Isolation of Subtype 3c, 3e and 3f-Like Hepatitis E Virus Strains Stably Replicating to High Viral Loads in an Optimized Cell Culture System
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
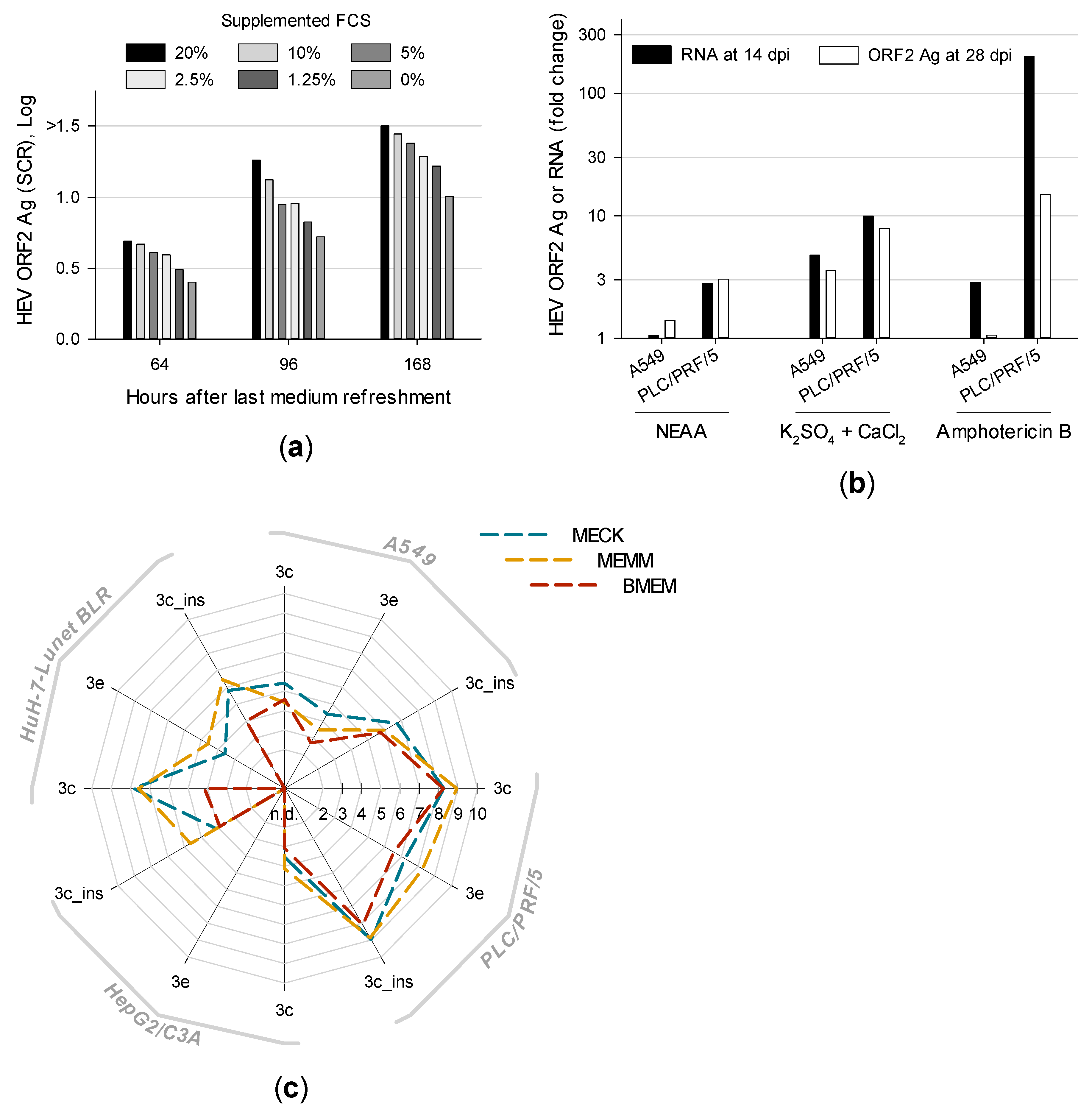
2.1.1. Cell Culture Media Composition
- BMEM:Eagle minimum essential medium (MEM) supplemented with 10% heat-inactivated fetal calf serum (FCS), 2 mM l-glutamine, 1% non-essential amino acids (NEAA), 100 U/mL penicillin G and 100 µg/mL streptomycin.
