Abstract
The history of giant viruses began in 2003 with the identification of Acanthamoeba polyphaga mimivirus. Since then, giant viruses of amoeba enlightened an unknown part of the viral world, and every discovery and characterization of a new giant virus modifies our perception of the virosphere. This notably includes their exceptional virion sizes from 200 nm to 2 µm and their genomic complexity with length, number of genes, and functions such as translational components never seen before. Even more surprising, Mimivirus possesses a unique mobilome composed of virophages, transpovirons, and a defense system against virophages named Mimivirus virophage resistance element (MIMIVIRE). From the discovery and isolation of new giant viruses to their possible roles in humans, this review shows the active contribution of the University Hospital Institute (IHU) Mediterranee Infection to the growing knowledge of the giant viruses’ field.
1. Introduction
For this 10th-year anniversary special issue of the journal Viruses, members or former members (mostly former PhD) of our team working at the University Hospital Institute (IHU) Mediterranee Infection prepared this review that presents the last 15 years of our work on giant viruses, from the discovery and description of Mimivirus to our latest studies, some under review and others not yet completed.
2. The History of Discovery of Giant Viruses and Their Genetic Mobile Elements
Our former laboratory, URMITE, a research unit on emerging infectious and tropical diseases, reference center for Rickettsia and rickettsial diseases, was traditionally expert in the isolation and study of fastidious and intracellular microorganisms. In the 1990s, we were the first French hospital laboratory to routinely identify rare bacteria using 16S ribosomal RNA (rRNA) gene sequencing [1]. In the same period, we began using amoebas as cell supports for the isolation of intracellular bacteria, mostly Legionella, but also as a means to isolate, from human and environmental samples, potential new agents responsible for pneumonia that would not grow on axenic culture media [2,3,4,5,6,7,8]. Amoeba co-culture and identification based on the 16S rRNA sequence led us to collaborate with Dr. Tim Rowbotham, the scientist who described the isolation of Legionella using amoeba co-culture [9]; in 1995, a post-doctoral student from his laboratory, Richard Birtles, came to Marseille to identify his collection of amoeba-associated bacterial isolates. Among the bacteria of this collection, most being identified as Legionella-like amoebal pathogens by 16S rRNA gene sequencing [10], there was a small Gram-positive coccoid bacterium provisionally named “Bradford coccus”. This bacterium was later resistant to molecular analysis and we decided that examination of the ultrastructure of Bradford coccus using electron microscopy might provide us with a solution to our technical problems, i.e., a particular cell wall that would protect DNA from extraction. During examination, we observed unexpected regular icosahedral bodies that were typical of a virus but with the size of a small bacterium (Figure 1). We observed for the first time a particle belonging to a new group of viruses, giant viruses, with its first member Acanthamoeba polyphaga mimivirus (APMV) [11]. Its size of ~600 nm with its surrounding fibrils, the size of its genome of 1.2 Mbp, and its coding capacity including genes/functions never observed in a virus let us imagine that this discovery could deeply modify the perception we had of the virosphere [12]. The next discoveries we made (as well as others) on the subject gradually confirmed this first impression (Table 1). However, at the stage of this manuscript, it should be noted that Mimivirus, seen as a unique circus freak in 2003, is now, 15 years later, one of the most diverse microorganisms, at least in sea water [13].
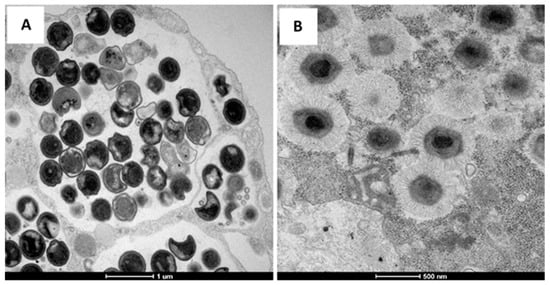
Figure 1.
Transmission electron microscopy of the Chlamydia bacterium (A) and Mimivirus (B).

Table 1.
