Interaction between Two Iridovirus Core Proteins and Their Effects on Ranavirus (RGV) Replication in Cells from Different Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Virus
2.2. Plasmid Construction
2.3. Yeast Two Hybrid (Y2H) Assays
2.4. Co-IP Assays
2.5. Western Blot Analysis
2.6. Fluorescence Microscopy
2.7. Quantitative Analysis of RGV Genomic DNA
3. Results
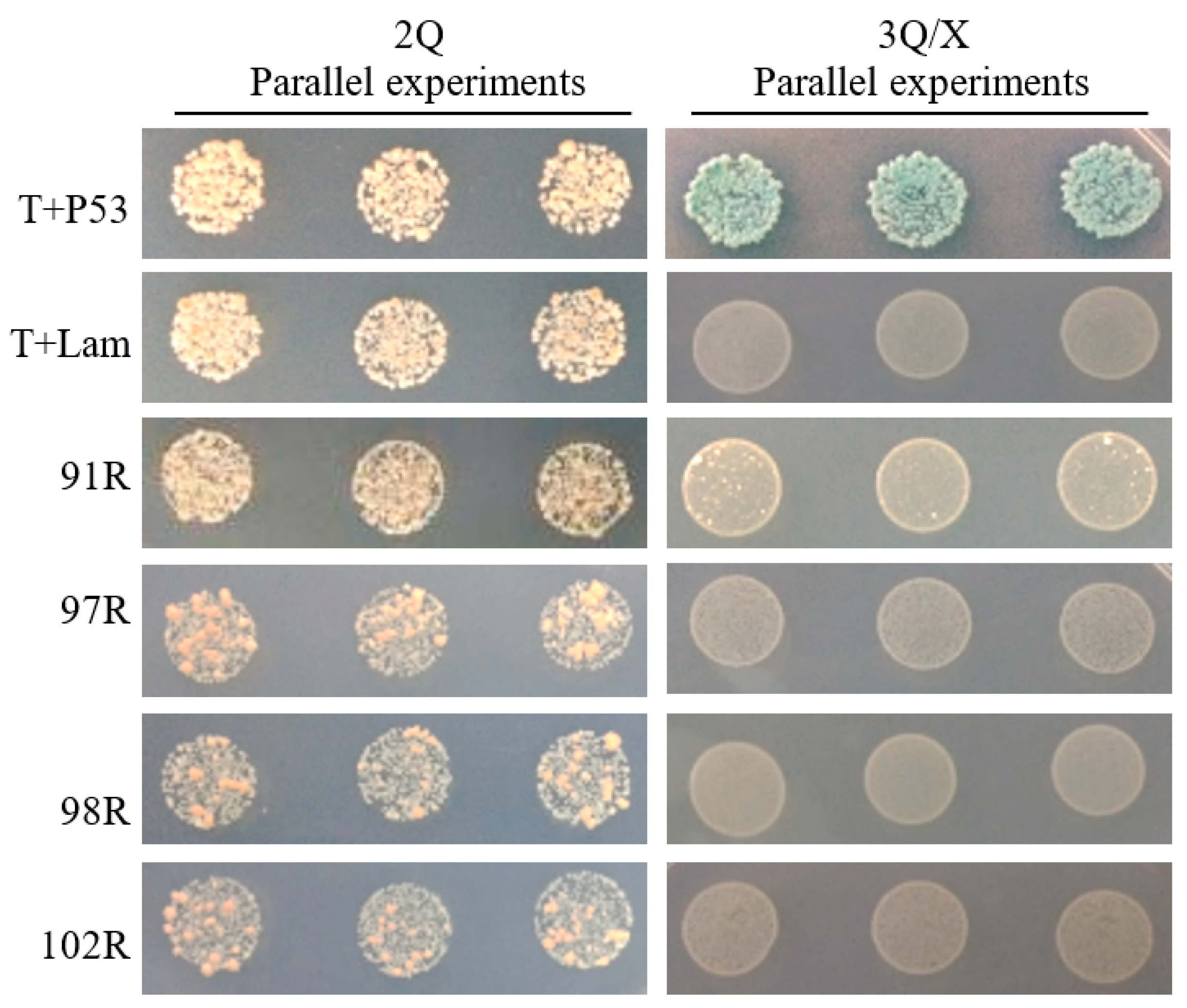
3.1. Interaction between RGV-63R and RGV-91R was First Screened by Y2H
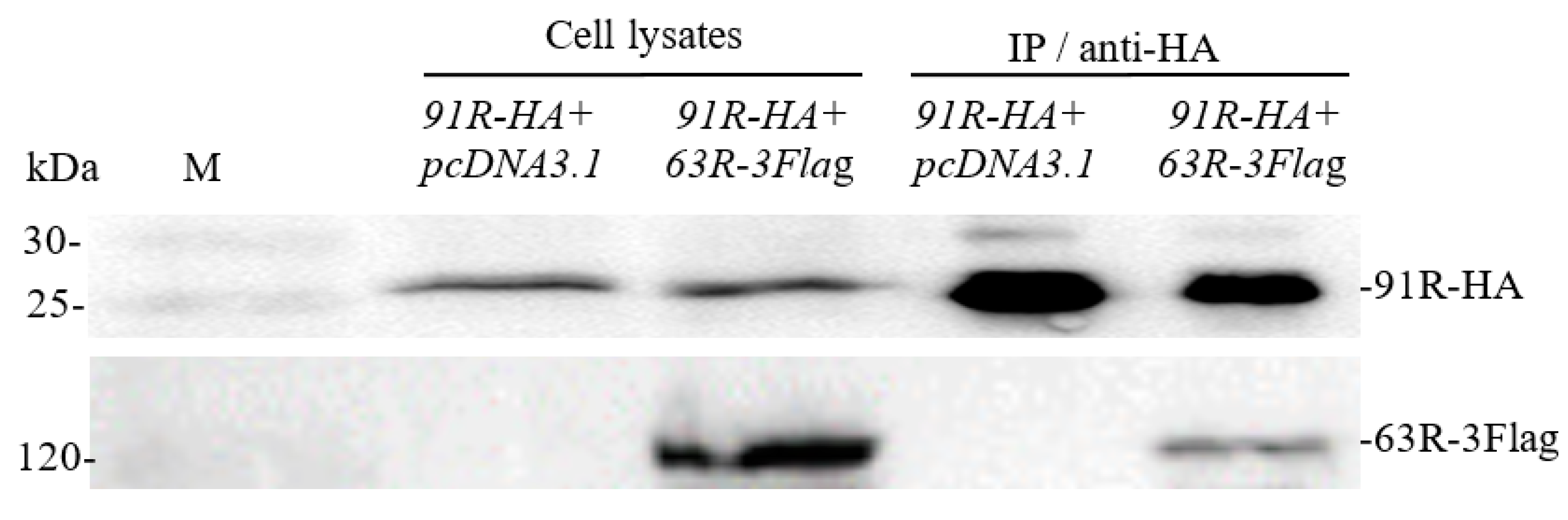
3.2. Confirmation of Interaction between RGV-63R and RGV-91R by Co-IP Followed by Western Blotting
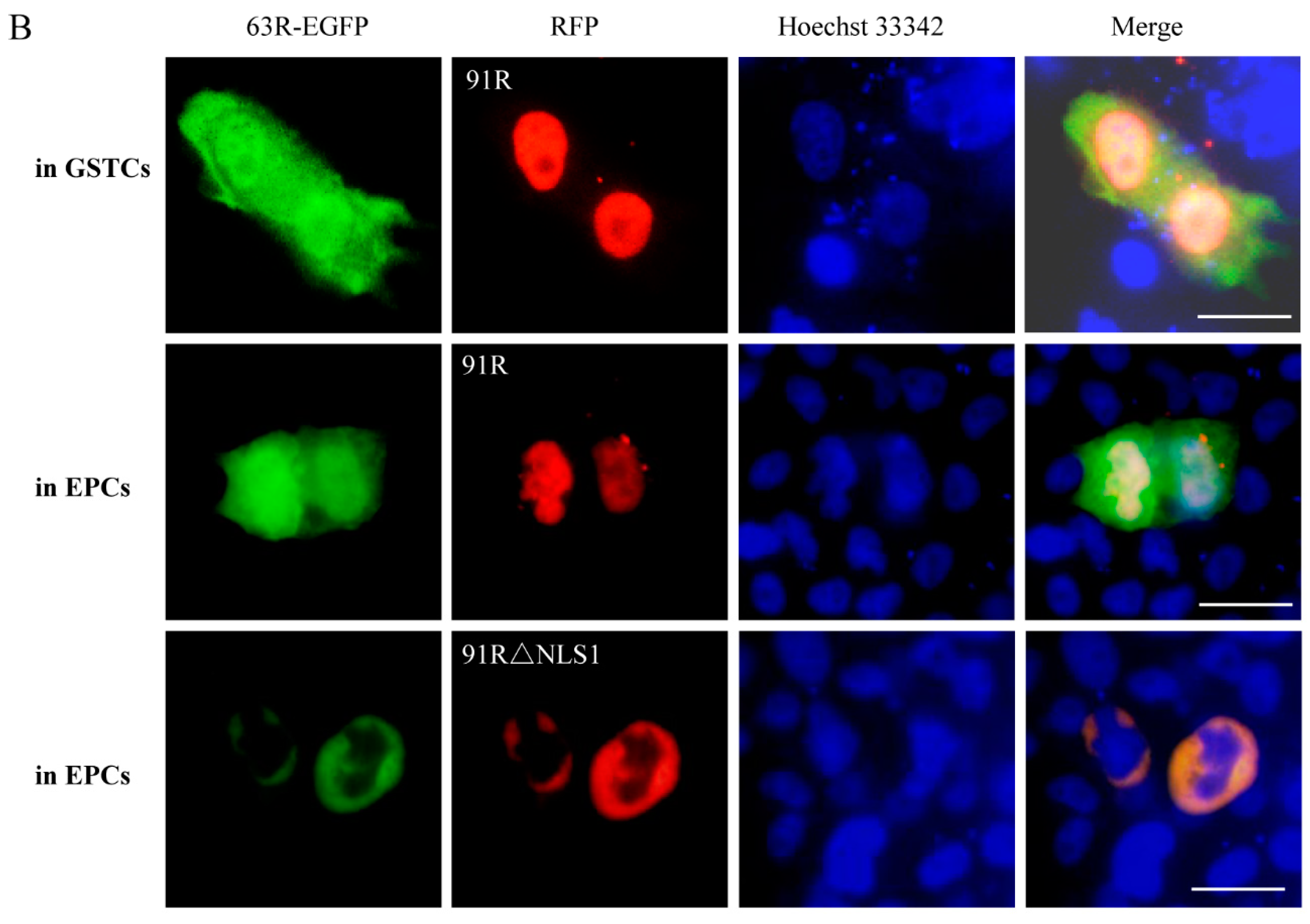
3.3. Localization of RGV-63R and RGV-91R in Cells of Different Species
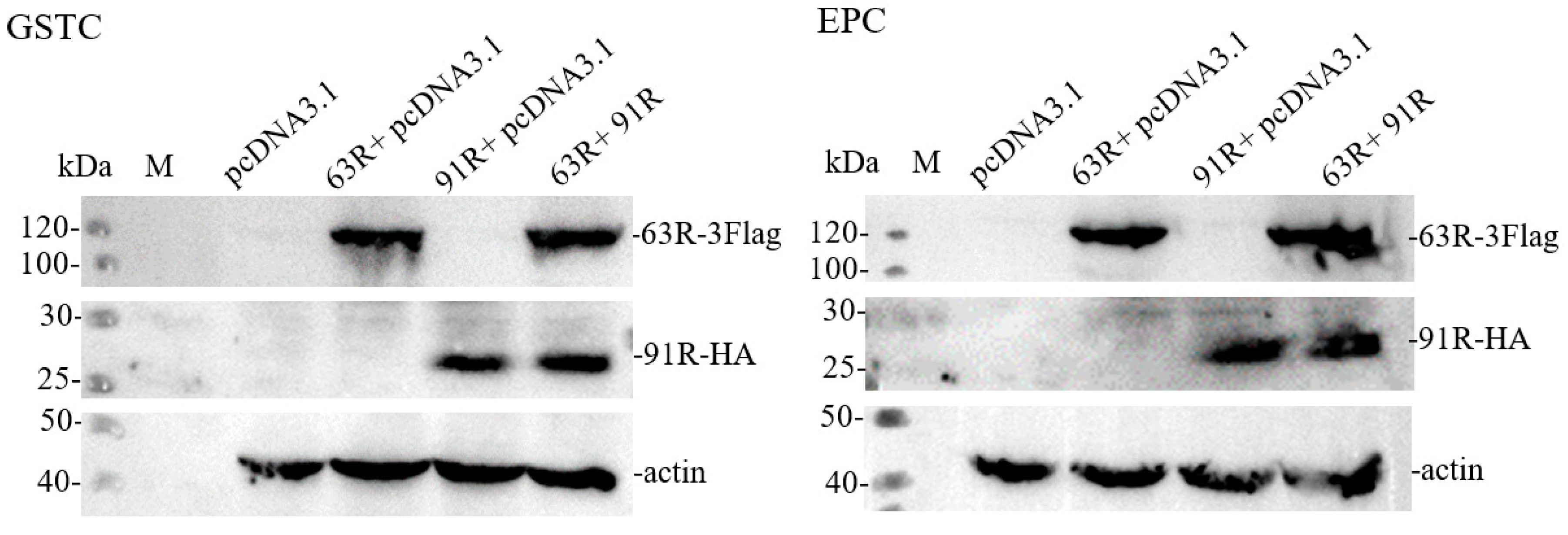
3.4. Detection of RGV-63R and RGV-91R Expressions by Western Blotting
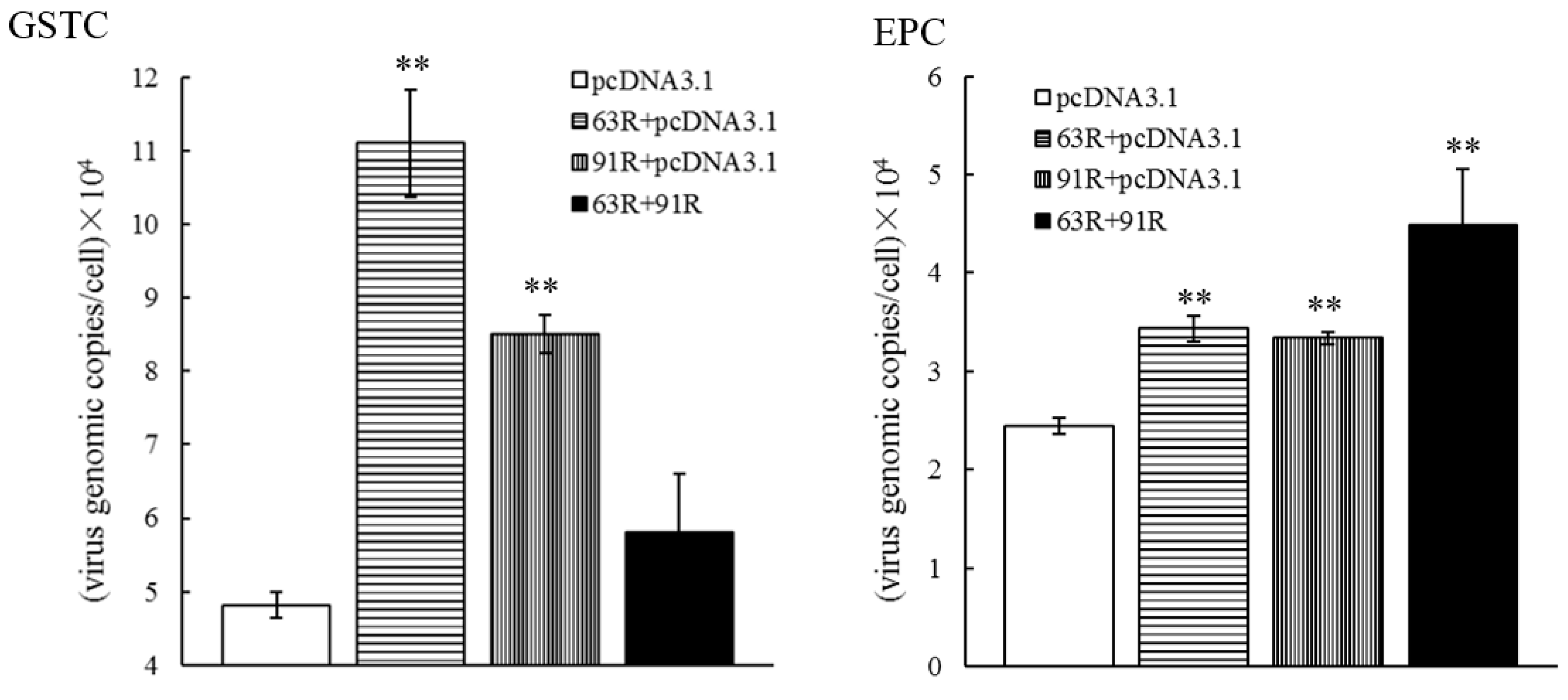
3.5. RGV-63R and RGV-91R Promote RGV Genome Replication
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Zhang, Q.Y.; Gui, J.F. Diversity, evolutionary contribution and ecological roles of aquatic viruses. Sci. China Life Sci. 2018, 61, 1486–1502. [Google Scholar] [CrossRef]
- Chinchar, V.G.; Hick, P.; Ince, I.A.; Jancovich, J.K.; Marschang, R.; Qin, Q.W.; Subramaniam, K.; Waltzek, T.B.; Whittington, R.; Williams, T.; Zhang, Q.Y. ICTV Report Consortium. ICTV Virus Taxonomy Profile: Iridoviridae. J. Gen. Virol. 2017, 98, 890–891. [Google Scholar] [PubMed]
- Zhang, Q.Y.; Gui, J.F. Virus genomes and virus-host interactions in aquaculture animals. Sci. China Life Sci. 2015, 58, 156–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.Y.; Gui, J.F. Atlas of Aquatic Viruses and Viral Diseases; Science Press: Beijing, China, 2012. [Google Scholar]