- MECK:BMEM additionally supplemented with 2.5 µg/mL amphotericin B, 10 mM CaCl2 and 10 mM K2SO4
- MEMM:BMEM additionally supplemented with 2.5 µg/mL amphotericin B and 30 mM MgCl2
- BMEM_G:Eagle MEM supplemented with 10% heat-inactivated FCS, 2 mM L-glutamine, 1% NEAA and 100 µg/mL gentamycin
- MEMM_G:BMEM_G additionally supplemented with 10 mM MgCl2
2.2. Viruses and Inocula
2.3. Virus Isolation and Passaging
2.4. HEV RNA Quantification
2.5. Detection of ORF2 Antigen
2.6. Immunofluorescence Microscopy
2.7. Whole Genome Sequencing
2.8. Phylogenetic Analysis
3. Results
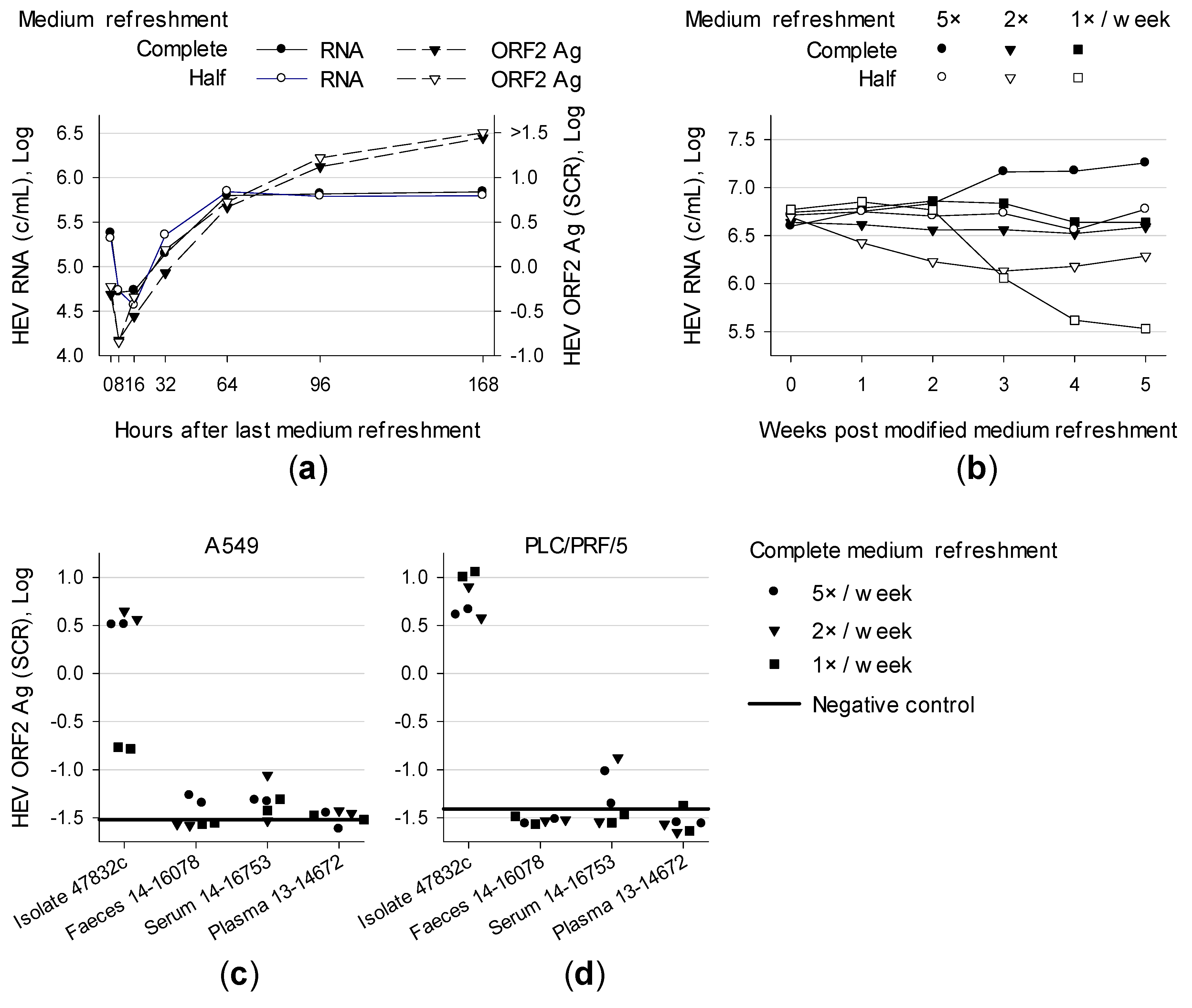
3.1. Effect of Medium Refreshment on Cells Infected With HEV Strain 47832c and Isolation of HEV Gt 3c Strain 14-16753
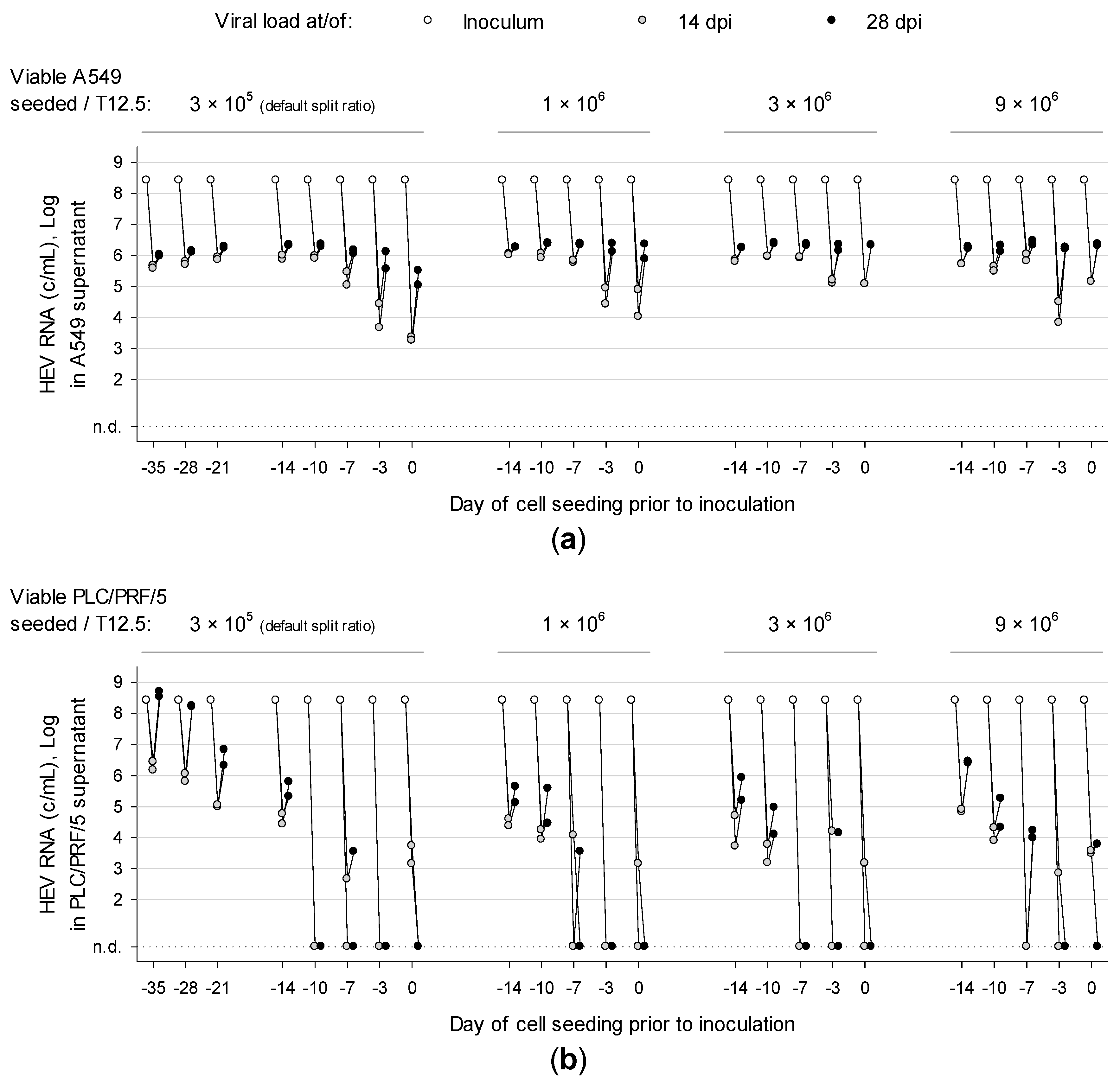
3.2. Overconfluently Grown Cells Seeded >1 Week Prior to Inoculation are More Permissive for Infection
3.3. Distinct Medium Supplements Enhance HEV Replication and Optimized Media Promote De Novo Isolation of HEV Gt 3e Strain 14-22707
3.4. First Isolation of HEV Gt 3f-like Strain 15-22016 Confirms PLC/PRF/5 as the Most Suitable Cell Line for Culturing HEV
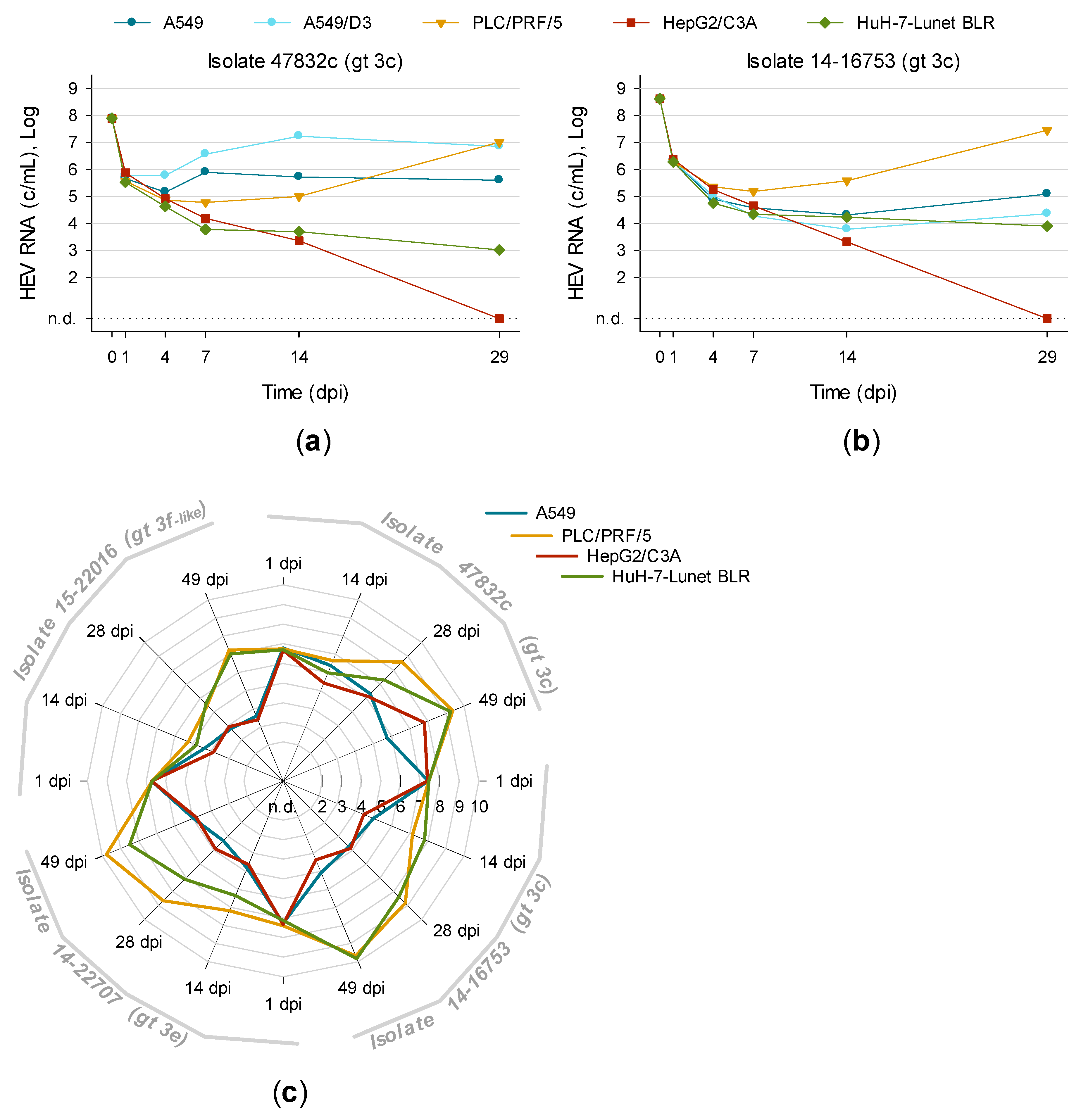
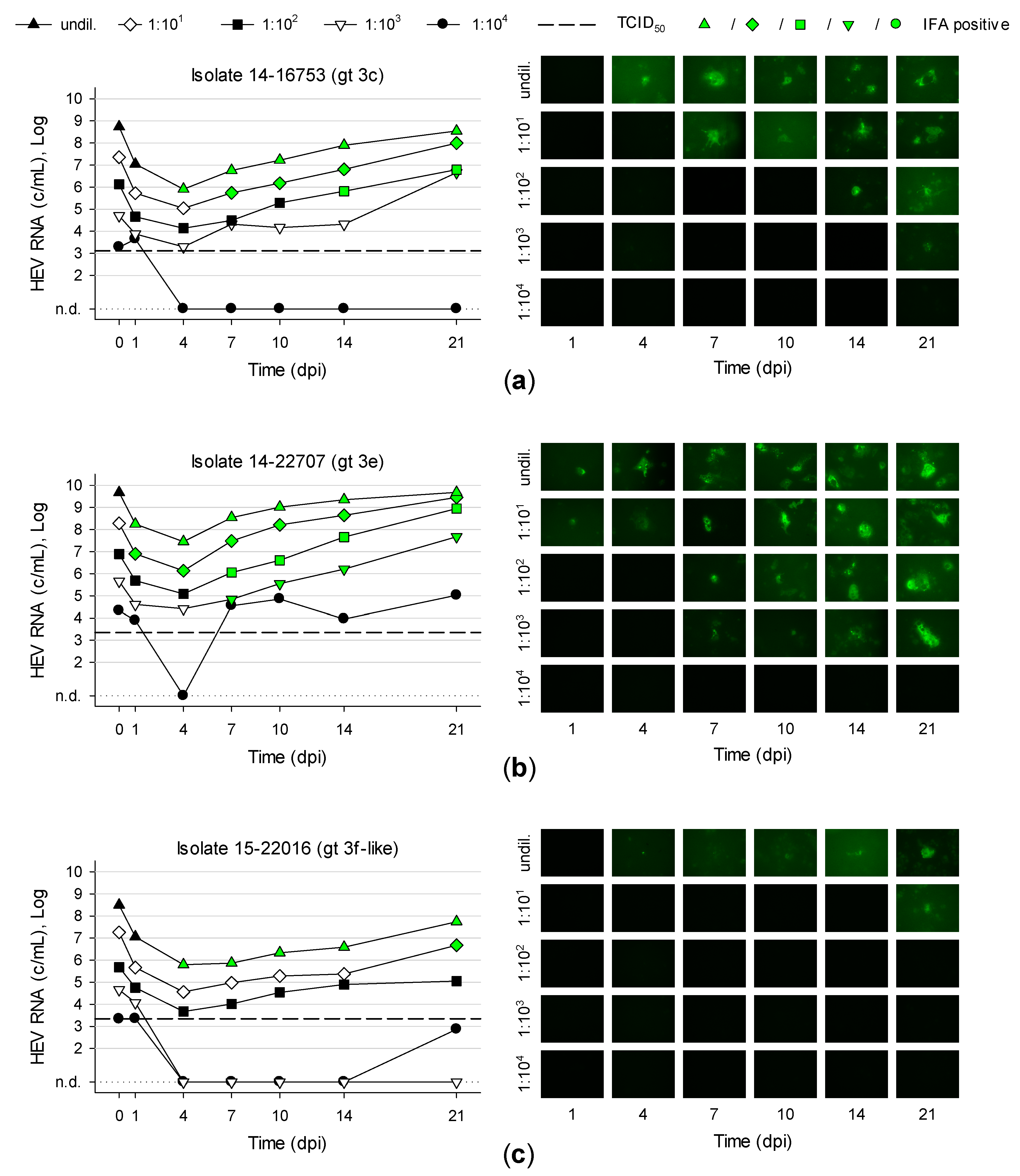
3.5. Titration and Growth of Newly Isolated HEV Strains Reveals Different Replication Kinetics
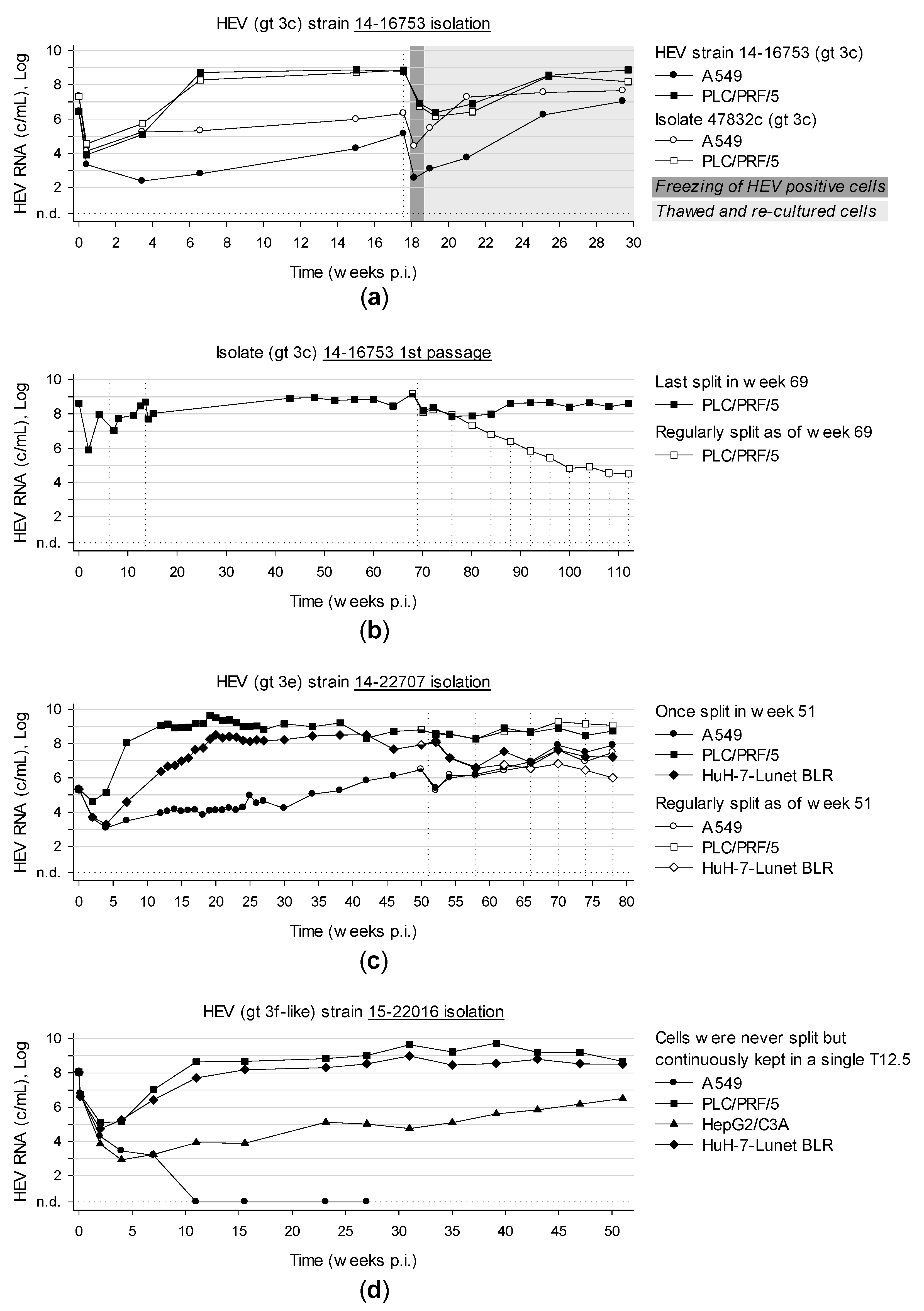
3.6. Long-Term Cultivation of Persistently Infected Cells Leads to Virtually Unlimited Production of HEV
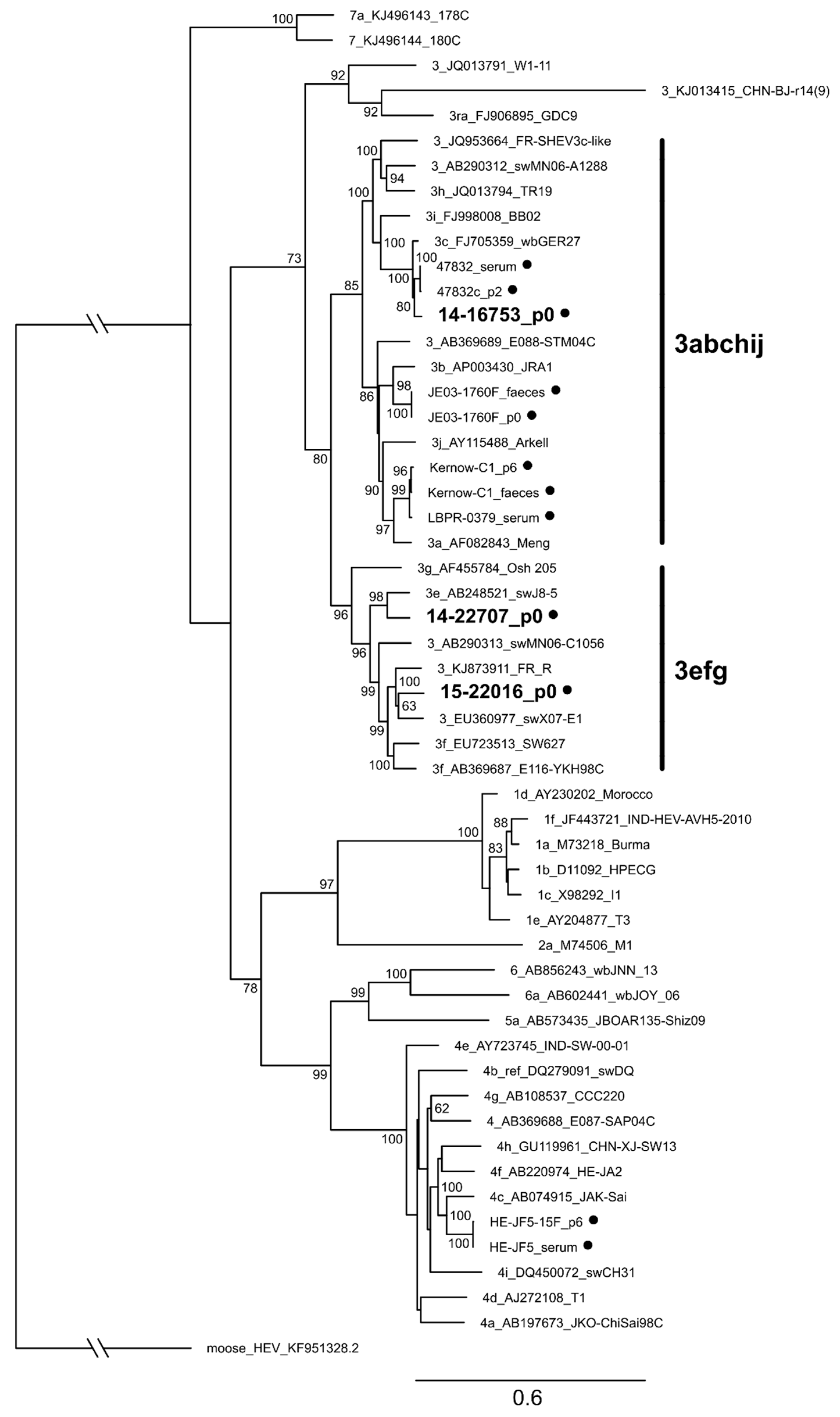
3.7. Characterization of Whole Genome Sequences of Novel Isolates Revealed No Insertion in ORF1
3.8. Optimized Method for Successful HEV Isolation from Serum
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Smith, D.B.; Simmonds, P.; Jameel, S.; Emerson, S.U.; Harrison, T.J.; Meng, X.-J.; Okamoto, H.; van der Poel, W.H.M.; Purdy, M.A. Consensus proposals for classification of the family Hepeviridae. J. Gen. Virol. 2014, 95, 2223–2232. [Google Scholar] [CrossRef] [PubMed]
- Sridhar, S.; Yip, C.C.Y.; Wu, S.; Cai, J.; Zhang, A.J.-X.; Leung, K.-H.; Chung, T.W.H.; Chan, J.F.W.; Chan, W.-M.; Teng, J.L.L.; et al. Rat Hepatitis E Virus as Cause of Persistent Hepatitis after Liver Transplant. Emerg. Infect. Dis. 2018, 24, 2241–2250. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.B.; Simmonds, P.; Izopet, J.; Oliveira-Filho, E.F.; Ulrich, R.G.; Johne, R.; Koenig, M.; Jameel, S.; Harrison, T.J.; Meng, X.