Input of the University Hospital Institute (IHU) Mediterranee Infection to the knowledge of giant viruses. (* First described by the laboratory; UPC: unpublished under characterization).
We firstly started to look specifically for giant viruses using amoeba co-culture, and we could identify three different lineages in the Mimivirus family: lineage of Mimivirus stricto sensu (lineage A), lineage of Moumouvirus (lineage B), and lineage of CE11 (lineage C) [14]. This lineage C was later named the Megavirus lineage [48,49]. During the search for giant virus isolates, we succeeded in isolating a new giant virus, Marseillevirus [15] and a curious Mimivirus with slow multiplication and particles seeming larger than the original APMV we named Mamavirus. Electron microscopy observations showed that it appeared larger due to particles with abnormal layers of particle wall, and a small virus of 50 nm in diameter was observed as developing within its viral factory [34]. This small virus we named Sputnik was in fact a virus that infected the APMV virus factory, and we further defined it as a virophage by analogy with bacteriophages that are virus-infecting bacteria. Subsequently, we were able to identify that the genome of a virophage could be integrated into the genome of a giant virus and we named it a provirophage [35]. In the same study, while searching for other mobile elements in giant viruses, we also detected transposons we named transpovirons. In later works, we searched for new strains of virophages using reporter giant viruses allowing the isolation of Sputnik 3 [25] and Guarani virophage [37]. One of the new virophages named Zamilon had the originality to infect lineage B and C of Mimivirus but not lineage A. We imagined that this resistance could be due to a CRISPR/Cas system, a system allowing bacteria to resist phages [50]. However, analysis of the APMV genome did not reveal any significant homology with enzymes of a CRISPR/Cas system. As a CRISPR/Cas system consists of enzymes but also of repeats and integration of the targeted bacteriophage sequence, we then searched for repeats and virophage sequences in the APMV genome. The genome of Zamilon was fragmented into short fragments of 40 nucleotides using a sliding window of 10 nucleotides (nts), and all fragments were compared using BLAST software against the respective APMV genomes. We were surprised to find, in the lineage A Mimiviruses, four repeats of 15 nts corresponding to Zamilon. Observation in the vicinity of these repeats of nuclease and helicase genes allowed us to suspect that this operon that we named MIMIVIRE for Mimivirus resistance element could be the defense mechanism of lineage A Mimivirus againt the Zamilon virophage. Silencing of this operon confirmed this hypothesis [38]. We were recently able to isolate a Mimivirus isolate of lineage A with a modified MIMIVIRE operon that is susceptible to Zamilon; via transient knock-out (KO) of the MIMIVIRE operon, we could restore susceptibility of APMV to the Zamilon virophage [31].
If metagenomics, especially with last-generation high-throughput sequencing, recently demonstrated an extraordinary potential to discover new families of giant viruses [51,52], we continue to believe that virus isolation attempts are not an outdated strategy. Following the discovery of Mimivirus, our laboratory, with the successful collaboration of the Virology Laboratory of the Federal University de Minas Gerais, continued its efforts to isolate new giant/protozoa-associated viruses. This search followed three major axes that we developed: building high-throughput isolation techniques, diversifying the amoeba supports, and searching in original biotopes. This was efficient for the discovery of new viruses like Faustovirus, Kaumoebavirus, Pacmanvirus, Orpheovirus, Cedratvirus, and Tupanvirus [18,19,20,21,22,23]. We also managed to isolate members of families described by our colleagues such as Pandoraviruses or Pithoviruses [16,17,53,54]. Moreover, we are currently analyzing new viruses we recently isolated, all of them representing new families, including Yasminevirus and Fadolivirus, the first isolated members of the putative family of Klosneuvirinae [51], as well as viruses never detected to date using metagenomics such as Clandestinovirus and Usurpativirus (Unpublished under characterization) (Figure 2).