- Gui, L.; Zhang, Q.Y. A brief review on aquatic animal virology researches in China. J. Fisheries China 2019, 43, 168–187. [Google Scholar]
- Stohr, A.C.; Lopez-Bueno, A.; Blahak, S.; Caeiro, M.F.; Rosa, G.M.; Alves de Matos, A.P.; Martel, A.; Alejo, A.; Marschang, R.E. Phylogeny and differentiation of reptilian and amphibian ranaviruses detected in Europe. PLoS ONE 2015, 10, e0118633. [Google Scholar] [CrossRef] [PubMed]
- Ke, F.; Gui, J.F.; Chen, Z.Y.; Li, T.; Lei, C.K.; Wang, Z.H.; Zhang, Q.Y. Divergent transcriptomic responses underlying the ranaviruses-amphibian interaction processes on interspecies infection of Chinese giant salamander. BMC Genomics 2018, 19, 211. [Google Scholar] [CrossRef]
- Robert, J.; Jancovich, J.K. Recombinant ranaviruses for studying evolution of host-pathogen interactions in ectothermic vertebrates. Viruses 2016, 8, 187. [Google Scholar] [CrossRef]
- Ke, F.; Zhang, Q.Y. Aquatic animal viruses mediated immune evasion in their host. Fish. Shellfish Immunol. 2019, 86, 1096–1105. [Google Scholar] [CrossRef]
- Gui, L.; Chinchar, V.G.; Zhang, Q.Y. Molecular basis of pathogenesis of emerging viruses infecting aquatic animals. Aquaculture Fisheries 2018, 3, 1–5. [Google Scholar] [CrossRef]
- Liu, J.; Yu, C.; Gui, J.F.; Pang, D.W.; Zhang, Q.Y. Real-time dissecting the entry and intracellular dynamics of single reovirus particle. Front. Microbiol. 2018, 9, 2797. [Google Scholar] [CrossRef] [PubMed]
- Corthell, J.T. Basic Molecular Protocols in Neuroscience: Tips, Tricks, and Pitfalls; Elsevier Science: Amsterdam, The Netherland, 2014. [Google Scholar]
- Striebinger, H.; Koegl, M.; Bailer, S.M. A high-throughput yeast two-hybrid protocol to determine virus-host protein interactions. In Virus-Host Interactions. Methods in Molecular Biology (Methods and Protocols); Bailer, S., Lieber, D., Eds.; Humana Press: Totowa, NJ, USA, 2013; vol 1064, pp. 1–15. [Google Scholar]