-J.; et al. Proposed reference sequences for hepatitis E virus subtypes. J. Gen. Virol. 2016, 97, 537–542. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.-H.; Tan, B.-H.; Teo, E.C.-Y.; Lim, S.-G.; Dan, Y.-Y.; Wee, A.; Aw, P.P.K.; Zhu, Y.; Hibberd, M.L.; Tan, C.-K.; et al. Chronic Infection With Camelid Hepatitis E Virus in a Liver Transplant Recipient Who Regularly Consumes Camel Meat and Milk. Gastroenterology 2016, 150, 355–357. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, M.C.; Scobie, L.; Crossan, C.L.; Dalton, H.; Hayes, P.C.; Simpson, K.J. Review article: hepatitis E-a concise review of virology, epidemiology, clinical presentation and therapy. Aliment. Pharmacol. Ther. 2017. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lau, S.K.P.; Teng, J.L.L.; Cao, K.-Y.; Wernery, U.; Schountz, T.; Chiu, T.H.; Tsang, A.K.L.; Wong, P.-C.; Wong, E.Y.M.; et al. New Hepatitis E Virus Genotype in Bactrian Camels, Xinjiang, China, 2013. Emerg. Infect. Dis. 2016, 22, 2219–2221. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Meng, X.-J. Molecular biology and replication of hepatitis E virus. Emerg. Microbes Infect. 2012, 1, e17. [Google Scholar] [CrossRef]
- Montpellier, C.; Wychowski, C.; Sayed, I.M.; Meunier, J.-C.; Saliou, J.-M.; Ankavay, M.; Bull, A.; Pillez, A.; Abravanel, F.; Helle, F.; et al. Hepatitis E Virus Lifecycle and Identification of 3 Forms of the ORF2 Capsid Protein. Gastroenterology 2018. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Ying, D.; Lhomme, S.; Tang, Z.; Walker, C.M.; Xia, N.; Zheng, Z.; Feng, Z. Origin, antigenicity, and function of a secreted form of ORF2 in hepatitis E virus infection. Proc. Natl. Acad. Sci. USA 2018, 115, 4773–4778. [Google Scholar] [CrossRef]
- Ding, Q.; Heller, B.; Capuccino, J.M.V.; Song, B.; Nimgaonkar, I.; Hrebikova, G.; Contreras, J.E.; Ploss, A. Hepatitis E virus ORF3 is a functional ion channel required for release of infectious particles. Proc. Natl. Acad. Sci. USA 2017, 114, 1147–1152. [Google Scholar] [CrossRef]
- Nair, V.P.; Anang, S.; Subramani, C.; Madhvi, A.; Bakshi, K.; Srivastava, A.; Shalimar; Nayak, B.; Ct, R.K.; Surjit, M. Endoplasmic Reticulum Stress Induced Synthesis of a Novel Viral Factor Mediates Efficient Replication of Genotype-1 Hepatitis E Virus. PLoS Pathog. 2016, 12, e1005521. [Google Scholar] [CrossRef] [PubMed]
