DWV-A Lethal to Honey Bees (Apis mellifera): A Colony Level Survey of DWV Variants (A, B, and C) in England, Wales, and 32 States across the US
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Sample Processing
2.3. RT-qPCR
2.4. Analysis of the Results
3. Results
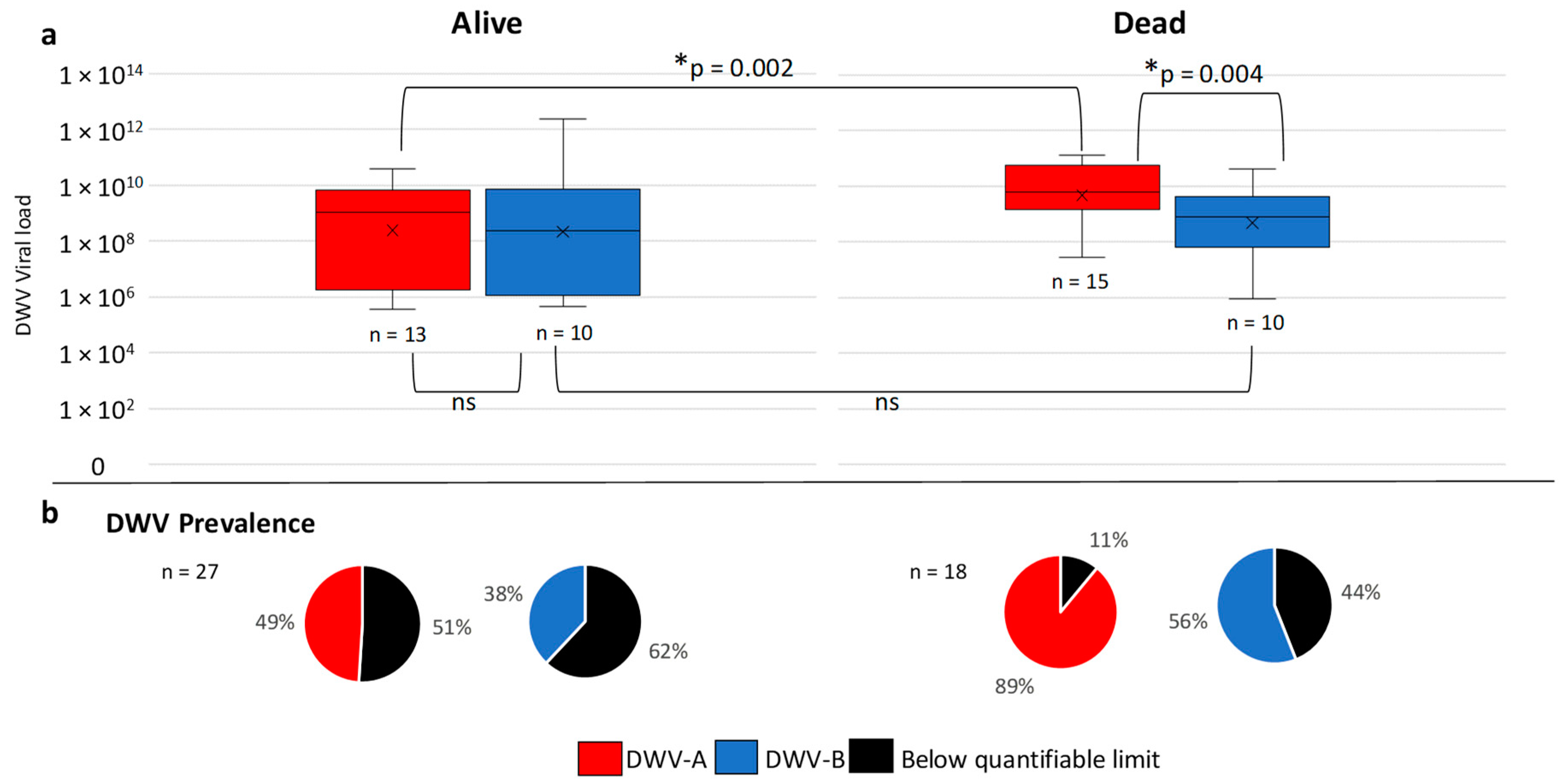
3.1. DWV Variant Prevalence and Viral Load Between Spring and Autumn in Apiaries in England and Wales
3.2. English and Welsh Colony Level DWV Analysis
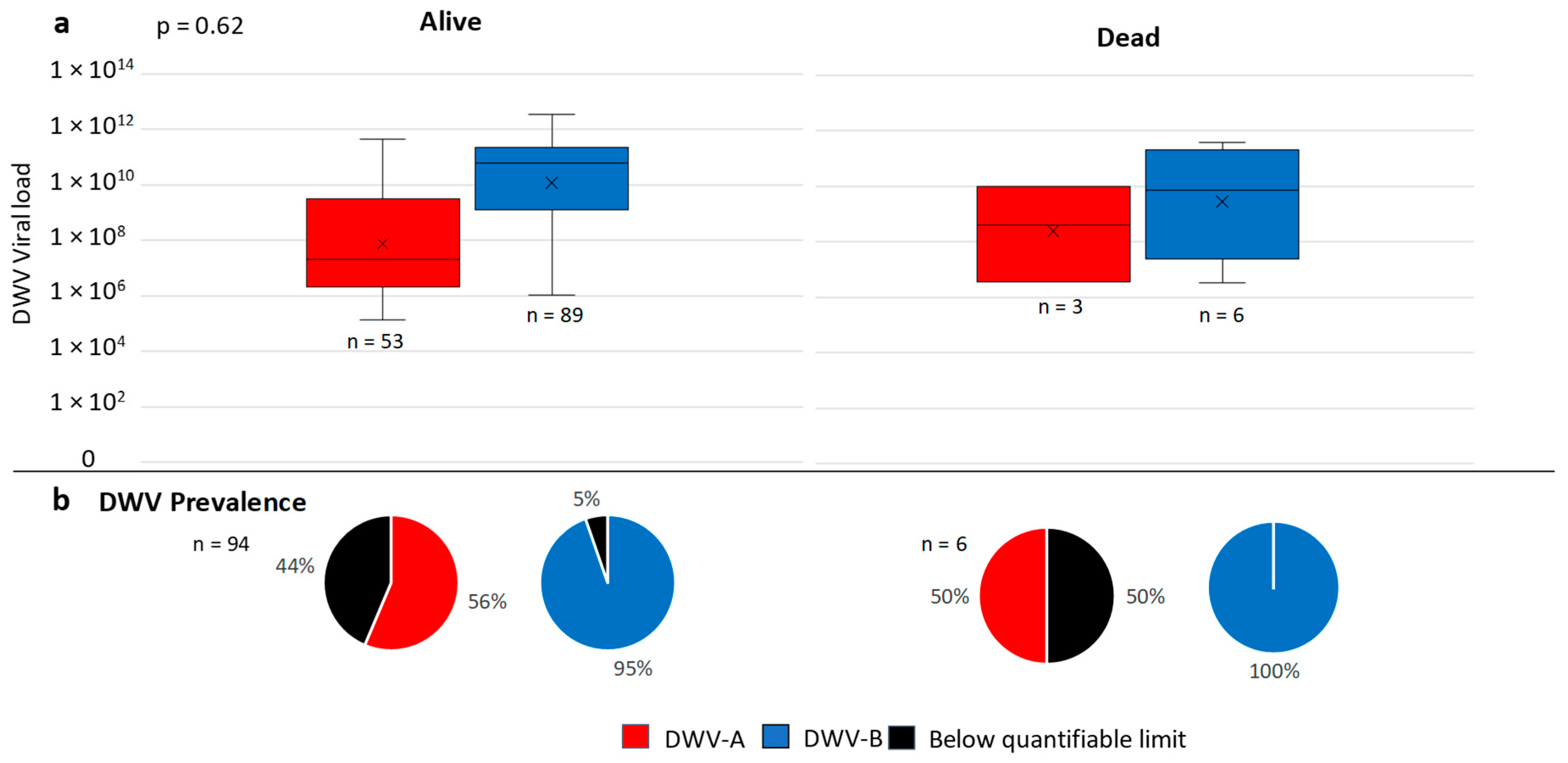
3.3. Apiary Level Analysis of Pre-Winter United States Honey Bee Colonies
3.4. Overwinter Colony Mortality
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Potts, S.G.; Biesmeijer, J.C.; Kremen, C.; Neumann, P.; Schweiger, O.; Kunin, W.E. Global pollinator declines: Trends, impacts and drivers. Trends Ecol. Evol. 2010, 25, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.J.; Highfield, A.C.; Brettell, L.; Villalobos, E.M.; Budge, G.E.; Powell, M.; Nikaido, S.; Schroeder, D.C. Global honey bee viral landscape altered by a parasitic mite. Science 2012, 336, 1304–1306. [Google Scholar] [CrossRef]
- Wilfert, L.; Long, G.; Leggett, H.C.; Schmid-Hempel, P.; Butlin, R.; Martin, S.J.M.; Boots, M. Deformed wing virus is a recent global epidemic in honeybees driven by Varroa mites. Science 2016, 351, 594–597. [Google Scholar] [CrossRef]
- Martin, S.J.; Brettell, L.E. Deformed wing virus in honey bees and other insects. Annu. Rev. Virol. 2019. [Google Scholar] [CrossRef]
- Anderson, D.L.; Trueman, J.W.H. Varroa jacobsoni (Acari: Varroidae) is more than one species. Exp. Appl. Acarol. 2000, 24, 165–189. [Google Scholar] [CrossRef] [PubMed]
- Sammataro, D.; Gerson, U.; Needham, G. Parasitic mites of honey bees: Life history, implications, and impact. Annu. Rev. Entomol. 2000, 45, 519–548. [Google Scholar] [CrossRef] [PubMed]
- Danka, R.G.; Rinderer, T.E.; Kuznetsov, V.N.; Delatte, G.T. A USDA-ARS project to evaluate resistance to Varroa jacobsoni by honey bees of Far-Eastern Russia. Am. Bee J. 1995, 135, 746–748. [Google Scholar]