- Ozsahin, E.; van Oers, M.M.; Nalcacioglu, R.; Demirbag, Z. Protein–protein interactions among the structural proteins of Chilo iridescent virus. J. Gen. Virol. 2018, 99, 851–859. [Google Scholar] [CrossRef] [PubMed]
- Halperin, S.O.; Tou, C.J.; Wong, E.B.; Modavi, C.; Schaffer, D.V.; Dueber, J.E. CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window. Nature 2018, 560, 248–252. [Google Scholar] [CrossRef]
- Gardner, A.F.; Kelman, Z. DNA polymerases in biotechnology. Front. Microbiol. 2014, 5, 659. [Google Scholar] [CrossRef]
- Czarnecki, M.W.; Traktman, P. The vaccinia virus DNA polymerase and its processivity factor. Virus Res. 2017, 234, 193–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zarrouk, K.; Piret, J.; Boivin, G. Herpesvirus DNA polymerases: structures, functions and inhibitors. Virus Res. 2017, 234, 177–192. [Google Scholar] [CrossRef]
- Chinchar, V.G.; Waltzek, T.B. Ranaviruses: not just for frogs. PLoS Pathog. 2014, 10, e1003850. [Google Scholar] [CrossRef]
- Chinchar, V.G.; Waltzek, T.B.; Subramaniam, K. Ranaviruses and other members of the family Iridoviridae: Their place in the virosphere. Virology 2017, 511, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Maga, G.; Villani, G.; Crespan, E.; Wimmer, U.; Ferrari, E.; Bertocci, B.; Hubscher, U. 8-oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature 2007, 447, 606–608. [Google Scholar] [CrossRef] [PubMed]
- Dieckman, L.M.; Freudenthal, B.D.; Washington, M.T. PCNA structure and function: insights from structures of PCNA complexes and post-translationally modified PCNA. In The Eukaryotic Replisome: A Guide to Protein Structure and Function; MacNeill, S., Ed.; Springer: Dordrecht, The Netherlands, 2012; pp. 281–299. [Google Scholar]