- Adlhoch, C.; Avellon, A.; Baylis, S.A.; Ciccaglione, A.R.; Couturier, E.; de Sousa, R.; Epstein, J.; Ethelberg, S.; Faber, M.; Feher, A.; et al. Hepatitis E virus: Assessment of the epidemiological situation in humans in Europe, 2014/15. J. Clin. Virol. 2016, 82, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Meister, T.L.; Bruening, J.; Todt, D.; Steinmann, E. Cell culture systems for the study of hepatitis E virus. Antivir. Res. 2019. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Takahashi, M.; Kusano, E.; Okamoto, H. Development and evaluation of an efficient cell-culture system for Hepatitis E virus. J. Gen. Virol. 2007, 88, 903–911. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Takahashi, M.; Takahashi, H.; Ichiyama, K.; Hoshino, Y.; Nagashima, S.; Mizuo, H.; Okamoto, H. Development and characterization of a genotype 4 hepatitis E virus cell culture system using a HE-JF5/15F strain recovered from a fulminant hepatitis patient. J. Clin. Microbiol. 2009, 47, 1906–1910. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Tanaka, T.; Takahashi, H.; Hoshino, Y.; Nagashima, S.; Jirintai; Mizuo, H.; Yazaki, Y.; Takagi, T.; Azuma, M.; et al. Hepatitis E Virus (HEV) strains in serum samples can replicate efficiently in cultured cells despite the coexistence of HEV antibodies: characterization of HEV virions in blood circulation. J. Clin. Microbiol. 2010, 48, 1112–1125. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Tanaka, T.; Jirintai, S.; Nagashima, S.; Takahashi, M.; Nishizawa, T.; Mizuo, H.; Yazaki, Y.; Okamoto, H. A549 and PLC/PRF/5 cells can support the efficient propagation of swine and wild boar hepatitis E virus (HEV) strains: demonstration of HEV infectivity of porcine liver sold as food. Arch. Virol. 2012, 157, 235–246. [Google Scholar] [CrossRef]
- Schielke, A.; Filter, M.; Appel, B.; Johne, R. Thermal stability of hepatitis E virus assessed by a molecular biological approach. Virol. J. 2011, 8, 487. [Google Scholar] [CrossRef]
- Berto, A.; van der Poel, W.H.M.; Hakze-van der Honing, R.; Martelli, F.; La Ragione, R.M.; Inglese, N.; Collins, J.; Grierson, S.; Johne, R.; Reetz, J.; et al. Replication of hepatitis E virus in three-dimensional cell culture. J. Virol. Methods 2013, 187, 327–332. [Google Scholar] [CrossRef]