- Ryabov, E.V.; Wood, G.R.; Fannon, J.M.; Moore, J.D.; Bull, J.C.; Chandler, D.; Mead, A.; Burroughs, N.; Evans, D.J. A virulent strain of deformed wing virus (DWV) of honeybees (Apis mellifera) prevails after Varroa destructor-mediated, or in vitro, transmission. PLoS Pathog. 2014, 10, e1004230. [Google Scholar] [CrossRef]
- Loope, K.J.; Baty, J.W.; Lester, P.J.; Wilson Rankin, E.E. Pathogen shifts in a honeybee predator following the arrival of the Varroa mite. Proc. R. Soc. B 2019, 286, 20182499. [Google Scholar] [CrossRef]
- Schroeder, D.C.; Martin, S.J. Deformed wing virus: The main suspect in unexplained honeybee deaths worldwide. Virulence 2012, 3, 589–591. [Google Scholar] [CrossRef]
- Francis, R.M.; Nielsen, S.L.; Kryger, P. Varroa-virus interaction in collapsing honey bee colonies. PLoS ONE 2013, 8, e57540. [Google Scholar] [CrossRef] [PubMed]
- Ongus, J.R.; Peters, D.; Bonmatin, J.M.; Bengsch, E.; Vlak, J.M.; van Oers, M.M. Complete sequence of a picorna-like virus of the genus Iflavirus replicating in the mite Varroa destructor. J. Gen. Virol. 2004, 85, 3747–3755. [Google Scholar] [CrossRef] [PubMed]
- Baker, A.C.; Schroeder, D.C. The use of RNA-dependent RNA polymerase for the taxonomic assignment of Picorna-like viruses (order Picornavirales) infecting Apis mellifera L. populations. Virol. J. 2008, 5, 10. [Google Scholar] [CrossRef] [PubMed]
- Mordecai, G.J.; Wilfert, L.; Martin, S.J.; Jones, I.M.; Schroeder, D.C. Diversity in a honey bee pathogen: First report of a third master variant of the Deformed Wing Virus quasispecies. ISME J. 2016, 10, 1264–1273. [Google Scholar] [CrossRef] [PubMed]
- Mordecai, G.J.; Brettell, L.E.; Martin, S.J.; Dixon, D.; Jones, I.M.; Schroeder, D.C. Superinfection exclusion and the long-term survival of honey bees in Varroa-infested colonies. ISME J. 2016, 10, 1182–1191. [Google Scholar] [CrossRef] [PubMed]