- Baple, E.L.; Chambers, H.; Cross, H.E.; Fawcett, H.; Nakazawa, Y.; Chioza, B.A.; Harlalka, G.V.; Mansour, S.; Sreekantan-Nair, A.; Patton, M.A.; et al. Hypomorphic PCNA mutation underlies a human DNA repair disorder. J. Clin. Invest. 2014, 124, 3137–3146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eaton, H.E.; Metcalf, J.; Penny, E.; Tcherepanov, V.; Upton, C.; Brunetti, C.R. Comparative genomic analysis of the family Iridoviridae: re-annotating and defining the core set of iridovirus genes. Virol. J. 2007, 4, 11. [Google Scholar] [CrossRef]
- Zhao, Z.; Ke, F.; Gui, J.F.; Zhang, Q.Y. Characterization of an early gene encoding for dUTPase in Rana grylio virus. Virus Res. 2007, 123, 128–137. [Google Scholar] [CrossRef] [PubMed]
- Ke, F.; Zhao, Z.; Zhang, Q.Y. Cloning, expression and subcellular distribution of a Rana grylio virus late gene encoding ERV1 homologue. Mol. Biol. Rep. 2009, 36, 1651–1659. [Google Scholar] [CrossRef]
- Lei, X.Y.; Ou, T.; Zhu, R.L.; Zhang, Q.Y. Sequencing and analysis of the complete genome of Rana grylio virus (RGV). Arch. Virol. 2012a, 157, 1559–1564. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.Y.; Ou, T.; Zhang, Q.Y. Rana grylio virus (RGV) 50L is associated with viral matrix and exhibited two distribution patterns. PLoS ONE 2012b, 7, e43033. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.D.; Chen, Z.Y.; Huang, X.; Gao, X.C.; Zhang, Q.Y. Establishment of three cell lines from Chinese giant salamander and their sensitivities to the wild-type and recombinant ranavirus. Vet. Res. 2015, 46, 58. [Google Scholar] [CrossRef]
- Zeng, X.T.; Gao, X.C.; Zhang, Q.Y. Rana grylio virus 43R encodes an envelope protein involved in virus entry. Virus Genes 2018, 54, 779–791. [Google Scholar] [CrossRef]