- Baez, P.A.; Lopez, M.C.; Duque-Jaramillo, A.; Pelaez, D.; Molina, F.; Navas, M.-C. First evidence of the Hepatitis E virus in environmental waters in Colombia. PLoS ONE 2017, 12, e0177525. [Google Scholar] [CrossRef]
- Shukla, P.; Nguyen, H.T.; Torian, U.; Engle, R.E.; Faulk, K.; Dalton, H.R.; Bendall, R.P.; Keane, F.E.; Purcell, R.H.; Emerson, S.U. Cross-species infections of cultured cells by hepatitis E virus and discovery of an infectious virus-host recombinant. Proc. Natl. Acad. Sci. USA 2011, 108, 2438–2443. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.; Torian, U.; Faulk, K.; Mather, K.; Engle, R.E.; Thompson, E.; Bonkovsky, H.L.; Emerson, S.U. A naturally occurring human/hepatitis E recombinant virus predominates in serum but not in faeces of a chronic hepatitis E patient and has a growth advantage in cell culture. J. Gen. Virol. 2012, 93, 526–530. [Google Scholar] [CrossRef]
- Johne, R.; Reetz, J.; Ulrich, R.G.; Machnowska, P.; Sachsenroder, J.; Nickel, P.; Hofmann, J. An ORF1-rearranged hepatitis E virus derived from a chronically infected patient efficiently replicates in cell culture. J. Viral Hepat. 2014, 21, 447–456. [Google Scholar] [CrossRef]
- Shiota, T.; Li, T.-C.; Yoshizaki, S.; Kato, T.; Wakita, T.; Ishii, K. The hepatitis E virus capsid C-terminal region is essential for the viral life cycle: implication for viral genome encapsidation and particle stabilization. J. Virol. 2013, 87, 6031–6036. [Google Scholar] [CrossRef]
- Gouttenoire, J.; Pollán, A.; Abrami, L.; Oechslin, N.; Mauron, J.; Matter, M.; Oppliger, J.; Szkolnicka, D.; Dao Thi, V.L.; van der Goot, F.G.; et al. Palmitoylation mediates membrane association of hepatitis E virus ORF3 protein and is required for infectious particle secretion. PLoS Pathog. 2018, 14, e1007471. [Google Scholar] [CrossRef]
- Sridhar, S. Use of S17 fragment containing hepatitis E virus infectious clones in cell culture experiments: The fine print does matter. J. Viral Hepat. 2018, 25, 1105. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, Q.; Xia, N. Prophylactic Hepatitis E Vaccine. Adv. Exp. Med. Biol. 2016, 948, 223–246. [Google Scholar] [CrossRef] [PubMed]
- Cook, N.; D’Agostino, M.; Johne, R. Potential Approaches to Assess the Infectivity of Hepatitis E Virus in Pork Products: A Review. Food Environ. Virol. 2017. [Google Scholar] [CrossRef]