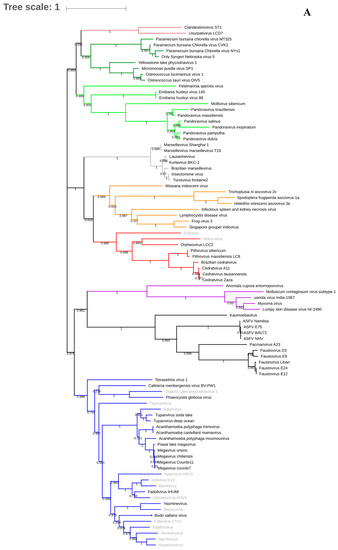
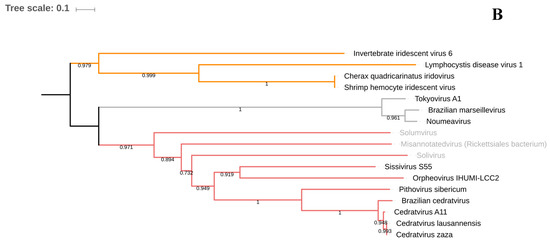
Figure 2.
Phylogenetic trees based on DNA polymerase B of giant viruses (A) and on VETF gene to illustrate the clustering of Pithoviridae and Sissivirus (B). The viruses described only in metagenomics are labeled in gray. The analysis was performed using Muscle and FastTree, applying the maximum-likelihood method with 1000 bootstrap replicates.
3. History and Revolution of the Giant Viruses’ Isolation Process through the Years
Many studies confirmed giant virus ubiquity and diversity in the environment and in humans [55,56,57,58]. Therefore, isolating the viral particle remains crucial in order to access its genetic content and perform further analysis allowing a better understanding of this new emerging world. Their natural host remains unknown, and co-culture on amoeba remains the only key engine for isolating these viruses. This strategy, although it allowed the isolation of the first Acanthamoeba polyphaga mimivirus [11], remained inefficient for many years before it could isolate other giant viruses, suggesting the need to improve, standardize, and automate the co-culture to a level that would allow more isolates to be produced. In this review, we summarize the main improvements brought to the giant virus isolation strategies in our lab over the years along with a brief descriptive timeline of the main modifications that marked the amoebal co-culture and giant virus isolation (Table 2).

Table 2.
Evolution of the co-culture strategies over the years: tools, host panels, and sample diversity.
Two cases are discussed regarding this issue; one is related to the samples and the cell hosts, and the other is related to the tools used to reduce the time of co-culture and enhance the isolation rate.
The “amoebal enrichment method” in shell vials was first implemented by T.J. Rowbotham to isolate Legionella species [9]. This strategy allowed the isolation of Acanthamoeba polyphaga mimivirus in 2003 [11] and was adopted for future giant virus isolation processes. However, this technique remained fastidious for many years due to the lack of expertise in the field. Many attempts were made to improve the isolation strategy starting with the diversification of the samples where giant viruses were reportedly isolated from various environments such as permafrosts, lakes, sewage, soil, water, human samples, and many others [19,39,45,53,59]. Other improvements involved the pre-treatment of the samples prior to co-culture by means of filtration, precipitation, pre-enrichment, and antibiotic and antifungal treatments. Many isolates were retrieved from treated samples notably Pithovirus sibericum [53], Mollivirus sibericum [60], Pandoravirus salinus and dulcis [54], Megavirus chilensis [48], Samba virus [61], and others. These improvements are fairly detailed in a previous review written by Bou Khalil et al. [62]. Many studies later proved that enlarging the panel of amoeba used for co-culture allowed the recovery of different isolates, whereby a single giant virus can be positive for a certain amoeba but negative for another [63]. For many years, we used Acanthamoeba polyphaga as a cell support for the isolation procedure, but we never managed to isolate new viruses other than Mimivirus or Marseillevirus, whereas other teams using Acanthamoeba castellanii successively isolated Pandoravirus, Mollivirus, and Pithovirus. For this, and after using diverse cell hosts, we managed to isolate many new strains of giant viruses [21,22,23]. This concept also allowed the isolation of the first Faustovirus on Vermamoeba vermiformis [18] and recently the isolation of Orpheovirus IHU-MI [21].
