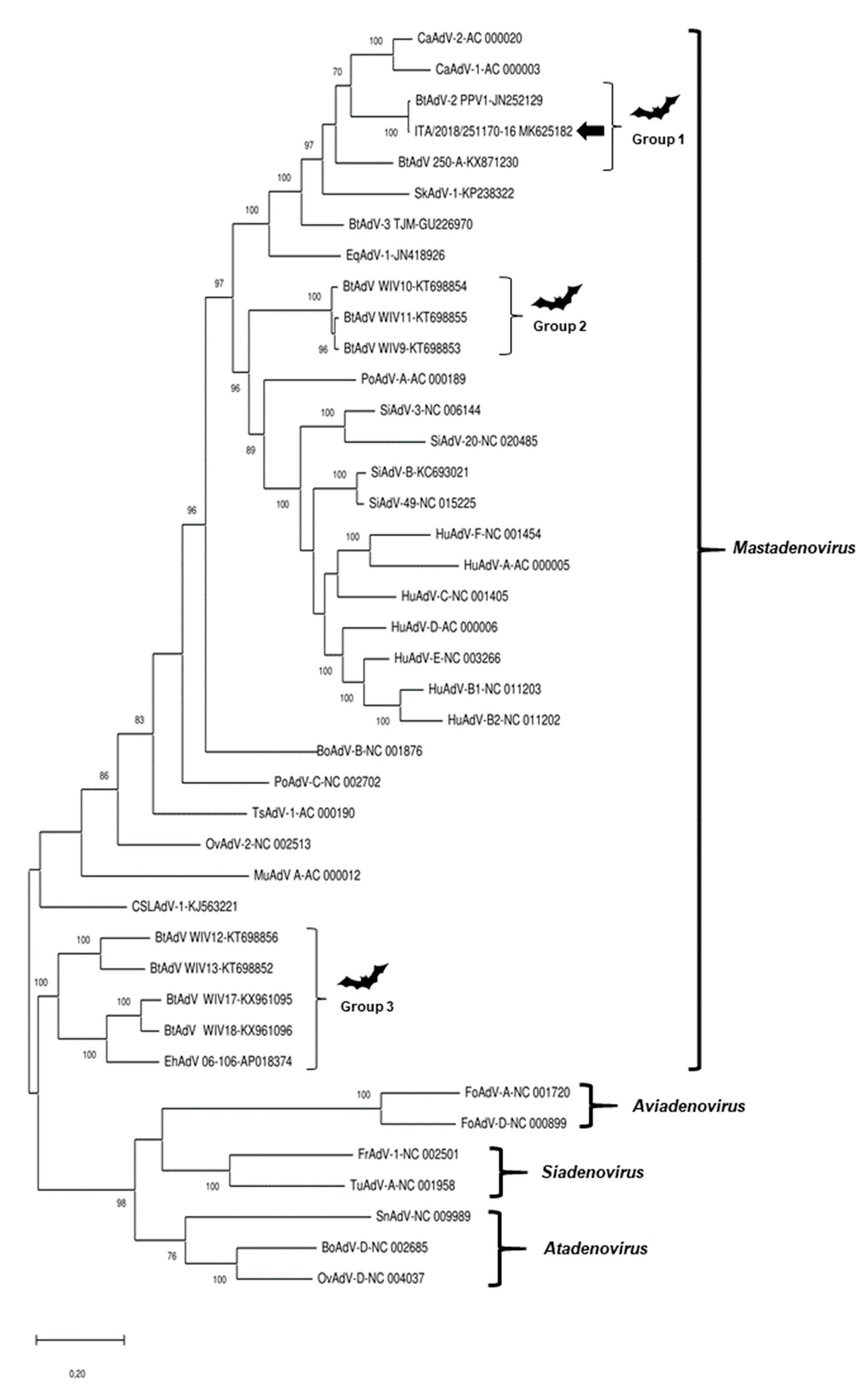

| Strain | Species | GenBank Accession No. | Reference |
|---|---|---|---|
| TJM | Bat mastadenovirus A | GU226970 | [25] |
| PPV1 | Bat mastadenovirus B | JN252129 | [24] |
| WIV9 | Bat mastadenovirus C | KT698853 | [21] |
| WIV12 | Bat mastadenovirus D | KT698856 | [2] |
| WIV13 | Bat mastadenovirus E | KT698852 | [2] |
| WIV17 | Bat mastadenovirus F | KX961095 | [2] |
| 250-A | Bat mastadenovirus G | KX871230 | [22] |
| EhAdV 06-106 | Bat mastadenovirus H * | JX885602 | [23] |
| Results of FASTA Interrogation | Pairwise Identity among the Italian Viruses | ||||||
|---|---|---|---|---|---|---|---|
| Strain (Id lab) | Bat species | Identity/p-value | GenBank accession no. | Name/species | Country | Identity | Strain |
| PA2/16 HS (189/18-2) | Hypsugo savii | 99.5%/1.7E-106 | JX065122 | Hsa_070703-2/Hypsugo savii | Spain (2007) | 99.5% | PA42, PA46, PA16-350/15 |
| PA42/17 HS (189/18-42) | Hypsugo savii | 100.0%/8.2E-106 | JX065122 | Hsa_070703-2/Hypsugo savii | Spain (2007) | 99.5–100.0% | PA2, PA46, PA16-350/15 |
| PA46/17 PK (189/18-46) | Pipistrellus kuhlii | 100.0%/7.1E-106 | JX065122 | Hsa_070703-2/Hypsugo savii | Spain (2007) | 99.5–100.0% | PA2, PA42, PA16-350/15 |
| PA16-350/15 PS (350/15-16) | Pipistrellus spp. | 99.5%/1.5E-107 | JX065122 | Hsa_070703-2/Hypsugo savii | Spain (2007) | 99.5% | PA2, PA42, PA46 |
| PA15/16 PS (189/18-15) | Pipistrellus spp. | 76.1%/1.1E-45 | KM043095 | 140/07/Pipistrellus nathusii | Germany (2007) | 86.2–86.7% | PA24, PA74 |
| PA24/16 PS (189/18-24) | Pipistrellus spp. | 76.1%/1.1E-40 | KM043095 | 140/07/Pipistrellus nathusii | Germany (2007) | 99.5% | PA74 |
| PA74/16 PK (189/18-74) | Pipistrellus kuhlii | 76.1%/5.1E-42 | KM043095 | 140/07/Pipistrellus nathusii | Germany (2007) | 99.5% | PA24 |
| PA21/16 PS (189/18-21) | Pipistrellus spp. | 74.0%/4.2E-51 | MF175113 | AAU11020/Arctocephalus australis | Peru (2009) | 52.1–52.5% | PA150, PA151 |
| PA22/16 PS (189/18-22) | Pipistrellus spp. | 100.0%/4.7E-82 | KM043100 | 09/09/Pipistrellus kuhlii | Germany (2009) | 100.0% | PA119 |