- Friebe, P.; Boudet, J.; Simorre, J.-P.; Bartenschlager, R. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 2005, 79, 380–392. [Google Scholar] [CrossRef] [PubMed]
- Schemmerer, M.; Apelt, S.; Trojnar, E.; Ulrich, R.G.; Wenzel, J.J.; Johne, R. Enhanced Replication of Hepatitis E Virus Strain 47832c in an A549-Derived Subclonal Cell Line. Viruses 2016, 8, 267. [Google Scholar] [CrossRef]
- Wenzel, J.J.; Preiss, J.; Schemmerer, M.; Huber, B.; Plentz, A.; Jilg, W. Detection of hepatitis E virus (HEV) from porcine livers in Southeastern Germany and high sequence homology to human HEV isolates. J. Clin. Virol. 2011, 52, 50–54. [Google Scholar] [CrossRef] [PubMed]
- Riddell, M.A.; Li, F.; Anderson, D.A. Identification of immunodominant and conformational epitopes in the capsid protein of hepatitis E virus by using monoclonal antibodies. J. Virol. 2000, 74, 8011–8017. [Google Scholar] [CrossRef]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000, 132, 365–386. [Google Scholar] [PubMed]
- Sobhy, H.; Colson, P. Gemi: PCR primers prediction from multiple alignments. Comp. Funct. Genom. 2012, 2012, 783138. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Norder, H.; Uhlhorn, H.; Belák, S.; Widén, F. Novel hepatitis E like virus found in Swedish moose. J. Gen. Virol. 2014, 95, 557–570. [Google Scholar] [CrossRef] [PubMed]
- Inoue, J.; Nishizawa, T.; Takahashi, M.; Aikawa, T.; Mizuo, H.; Suzuki, K.; Shimosegawa, T.; Okamoto, H. Analysis of the full-length genome of genotype 4 hepatitis E virus isolates from patients with fulminant or acute self-limited hepatitis E. J. Med. Virol. 2006, 78, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Tanaka, T.; Azuma, M.; Kusano, E.; Aikawa, T.; Shibayama, T.; Yazaki, Y.; Mizuo, H.; Inoue, J.; Okamoto, H. Prolonged fecal shedding of hepatitis E virus (HEV) during sporadic acute hepatitis E: Evaluation of infectivity of HEV in fecal specimens in a cell culture system. J. Clin. Microbiol. 2007, 45, 3671–3679. [Google Scholar] [CrossRef] [PubMed]
- Shukla, P.; Nguyen, H.T.; Faulk, K.; Mather, K.; Torian, U.; Engle, R.E.; Emerson, S.U. Adaptation of a genotype 3 hepatitis E virus to efficient growth in cell culture depends on an inserted human gene segment acquired by recombination. J. Virol. 2012, 86, 5697–5707. [Google Scholar] [CrossRef]