From a different perspective, Boughalmi et al. introduced in 2012 a novel way of detecting amoebal lysis with the naked eye using co-culture on agar plates [24]. This technique allowed the isolation of new representatives of the Mimiviridae and Marseilleviridae families. However, it was still fastidious and limited to the use of protozoa growing on agar plates, which increased the risk of cross-contamination between samples. In 2013, the co-culture on amoeba was adapted to the use of microplates. Antibiotics and antifungal were also used in order to eliminate contaminants coming from the sample [64]. However, until that time, the isolation strategy was still fastidious, highly time-consuming, and operator-dependent, and it presented a low yield compared to the ubiquity and diversity of giant viruses in the environment reported by metagenomic studies [55,56,57,58]. This suggested the need for new automated tools to be used for the rapid isolation and identification of giant viruses. Therefore, in 2016, Bou Khalil et al. introduced flow cytometry in liquid medium to the isolation process. This tool allowed the automated detection of cell burst, the presumptive identification of giant viruses based on their DNA content [26], and the sorting of viral mixtures [27]. This technique also allowed the use of highly motile protozoa as cell hosts for the co-culture. In addition, the enrichment steps were optimized and coupled with new targeted antibiotic and antifungal mixtures, which dramatically reduced the time for each step. However, flow cytometry is a blind technique and presents a high risk of contamination. To overcome these limitations, we recently developed a new isolation strategy based on an automated high-content screening of co-culture and rapid scanning electron microscopy identification [65]). These new tools will expand the research spectrum in terms of varying samples and hosts for a more efficient hunt of giant viruses. To summarize, the co-culture process witnessed many improvements and modifications through the years, where some were fruitful and others remained less useful. However, we could learn much from this chronological overview to summarize a reliable strategy that could be adapted and standardized by all trackers of giant viruses by applying the same protocols of sample preparation and using a large panel of potential protozoa capable of harvesting or producing giant viruses. Automation of detection was one of the biggest achievements realized in our lab that revolutionized the giant virus isolation step and yielded hundreds of new strains. This process will continue to be developed and improved in our lab in order to get artificial intelligence software for correlative microscopy capable of easily identifying new isolates.
4. Mimiviridae
Following the outstanding discovery and characterization of Acanthamoeba polyphaga mimivirus (APMV), a new family, named Mimiviridae, was created to encompass this virus and other new members that also present similar morphological and genetic features [11,12,66]. Currently, this family includes two genera recognized by the International Committee of Taxonomy of Viruses (ICTV): Mimivirus and Cafeteriavirus. The IHU Mediterranee Infection team isolated hundreds of mimiviruses from a plethora of environmental and clinical samples, which increased knowledge about the exceptional diversity of this group of viruses [66]. Currently available data suggest the division of mimiviruses into at least three lineages (A, B, and C) [40,48,61,63,66,67,68,69]. The genus Cafeteriavirus comprises a single species, Cafeteria roenbergensis virus (CRoV), in which particles infect marine flagellates [70]. Both mimiviruses and CRoV were described to be parasited by virophages, and complex relationships were described, involving protists, giant viruses, an exclusive mobilome, and virophages [35,38,71].
Recently, phylogenomic analyses of viruses, previously considered as Phycodnaviridae, showed that they are actually closer to other mimiviruses than phycodnaviruses [72,73]. The description of this group of viruses, now denominated as extended mimiviruses, represented the beginning of a substantial expansion of Mimiviridae that was followed by other important discoveries. In 2017, metagenomic data identified the genome of four different Mimiviridae, with an expanded complement of translation factors, named Klosneuvirus [51]. In 2018, a member of Klosneuvirus group, named Bodo saltans virus, was isolated for the first time from a kinetoplastid protozoan [74]. We recently isolated two members of this Klosneuvirinae subfamily that are currently under characterization, Fadolivirus and Yasminevirus.