- Wang, J.; Gui, L.; Chen, Z.Y.; Zhang, Q.Y. Mutations in the C-terminal region affect subcellular localization of crucian carp herpesvirus (CaHV) GPCR. Virus Genes 2016, 52, 484–494. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.Y.; Li, T.; Gao, X.C.; Wang, C.F.; Zhang, Q.Y. Protective immunity induced by DNA vaccination against ranavirus infection in chinese giant salamander Andrias davidianus. Viruses 2018, 10, 52. [Google Scholar] [CrossRef]
- Peters, R. Translocation through the nuclear pore: Kaps pave the way. Bioessays 2009, 31, 466–477. [Google Scholar] [CrossRef]
- Chinchar, V.G.; Yu, K.H.; Jancovich, J.K. The molecular biology of frog virus 3 and other iridoviruses infecting cold-blooded vertebrates. Viruses 2011, 3, 1959–1985. [Google Scholar] [CrossRef]
- Sun, W. Molecular cloning and characterization of two novel viral genes, 3β-HSD and PCNA from Rana grylio virus. Ph.D. Thesis, University of Chinese Academy of Sciences, Wuhan, China, 2007. [Google Scholar]
- Li, W.; Zhang, X.; Weng, S.P.; Zhao, G.X.; He, J.G.; Dong, C.F. Virion-associated viral proteins of a Chinese giant salamander (Andrias davidianus) iridovirus (genus Ranavirus) and functional study of the major capsid protein (MCP). Vet. Microbiol. 2014, 172, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.G.; Guo, M.L.; Ji, H.S.; Yan, Y.; Ouyang, Z.L.; Huang, X.H.; Hang, Y.H.; Qin, Q.W. Grouper translationally controlled tumor protein prevents cell death and inhibits the replication of Singapore grouper iridovirus (SGIV). Fish. Shellfish Immunol. 2012, 33, 916–925. [Google Scholar] [CrossRef] [PubMed]
- Lodermeyer, V.; Ssebyatika, G.; Passos, V.; Ponnurangam, A.; Malassa, A.; Ewald, E.; Stürzel, C.M.; Kirchhoff, F.; Rotger, M.; Falk, C.S.; et al. The antiviral activity of the cellular glycoprotein LGALS3BP/90K is species specific. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [PubMed]