- Huang, R.; Li, D.; Wei, S.; Li, Q.; Yuan, X.; Geng, L.; Li, X.; Liu, M. Cell culture of sporadic hepatitis E virus in China. Clin. Diagn. Lab. Immunol. 1999, 6, 729–733. [Google Scholar] [PubMed]
- Roethl, E.; Gassner, M.; Krenn, B.M.; Romanovskaya-Romanko, E.A.; Seper, H.; Romanova, J.; Nakowitsch, S.; Sturlan, S.; Wolschek, M.; Sirotkin, A.; et al. Antimycotic-antibiotic amphotericin B promotes influenza virus replication in cell culture. J. Virol. 2011, 85, 11139–11145. [Google Scholar] [CrossRef] [PubMed]
- Holla, P.; Ahmad, I.; Ahmed, Z.; Jameel, S. Hepatitis E virus enters liver cells through a dynamin-2, clathrin and membrane cholesterol-dependent pathway. Traffic 2015, 16, 398–416. [Google Scholar] [CrossRef]
- Yin, X.; Ambardekar, C.; Lu, Y.; Feng, Z. Distinct Entry Mechanisms for Nonenveloped and Quasi-Enveloped Hepatitis E Viruses. J. Virol. 2016, 90, 4232–4242. [Google Scholar] [CrossRef]
- Kamar, N.; Izopet, J.; Tripon, S.; Bismuth, M.; Hillaire, S.; Dumortier, J.; Radenne, S.; Coilly, A.; Garrigue, V.; D’Alteroche, L.; et al. Ribavirin for chronic hepatitis E virus infection in transplant recipients. N. Engl. J. Med. 2014, 370, 1111–1120. [Google Scholar] [CrossRef] [PubMed]
- Ikram, A.; Hakim, M.S.; Zhou, J.-H.; Wang, W.; Peppelenbosch, M.P.; Pan, Q. Genotype-specific acquisition, evolution and adaptation of characteristic mutations in hepatitis E virus. Virulence 2017, 9, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Debing, Y.; Gisa, A.; Dallmeier, K.; Pischke, S.; Bremer, B.; Manns, M.; Wedemeyer, H.; Suneetha, P.V.; Neyts, J. A mutation in the hepatitis E virus RNA polymerase promotes its replication and associates with ribavirin treatment failure in organ transplant recipients. Gastroenterology 2014, 147, 1008–1011. [Google Scholar] [CrossRef] [PubMed]
- Dao Thi, V.L.; Debing, Y.; Wu, X.; Rice, C.M.; Neyts, J.; Moradpour, D.; Gouttenoire, J. Sofosbuvir Inhibits Hepatitis E Virus Replication In Vitro and Results in an Additive Effect When Combined With Ribavirin. Gastroenterology 2016, 150, 82–85. [Google Scholar] [CrossRef]
- Biliotti, E.; Franchi, C.; Spaziante, M.; Garbuglia, A.R.; Volpicelli, L.; Palazzo, D.; de Angelis, M.; Esvan, R.; Taliani, G. Autochthonous acute hepatitis E: Treatment with sofosbuvir and ribavirin. Infection 2018. [Google Scholar] [CrossRef]