- McMahon, D.P.; Natsopoulou, M.E.; Doublet, V.; Fürst, M.; Weging, S.; Brown, M.J.; Gogol-Döring, A.; Paxton, R.J. Elevated virulence of an emerging viral genotype as a driver of honeybee loss. Proc. R. Soc. Lond. B Biol. Sci. 2016, 283. [Google Scholar] [CrossRef]
- Tehel, A.; Vu, Q.; Bigot, D.; Gogol-Döring, A.; Koch, P.; Jenkins, C.; Doublet, V.; Theodorou, P.; Paxton, R. The Two Prevalent Genotypes of an Emerging Infectious Disease, Deformed Wing Virus, Cause Equally Low Pupal Mortality and Equally High Wing Deformities in Host Honey Bees. Viruses 2019, 11, 114. [Google Scholar] [CrossRef]
- Gisder, S.; Möckel, N.; Eisenhardt, D.; Genersch, E. In vivo evolution of viral virulence: Switching of deformed wing virus between hosts results in virulence changes and sequence shifts. Environ. Microbiol. 2018, 20, 4612–4628. [Google Scholar] [CrossRef]
- Highfield, A.C.; El Nagar, A.; Mackinder, L.C.; Laure, M.L.N.; Hall, M.J.; Martin, S.J.; Schroeder, D.C. Deformed wing virus implicated in overwintering honeybee colony losses. Appl. Environ. Microbiol. 2009, 75, 7212–7220. [Google Scholar] [CrossRef]
- Kevill, J.L.; Highfield, A.; Mordecai, G.J.; Martin, S.J.; Schroeder, D.C. ABC assay: Method development and application to quantify the role of three DWV master variants in overwinter colony losses of European honey bees. Viruses 2017, 9, 314. [Google Scholar] [CrossRef]
- Allsopp, M.H. Analysis of Varroa Destructor Infestation of Southern African Honeybee Populations. Ph.D. Thesis, University of Pretoria, Pretoria, South Africa, 2006. [Google Scholar]
- Roberts, J.M.; Anderson, D.L.; Durr, P.A. Metagenomic analysis of Varroa-free Australian honey bees (Apis mellifera) shows a diverse Picornavirales virome. J. Gen. Virol. 2018, 99, 818–826. [Google Scholar] [CrossRef]
- Bee Informed Partnership. 2016–2017 Loss Results. Available online: https://beeinformed.org/2017/05/25/2016-2017-loss-results-thank-you-to-all-survey-participants/ (accessed on 1 June 2018).
- BBKA. Honey Bee Colony Losses in England. Winter Survival Survey 2017. Available online: https://www.bbka.org.uk/winter-honey-bee-losses-in-england (accessed on 1 June 2018).