- Eunhye, K.; Hyunjhung, J.; Joohee, K.; Joohee, K.; Unjoo, P.; Seunghyun, J.; Areum, K.; Sinae, K.; Tan, T.N.; Yongsun, K.; et al. Species specific antiviral activity of porcine interferon-α8 (IFNα8). Immune Netw. 2017, 17, 424–436. [Google Scholar]

| Name | Sequence (5’ to 3’) a | Usage |
|---|---|---|
| 63R-F | GGGAATTCCATATGGATCTCTTTGTGTACCAGTG (NdeI) | pGBKT7-63R/Y2H |
| 63R-R | CCGGAATTCTTACTTTTTCTTGAACGACA (EcoRI) | |
| 1R-F | CCGGAATTCATGGCATTCTCGGCAGAAGA (EcoRI) | pGADT7-1R/Y2H |
| 1R-R | CCGCTCGAGTCATAGGGGGGTAAACTTCC (XhoI) | |
| 2L-F | CCGGAATTCATGTCCATCATCGGAGCGAC (EcoRI) | pGADT7-2L/Y2H |
| 2L-R | CGCGGATCCTTACCATCTCACTGTAGAGA (BamHI) | |
| 9R-F | GGGAATTCCATATGGAAATGTTTGCATCTAAATC (NdeI) | pGADT7-9R/Y2H |
| 9R-R | CCGGAATTCTCATCGCCACTCAAAGGATT (EcoRI) | |
| 10L-F | CCGGAATTCATGGACACATCACCCTACGA (EcoRI) | pGADT7-10L/Y2H |
| 10L-R | CCGCTCGAGTCAGGCAAACTTGCCCCTCC (XhoI) | |
| 13L-F | GGGAATTCCATATGTGCTCCAAACTCGTAGAGAT (NdeI) | pGADT7-13L/Y2H |
| 13L-R | CCGGAATTCTTAGAAACCCATGGTCTCGA (EcoRI) | |
| 16R-F | CCGGAATTCATGGAACAAGTACCCATAAA (EcoRI) | pGADT7-16R/Y2H |
| 16R-R | CCGCTCGAGCTAATCGTCCAAGTCCGACT (XhoI) | |
| 21R-F | GGGAATTCCATATGGCTACAAATTACTGTGACGA (NdeI) | pGADT7-21R/Y2H |
| 21R-R | CCGGAATTCTTACATCGTGAAGCTCTCAA (EcoRI) | |
| 23L-F | CCGGAATTCATGGAAACCATAGTGCTGGT (EcoRI) | pGADT7-23L/Y2H |
| 23L-R | CCGCTCGAGTTACGACGAGGACCCAAATG (XhoI) | |
| 24R-F | CCGGAATTCATGGCTAACGCTACCATAAA (EcoRI) | pGADT7-24R/Y2H |
| 24R-R | CGCGGATCCCTACTCTTGCTGCTCGGCTC (BamHI) | |
| 29R-F | GGGAATTCCATATGGCCAATTTTCTACAAGATGT (NdeI) | pGADT7-29R/Y2H |
| 29R-R | CCGGAATTCTCAATGACGCTCCTTGGCCC (EcoRI) | |
| 40R-F | CCGGAATTCATGCAAGTTTTTCTAGATTT (EcoRI) | pGADT7-40R/Y2H |
| 40R-R | CGCGGATCCTCACCTCCTCCTGCTCCTGC (BamHI) | |
| 44R-F | GGGAATTCCATATGAGAGTCGTGGTAAACGCAAA (NdeI) | pGADT7-44R/Y2H |