- Donnelly, M.C.; Imlach, S.N.; Abravanel, F.; Ramalingam, S.; Johannessen, I.; Petrik, J.; Fraser, A.R.; Campbell, J.D.M.; Bramley, P.; Dalton, H.R.; et al. Sofosbuvir and Daclatasvir Anti-Viral Therapy Fails to Clear HEV Viremia and Restore Reactive T Cells in a HEV/HCV Co-Infected Liver Transplant Recipient. Gastroenterology 2017, 152, 300–301. [Google Scholar] [CrossRef]
- Van der Valk, M.; Zaaijer, H.L.; Kater, A.P.; Schinkel, J. Sofosbuvir shows antiviral activity in a patient with chronic hepatitis E virus infection. J. Hepatol. 2017, 66, 242–243. [Google Scholar] [CrossRef] [PubMed]
- Glitscher, M.; Himmelsbach, K.; Woytinek, K.; Johne, R.; Reuter, A.; Spiric, J.; Schwaben, L.; Grünweller, A.; Hildt, E. Inhibition of Hepatitis E Virus Spread by the Natural Compound Silvestrol. Viruses 2018, 10, 301. [Google Scholar] [CrossRef] [PubMed]
- Todt, D.; Moeller, N.; Praditya, D.; Kinast, V.; Friesland, M.; Engelmann, M.; Verhoye, L.; Sayed, I.M.; Behrendt, P.; Dao Thi, V.L.; et al. The natural compound silvestrol inhibits hepatitis E virus (HEV) replication in vitro and in vivo. Antivir. Res. 2018, 157, 151–158. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schemmerer, M.; Johne, R.; Erl, M.; Jilg, W.; Wenzel, J.J. Isolation of Subtype 3c, 3e and 3f-Like Hepatitis E Virus Strains Stably Replicating to High Viral Loads in an Optimized Cell Culture System. Viruses 2019, 11, 483. https://doi.org/10.3390/v11060483
Schemmerer M, Johne R, Erl M, Jilg W, Wenzel JJ. Isolation of Subtype 3c, 3e and 3f-Like Hepatitis E Virus Strains Stably Replicating to High Viral Loads in an Optimized Cell Culture System. Viruses. 2019; 11(6):483. https://doi.org/10.3390/v11060483
Chicago/Turabian StyleSchemmerer, Mathias, Reimar Johne, Monika Erl, Wolfgang Jilg, and Jürgen J. Wenzel. 2019. "Isolation of Subtype 3c, 3e and 3f-Like Hepatitis E Virus Strains Stably Replicating to High Viral Loads in an Optimized Cell Culture System" Viruses 11, no. 6: 483. https://doi.org/10.3390/v11060483
APA StyleSchemmerer, M., Johne, R., Erl, M., Jilg, W., & Wenzel, J. J. (2019). Isolation of Subtype 3c, 3e and 3f-Like Hepatitis E Virus Strains Stably Replicating to High Viral Loads in an Optimized Cell Culture System. Viruses, 11(6), 483. https://doi.org/10.3390/v11060483