- Hayes, J., Jr.; Underwood, R.M.; Pettis, J. A survey of honey bee colony losses in the US, fall 2007 to spring 2008. PLoS ONE 2008, 3, e4071. [Google Scholar]
- Van Engelsdorp, D.; Hayes, J., Jr.; Underwood, R.M.; Pettis, J.S. A survey of honey bee colony losses in the United States, fall 2008 to spring 2009. J. Apic. Res. 2010, 49, 7–14. [Google Scholar] [CrossRef]
- Van Engelsdorp, D.; Hayes, J., Jr.; Underwood, R.M.; Caron, D.; Pettis, J. A survey of managed honey bee colony losses in the USA, fall 2009 to winter 2010. J. Apic. Res. 2011, 50, 1–10. [Google Scholar] [CrossRef]
- Van Engelsdorp, D.; Caron, D.; Hayes, J.; Underwood, R.; Henson, M.; Rennich, K.; Spleen, A.; Andree, M.; Snyder, R.; Lee, K.; et al. A national survey of managed honey bee 2010–11 winter colony losses in the USA: Results from the Bee informed Partnership. J. Apic. Res. 2012, 51, 115–124. [Google Scholar] [CrossRef]
- Ellis, J.D.; Evans, J.D.; Pettis, J. Colony losses, managed colony population decline, and Colony Collapse Disorder in the United States. J. Apic. Res. 2010, 49, 134–136. [Google Scholar] [CrossRef]
- Spleen, A.M.; Lengerich, E.J.; Rennich, K.; Caron, D.; Rose, R.; Pettis, J.S.; Henson, M.; Wilkes, J.T.; Wilson, M.; Stitzinger, J.; et al. A national survey of managed honey bee 2011–12 winter colony losses in the United States: Results from the Bee Informed Partnership. J. Apic. Res. 2013, 52, 44–53. [Google Scholar] [CrossRef]
- Steinhauer, N.A.; Rennich, K.; Wilson, M.E.; Caron, D.M.; Lengerich, E.J.; Pettis, J.S.; Rose, R.; Skinner, J.A.; Tarpy, D.R.; Wilkes, J.T.; et al. A national survey of managed honey bee 2012–2013 annual colony losses in the USA: Results from the Bee Informed Partnership. J. Apic. Res. 2014, 53, 1–18. [Google Scholar] [CrossRef]