| 44R-R | CCGGAATTCTCACATCAGAAGAGACACGT (EcoRI) | |
| 53R-F | CCGGAATTCATGGGAGCAGCGGAATCTAT (EcoRI) | pGADT7-53R/Y2H |
| 53R-R | CCGCTCGAGTTAACCCCTGTGGGCCGGAA (XhoI) | |
| 60R-F | GGGAATTCCATATGGCAATGGTTTCCAACGTAAA (NdeI) | pGADT7-60R/Y2H |
| 60R-R | CCGGAATTCCTACAGGCTCTTTAGGATAA (EcoRI) | |
| 63R-F | GGGAATTCCATATGGATCTCTTTGTGTACCAGTG (NdeI) | pGADT7-63R/Y2H |
| 63R-R2 | CCGGAATTCTTACTTTTTCTTGAACGACA (EcoRI) | |
| 65L-F | CGCGGATCCTGTCCAGGGGCATGACTACC (BamHI) | pGADT7-65L/Y2H |
| 65L-R | CCGCTCGAGTCACTTGAAGGCTATGGAAA (XhoI) | |
| 73L-F | CCGGAATTCATGTTTCCTCACGTCACCAT (EcoRI) | pGADT7-73L/Y2H |
| 73L-R | CGCGGATCCTTAGATGTCCAGGGGTTCGT (BamHI) | |
| 87L-F | CCGGAATTCATGGAAGGTTGGTTGGGAAA (EcoRI) | pGADT7-87L/Y2H |
| 87L-R | CCGCTCGAGCTAGACTCCCTTGGCATGAA (XhoI) | |
| 88R-F | CCGGAATTCATGTCTTTTCAGAGAGATTA (EcoRI) | pGADT7-88R/Y2H |
| 88R-R | CCGCTCGAGCTACCTGGTCCACCTCTTGC (XhoI) | |
| 91R-F | GGGAATTCCATATGCTGTGGGAAGCCGTAACAGA (NdeI) | pGADT7-91R/Y2H |
| 91R-R | CCGGAATTCTTAGCCCTCAAAGAGAGTCA (EcoRI) | |
| 92R-F | GGGAATTCCATATGAGCATCCCTACAGTCATAGC (NdeI) | pGADT7-92R/Y2H |
| 92R-R | CCGGAATTCTTACCGCACATTTCTAGACA (EcoRI) | |
| 95R-F | CCGGAATTCATGCACGGTTGCAATTGTAA (EcoRI) | pGADT7-95R/Y2H |
| 95R-R | CCGCTCGAGTCAGTTAAAAGTGCTCGTAT (XhoI) | |
| 97R-F | CCGGAATTCATGTCTTCTGTAACTGGTTC (EcoRI) | pGADT7-97R/Y2H |
| 97R-R | CCGCTCGAGGACCCATGACGGAAAAGACT (XhoI) | |
| 98R-F | CCGGAATTCATGGCAAACTTTGTGACAGA (EcoRI) | pGADT7-98R/Y2H |
| 98R-R | CCGCTCGAGTTAGGCTCTGACCACAAACA (XhoI) | |
| 101L-F | CCGGAATTCATGGATCCAGAAGGAATGAT (EcoRI) | pGADT7-101L/Y2H |
| 101L-R | CCGCTCGAGTCACAGCACCTTTCTCAGGT (XhoI) | |
| 102R-F | CCGGAATTCATGGGCATAAAAGGACTGAA (EcoRI) | pGADT7-102R/Y2H |
| 102R-R | CCGCTCGAGTCACTTGCGCTTGCACTTCT (XhoI) | |
| 63R-F2 | CCCAAGCTTATGGATCTCTTTGTGTACCA (HindIII) | pcDNA3.1-63R-3Flag |