A continuous search for giant virus relatives in extreme environments led the IHU/UFMG teams to the discovery of Tupanviruses. Isolated from samples collected in soda lake and oceanic sediments in Brazil, these viruses present similar characteristics with mimiviruses of amoebae, but also unique and distinctive features never observed before, including a tailed particle and the most complete set of translation-related genes (Figure 3) [19,75]. A new genus, named “Tupanvirus”, was recently proposed to include the species “Tupanvirus soda lake” and “Tupanvirus deep ocean” [76]. Tupanvirus presents a broad host-range, infecting not only Acanthamoeba genus, as described with other amoebal mimiviruses [77,78,79,80,81], but also Vermoameba vermiformis, Dyctiostelium discodeum, and Willeartia magna. A toxic profile induced by Tupanvirus was described, related to the shutdown of host and non-host rRNA [19,75]. Controversial and exciting topics regarding Mimiviridae were raised in recent years, including their potential pathogenicity to humans and their position in the tree/rhizome of life [80,81,82,83,84,85,86,87,88]. Mimivirus studies represent an open field for remarkable discoveries and fundamental debates regarding the origins of nucleocytoplasmic large DNA viruses (NCLDVs) and cellular organisms.
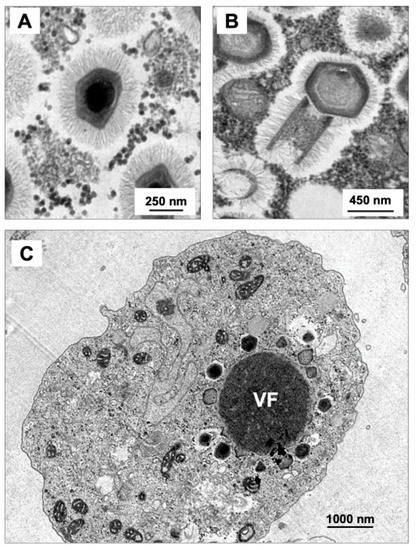
Figure 3.
Transmission electron microscopy of Mimivirus (A) and Tupanvirus particles (B). (C) Tupanvirus viral factory, occupying a large portion of Acanthamoeba castellanii cytoplasm. VF: viral factory.
5. Marseilleviridae
The Marseilleviridae family was the second family of giant viruses to be described [89]. Marseillevirus marseillevirus (MSRV), the prototype virus, was discovered in 2007 in our laboratory [15]. Its particle and genome are smaller than those of mimiviruses but still giant for viruses [90]. In 2018, 15 Marseillevirus isolates were described including six at the IHU Méditerranée Infection (Table 3) [14,15,32,42,91,92,93,94,95,96,97,98,99,100]. These viruses have diverse geographical origin (nine countries, five continents) and were retrieved from various environmental samples including waters, soils, insects, and mussels. They were isolated by co-culture with Acanthamoeba castellanii or Acanthamoeba polyphaga, and they were classified into five lineages.

Table 3.
List of the 15 published Marseillevirus isolates.
The analysis of their replication cycle showed different pathways to enter into amoebae including phagocytosis of vesicles containing hundreds of particles, and endocytosis of single particles [101]. The replicative cycle is completed 12 h post infection (p.i.) with an eclipse phase at 2 h p.i., before the appearance of a large viral factory in the host cytoplasm at 4 h p.i. where virion morphogenesis occurs, and a complete lysis of amoebae at 12 h p.i. with the release of the new viral progeny [101]. Marseilleviruses have an icosahedral capsid of about 250 nm, with small fibrils of 12 nm described for MSRV, enclosing circular double-stranded DNA genomes ranging in size from 346 to 386 kilobase pairs (kbp), predicted to encode between 386 and 491 proteins. Gene repertoires encompass the giant virus core genes along with genes unique to giant viruses including some involved in transcription and a few genes related to the translation process, paralogous genes, and large fractions of genes with no homolog outside the viral family or known function [90]. In addition, the genome of these viruses is characterized by a substantial level of mosaicism with sequences of different putative origins, including eukaryotic, bacterial, archaeal, and viral [64]. Transcriptome analysis of MSRV confirmed the existence of predicted genes and revealed a temporal expression pattern, but no correlation with conserved AT-rich putative promoter motifs present in single or multiple copies [102] over more than 50% of the genes [103].