- Lee, K.V.; Steinhauer, N.; Rennich, K.; Wilson, M.E.; Tarpy, D.R.; Caron, D.M.; Rose, R.; Delaplane, K.S.; Baylis, K.; Lengerich, E.J.; et al. A national survey of managed honey bee 2013–2014 annual colony losses in the USA. Apidologie 2015, 46, 292–305. [Google Scholar] [CrossRef]
- Berthoud, H.; Imdorf, A.; Haueter, M.; Radloff, S.; Neumann, P. Virus infections and winter losses of honey bee colonies (Apis mellifera). J. Apic. Res. 2010, 49, 60–65. [Google Scholar] [CrossRef]
- Dainat, B.; Evans, J.D.; Chen, Y.P.; Gauthier, L.; Neumann, P. Dead or alive: Deformed wing virus and Varroa destructor reduce the life span of winter honeybees. Appl. Environ. Microbiol. 2012, 78, 981–987. [Google Scholar] [CrossRef]
- Martin, S.J.; Ball, B.V.; Carreck, N.L. Prevalence and persistence of Deformed wing virus (DWV) in untreated or acaricide-treated Varroa destructor infested honey bee (Apis mellifera) colonies. J. Apic. Res. 2010, 49, 72–79. [Google Scholar] [CrossRef]
- Martin, S.J.; Ball, B.V.; Carreck, N.L. The role of Deformed wing virus in the initial collapse of Varroa infested honey bee colonies in the UK. J. Apic. Res. 2013, 52, 251–258. [Google Scholar] [CrossRef]
- Nazzi, F.; Brown, S.P.; Annoscia, D.; Del Piccolo, F.; Di Prisco, G.; Varricchio, P.; Della Vedova, G.; Cattonaro, F.; Caprio, E.; Pennacchio, F. Synergistic parasite-pathogen interactions mediated by host immunity can drive the collapse of honeybee colonies. PLoS Pathog. 2012, 8, e1002735. [Google Scholar] [CrossRef]
- Cornman, R.S.; Tarpy, D.R.; Chen, Y.; Jeffreys, L.; Lopez, D.; Pettis, J.S.; Evans, J.D. Pathogen webs in collapsing honey bee colonies. PLoS ONE 2012, 7, e43562. [Google Scholar] [CrossRef]