| PA119/17 PK (189/18-119) | Pipistrellus kuhlii | 100.0%/1.2E-81 | KM043100 | 09/09/Pipistrellus kuhlii | Germany (2009) | 100.0% | PA22 |
| PA90/17 PS (189/18-90) | Pipistrellus spp. | 91.3%/1.3E-72 | KM043095 | 140/07/Pipistrellus nathusii | Germany (2007) | 77.6% | PA22, PA119 |
| PA134/17 TT (189/18-134) | Tadarida teniotis | 77.6%/5.0E-47 | KY009649 | C052/Myotis fimbriatus | China (2015) | 70.3% | PA150 |
| PA150/17 PK (189/18-150) | Pipistrellus kuhlii | 99.1%/7.5E-102 | KM043108 | 202/09/Pipistrellus pipistrellus | Germany (2009) | 72.1–72.6% | PA2, PA42, PA46, PA151, PA16-350/15 |
| PA151/17 HS (189/18-151) | Hypsugo savii | 76.0%/5.8E-54 | KM043092 | 176/09/Pipistrellus pygmaeus | Germany (2009) | 98.6–99.1% | PA155, PA157 |
| PA155/17 PP (189/18-155) | Pipistrellus pipistrellus | 74.7%/5.0E-49 | KM043092 | 176/09/Pipistrellus pygmaeus | Germany (2009) | 98.6–99.5% | PA151, PA157 |
| PA157/17 HS (189/18-157) | Hysugo savii | 75.1%/6.7E-50 | KM043092 | 176/09/Pipistrellus pygmaeus | Germany (2009) | 99.1–99.5% | PA151, PA155 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diakoudi, G.; Lanave, G.; Moreno, A.; Chiapponi, C.; Sozzi, E.; Prosperi, A.; Larocca, V.; Losurdo, M.; Decaro, N.; Martella, V.; et al. Surveillance for Adenoviruses in Bats in Italy. Viruses 2019, 11, 523. https://doi.org/10.3390/v11060523
Diakoudi G, Lanave G, Moreno A, Chiapponi C, Sozzi E, Prosperi A, Larocca V, Losurdo M, Decaro N, Martella V, et al. Surveillance for Adenoviruses in Bats in Italy. Viruses. 2019; 11(6):523. https://doi.org/10.3390/v11060523
Chicago/Turabian StyleDiakoudi, Georgia, Gianvito Lanave, Ana Moreno, Chiara Chiapponi, Enrica Sozzi, Alice Prosperi, Vittorio Larocca, Michele Losurdo, Nicola Decaro, Vito Martella, and et al. 2019. "Surveillance for Adenoviruses in Bats in Italy" Viruses 11, no. 6: 523. https://doi.org/10.3390/v11060523
APA StyleDiakoudi, G., Lanave, G., Moreno, A., Chiapponi, C., Sozzi, E., Prosperi, A., Larocca, V., Losurdo, M., Decaro, N., Martella, V., Lavazza, A., & Lelli, D. (2019). Surveillance for Adenoviruses in Bats in Italy. Viruses, 11(6), 523. https://doi.org/10.3390/v11060523