| 63R-3Flag-R | CCGGAATTCTTACTTATCGTCGTCATCCTTGTAATCGATCTTATCGTCGTCATCCTTGTAATCTCCCTTATCGTCGTCATCCTTGTAATCCTTTTTCTTGAACGACACAA (EcoRI) | |
| 91R-F2 | CCCAAGCTTATGCTGTGGGAAGCCGTAAC (HindIII) | pcDNA3.1-91R-HA |
| 91R-HA-R | CCGGAATTCTTAAGCGTAATCTGGAACATCGTATGGGTACATGCCCTCAAAGAGAGTCACGG (EcoRI) | |
| 63R-R3 | CCGGAATTCGACTTTTTCTTGAACGACAC (EcoRI) | pEGFP-63R/ colocalization |
| 91R-F3 | CCCAAGCTTCAATGCTGTGGGAAGCCGTA (HindIII) | pDsRed2-91R/ colocalization |
| 91R-R2 | ACGCGTCGACTTAGCCCTCAAAGAGAGTCA (SalI) | |
| 91R△NLS1-R | CCCATGAGCCTCAGCGTCACGTAGCTGGTAAAGACCGATG | pDsRed2-91R△NLS1 |
| 91R△NLS1-F | CATCGGTCTTTACCAGCTACGTGACGCTGAGGCTCATGGG | |
| 91R△NLS2-R | GAAAAGGGTCCCATCTTGACCACTCCTCCGGACGCCACCA | pDsRed2-91R△NLS2 |
| 91R△NLS2-F | TGGTGGCGTCCGGAGGAGTGGTCAAGATGGGACCCTTTTC | |
| MCP-F | ATGGTTGTGGAGCAGGTG | qPCR |
| MCP-R | TGACGCAGGTGTAATTGGAG |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, X.-T.; Zhang, Q.-Y. Interaction between Two Iridovirus Core Proteins and Their Effects on Ranavirus (RGV) Replication in Cells from Different Species. Viruses 2019, 11, 416. https://doi.org/10.3390/v11050416
Zeng X-T, Zhang Q-Y. Interaction between Two Iridovirus Core Proteins and Their Effects on Ranavirus (RGV) Replication in Cells from Different Species. Viruses. 2019; 11(5):416. https://doi.org/10.3390/v11050416
Chicago/Turabian StyleZeng, Xiao-Tao, and Qi-Ya Zhang. 2019. "Interaction between Two Iridovirus Core Proteins and Their Effects on Ranavirus (RGV) Replication in Cells from Different Species" Viruses 11, no. 5: 416. https://doi.org/10.3390/v11050416
APA StyleZeng, X.-T., & Zhang, Q.-Y. (2019). Interaction between Two Iridovirus Core Proteins and Their Effects on Ranavirus (RGV) Replication in Cells from Different Species. Viruses, 11(5), 416. https://doi.org/10.3390/v11050416