- Traynor, K.S.; Rennich, K.; Forsgren, E.; Rose, R.; Pettis, J.; Kunkel, G.; Madella, S.; Evans, J.; Lopez, D. Multiyear survey targeting disease incidence in US honey bees. Apidologie 2016, 47, 325–347. [Google Scholar] [CrossRef]
- Glenny, W.; Cavigli, I.; Daughenbaugh, K.F.; Radford, R.; Kegley, S.E.; Flenniken, M.L. Honey bee (Apis mellifera) colony health and pathogen composition in migratory beekeeping operations involved in California almond pollination. PLoS ONE 2017, 12, e0182814. [Google Scholar] [CrossRef]
- Yue, C.; Genersch, E. RT-PCR analysis of Deformed wing virus in honeybees (Apis mellifera) and mites (Varroa destructor). J. Gen. Virol. 2005, 86, 3419–3424. [Google Scholar] [CrossRef]
- Genersch, E.; Von Der Ohe, W.; Kaatz, H.; Schroeder, A.; Otten, C.; Büchler, R.; Berg, S.; Ritter, W.; Mühlen, W.; Gisder, S.; et al. The German bee monitoring project: A long term study to understand periodically high winter losses of honey bee colonies. Apidologie 2010, 41, 332–352. [Google Scholar] [CrossRef]
- Zioni, N.; Soroker, V.; Chejanovsky, N. Replication of Varroa destructor virus 1 (VDV-1) and a Varroa destructor virus 1–Deformed wing virus recombinant (VDV-1–DWV) in the head of the honey bee. Virology 2011, 417, 106–112. [Google Scholar] [CrossRef]
- Siede, R.; König, M.; Büchler, R.; Failing, K.; Thiel, H.J. A real-time PCR based survey on acute bee paralysis virus in German bee colonies. Apidologie 2008, 39, 650–661. [Google Scholar] [CrossRef]
- Fujiyuki, T.; Ohka, S.; Takeuchi, H.; Ono, M.; Nomoto, A.; Kubo, T. Prevalence and phylogeny of Kakugo virus, a novel insect picorna-like virus that infects the honeybee (Apis mellifera L.), under various colony conditions. J. Virol. 2006, 80, 11528–11538. [Google Scholar] [CrossRef]
- Dunn, O.J. Multiple comparisons among means. J. Am. Stat. Assoc. 1961, 56, 52–64. [Google Scholar] [CrossRef]
- Ryabov, E.V.; Childers, A.K.; Chen, Y.; Madella, S.; Nessa, A.; Evans, J.D. Recent spread of Varroa destructor virus-1, a honey bee pathogen, in the United States. Sci. Rep. 2017, 7, 17447. [Google Scholar] [CrossRef]
- Natsopoulou, M.E.; McMahon, D.P.; Doublet, V.; Frey, E.; Rosenkranz, P.; Paxton, R.J. The virulent, emerging genotype B of Deformed wing virus is closely linked to overwinter honeybee worker loss. Sci. Rep. 2017, 7, 5242. [Google Scholar] [CrossRef]
- Tscherne, D.M.; Evans, M.J.; von Hahn, T.; Jones, C.T.; Stamataki, Z.; McKeating, J.A.; Lindenbach, B.D.; Rice, C.M. Superinfection exclusion in cells infected with hepatitis C virus. J. Virol. 2007, 81, 3693–3703. [Google Scholar] [CrossRef]
- Chen, Y.P.; Evans, J.; Feldlaufer, M. Horizontal and vertical transmission of viruses in the honey bee. Apis mellifera. J. Invertebr. Pathol. 2006, 92, 152–159. [Google Scholar] [CrossRef]
- Yue, C.; Schröder, M.; Bienefeld, K.; Genersch, E. Detection of viral sequences in semen of honeybees (Apis mellifera): Evidence for vertical transmission of viruses through drones. J. Invertebr. Pathol. 2006, 92, 105–108. [Google Scholar] [CrossRef]
- Yue, C.; Schröder, M.; Gisder, S.; Genersch, E. Vertical-transmission routes for Deformed wing virus of honeybees (Apis mellifera). J. Gen. Virol. 2007, 88, 2329–2336. [Google Scholar] [CrossRef]
- De Miranda, J.R.; Genersch, E. Deformed wing virus. J. Invertebr. Pathol. 2010, 103, 48–61. [Google Scholar] [CrossRef]
- Locke, B.; Semberg, E.; Forsgren, E.; De Miranda, J.R. Persistence of subclinical deformed wing virus infections in honeybees following Varroa mite removal and a bee population turnover. PLoS ONE 2017, 12, e0180910. [Google Scholar] [CrossRef] [PubMed]
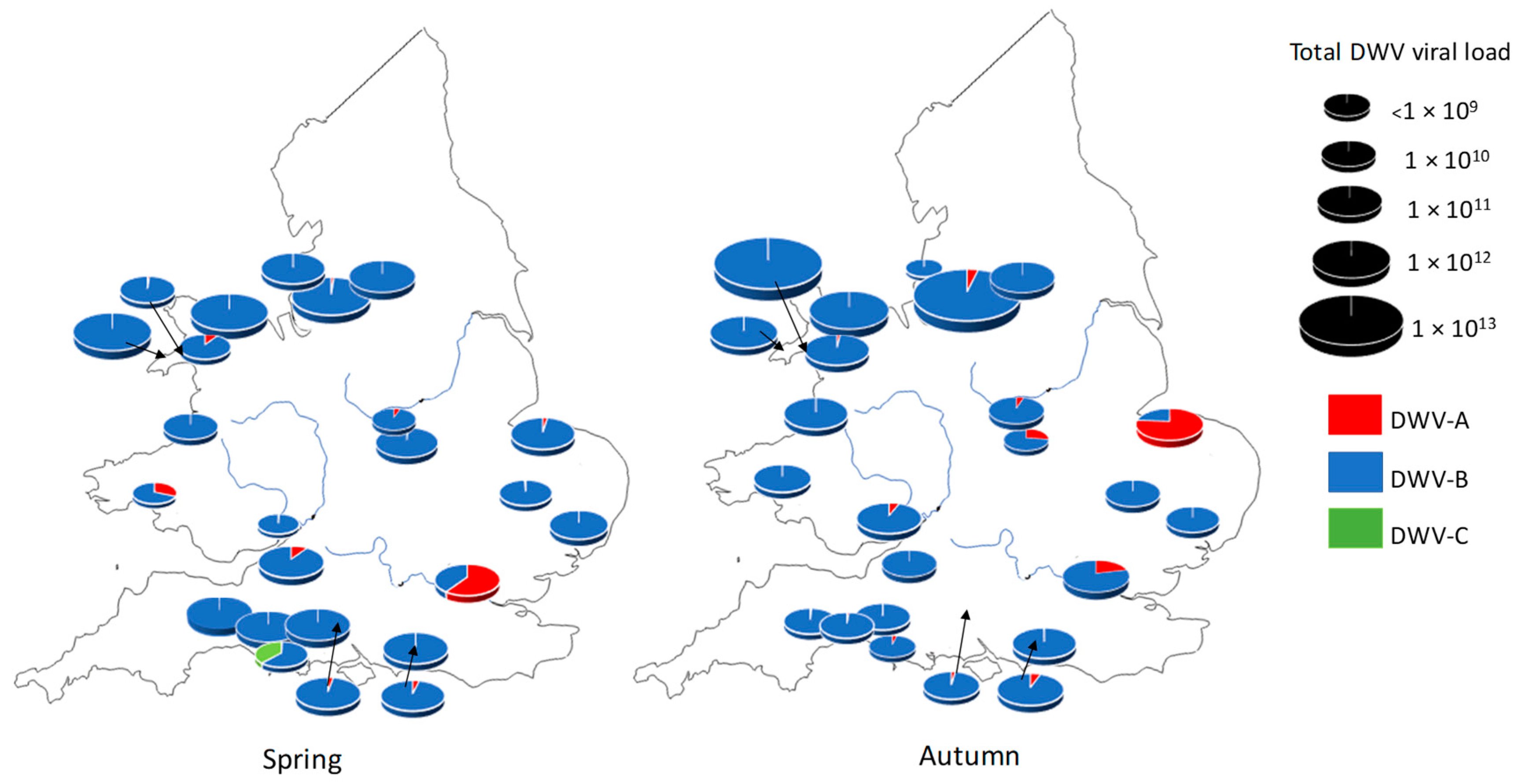
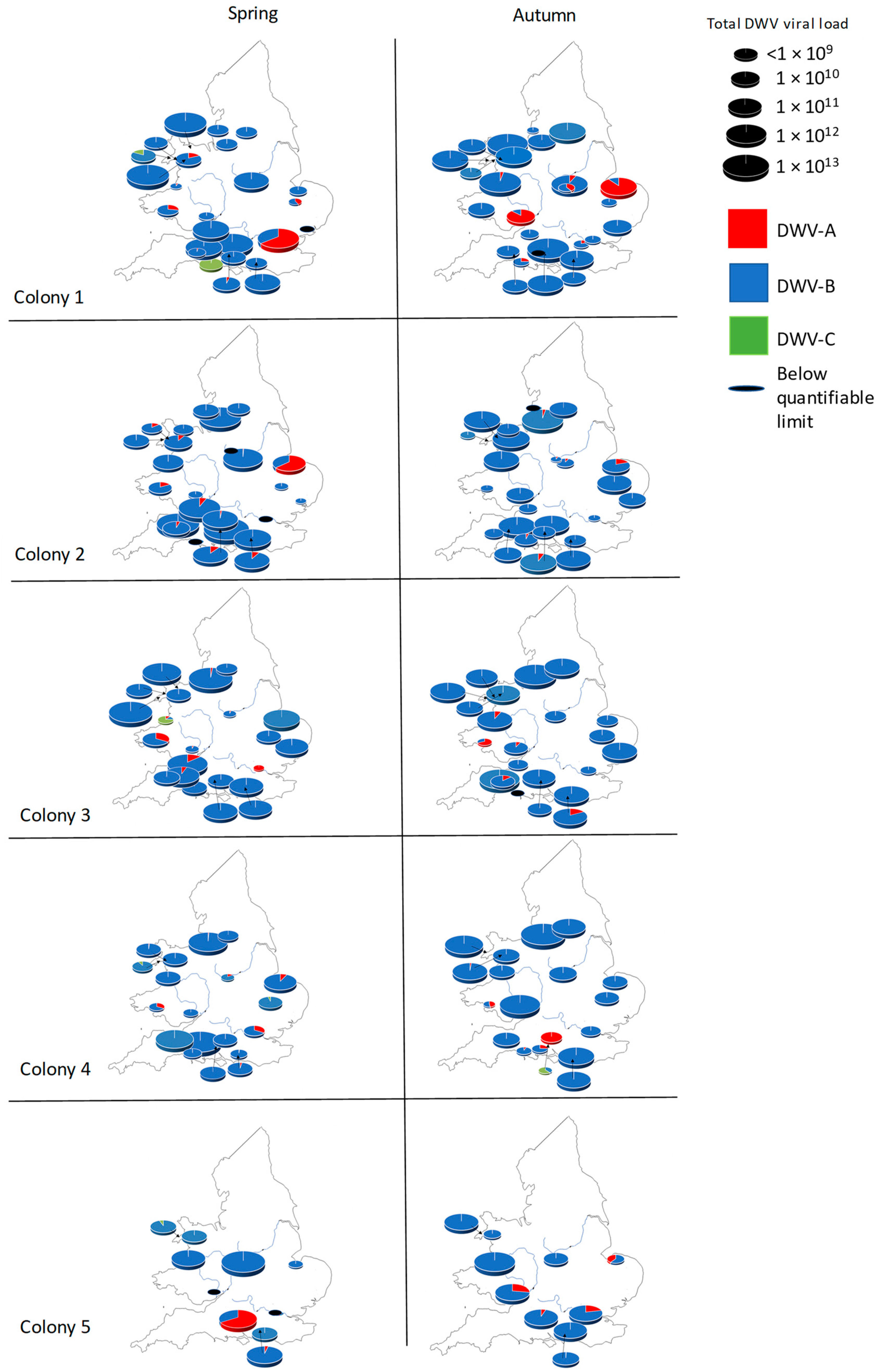
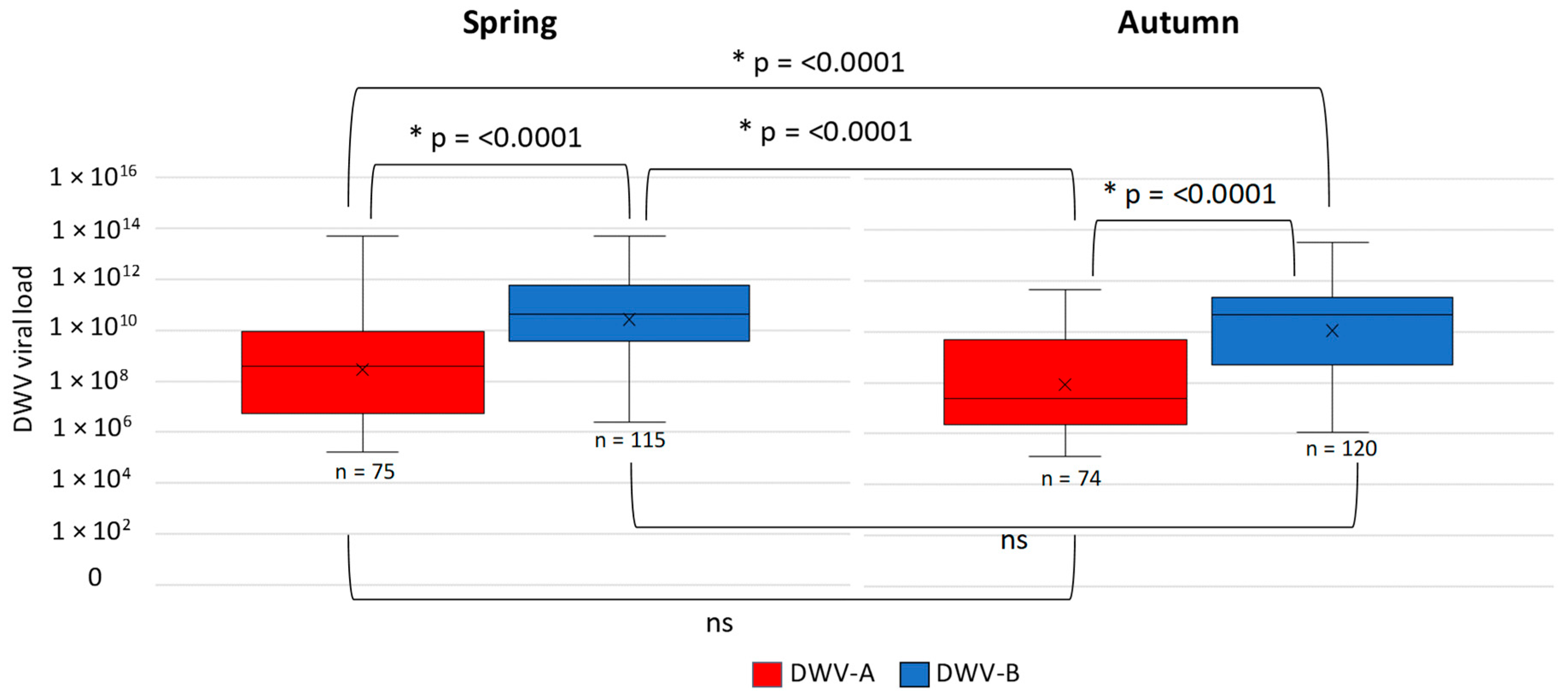
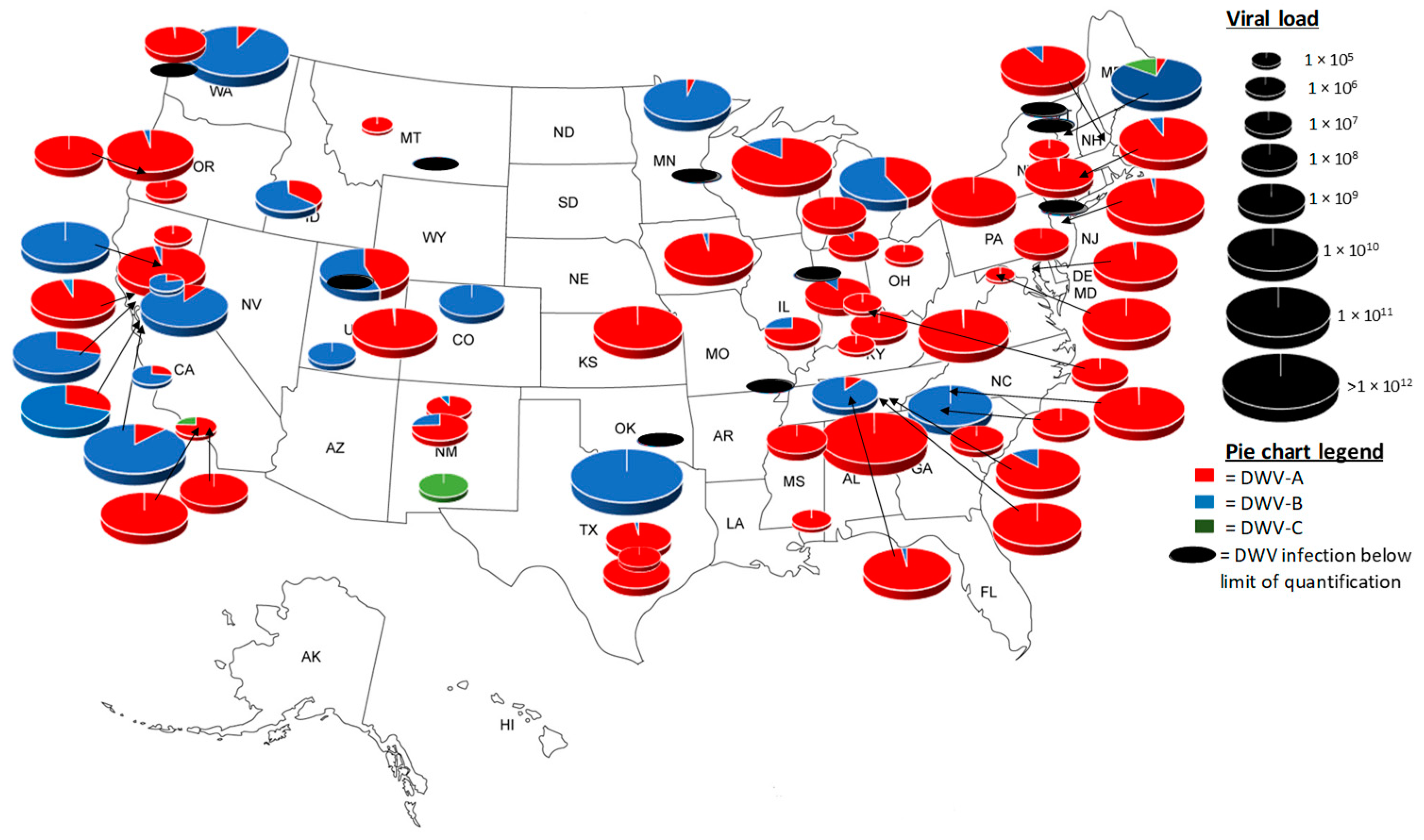
| Spring | SE Spring | Autumn | SE Autumn | |
|---|---|---|---|---|
| DWV-A | 3.89 × 1011 | 7.48 × 1010 | 2.46 × 1010 | 4.46 × 109 |
| DWV-B | 1.48 × 1012 | 2.26 × 1011 | 1.14 × 1012 | 2.01 × 1011 |
| DWV-C | 2.18 × 109 | 5.83 × 108 | 1.80 × 109 | 5.21 × 108 |
| Average Viral Load | SE | |
|---|---|---|
| DWV-A | 1.74 × 1010 | 2.16 × 109 |
| DWV-B | 7.12 × 1010 | 1.05 × 1010 |
| DWV-C | 1.99 × 108 | 5.53 × 107 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kevill, J.L.; de Souza, F.S.; Sharples, C.; Oliver, R.; Schroeder, D.C.; Martin, S.J. DWV-A Lethal to Honey Bees (Apis mellifera): A Colony Level Survey of DWV Variants (A, B, and C) in England, Wales, and 32 States across the US. Viruses 2019, 11, 426. https://doi.org/10.3390/v11050426
Kevill JL, de Souza FS, Sharples C, Oliver R, Schroeder DC, Martin SJ. DWV-A Lethal to Honey Bees (Apis mellifera): A Colony Level Survey of DWV Variants (A, B, and C) in England, Wales, and 32 States across the US. Viruses. 2019; 11(5):426. https://doi.org/10.3390/v11050426
Chicago/Turabian StyleKevill, Jessica L., Flaviane S. de Souza, Christopher Sharples, Randy Oliver, Declan C. Schroeder, and Stephen J. Martin. 2019. "DWV-A Lethal to Honey Bees (Apis mellifera): A Colony Level Survey of DWV Variants (A, B, and C) in England, Wales, and 32 States across the US" Viruses 11, no. 5: 426. https://doi.org/10.3390/v11050426
APA StyleKevill, J. L., de Souza, F. S., Sharples, C., Oliver, R., Schroeder, D. C., & Martin, S. J. (2019). DWV-A Lethal to Honey Bees (Apis mellifera): A Colony Level Survey of DWV Variants (A, B, and C) in England, Wales, and 32 States across the US. Viruses, 11(5), 426. https://doi.org/10.3390/v11050426