Moreover, marseilleviruses were detected in several samples collected from both symptomatic [45,46,47,101] and asymptomatic humans [42,43,104], being isolated once from a healthy man (see chapter on giant viruses in humans). Also, an up to 30-day-long persistence of MSRV in rats and mice was observed following intraperitoneal, intravenous, or airway inoculation [105]. The possible pathogenic nature of these viruses is still under investigation.
6. Faustoviruses and Asfarvirus-Related Giant Viruses of Amoeba
Faustoviruses were the first described giant viruses to infect an amoeba from a genus distinct from the commonly used Acanthamoeba species [18]. Considering the reported high abundance of Vermamoeba vermiformis in diverse environmental and human samples, (hospital water networks [106,107], drinking water [108], human stool samples [109], and contact lenses of keratitis patients [110,111]), its role as a reservoir for pathogenic agents was hypothesized. Our team, thus, incorporated this amoeba to the panel of cell supports in the high-throughput co-culture protocols allowing the successful isolation of Faustovirus E12, the prototype strain, from a sewage sample [18]. To date, we described and sequenced the genomes of 11 Faustovirus isolates classified in four lineages (Figure 4A). All isolates were recovered from sewage samples collected in different geographical locations (France, Senegal, Lebanon), but Faustovirus-like sequences were otherwise identified in diverse biomes including Culicoides guts, cattle sera, rodent organs, and human healthy and febrile patient sera [18,33,112,113].
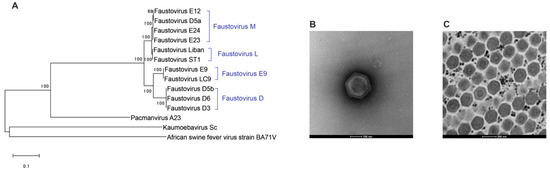
Figure 4.
(A) DNA-directed RNA polymerase beta subunit-based tree illustrating the clustering of Faustovirus isolates in four lineages, Kaumoebavirus, and Pacmanvirus. The maximum-likelihood method and Jones–Taylor–Thornton model for amino-acid substitution were used with 1000 bootstrap replicates. (B) Negative staining of Faustovirus ST1 purified suspension showing an icosahedral particle of 200 nm. (C) Electron microscopy of the honeycomb structure of Pacmanvirus A23 viral factory in Acanthamoeba castellanii.
Faustovirus E12 replication cycle in V. vermiformis lasts 18 to 20 h after individual viral particles’ phagocytosis by the amoeba. The eclipse phase occurs from 4 to 6 h p.i. along with a reorganization of the host nucleus. From 8 to 10 h p.i., a donut-shaped virus factory appears with newly formed viral particles, released through cell lysis in the last step of the cycle [18]. Viruses of this group form icosahedral virions of 200–240 nm in diameter containing a double-stranded DNA genome of 456 to 491 kilobase pairs (kbp) predicted to encode for 477 to 519 genes. A large proportion of these genes (~70%) encode for proteins whose functions are not yet identified due to the absence of known homologs. The comparative genomic study of nine Faustovirus isolates showed an open pan-genome of over 1000 genes and a stable core-genome of around 207 genes, 74% of which have best matches with members of the Asfarviridae family [114].
Among giant viruses, Faustoviruses are most closely related to the African swine fever virus (ASFV), a tick-borne virus responsible of highly epidemic hemorrhagic fever in domestic pigs. This is most clearly demonstrated when DNA polymerases are used for phylogenetic studies [84]. Interestingly, while these two viruses share a similar gene expression pattern and the highest proportion of homologous proteins, faustoviruses stand out with the size of their genomes, the complexity of their gene structure, and the double-protein shell that forms their capsid [115]. In Faustovirus E12, the gene coding for the major capsid protein (MCP) forming the external protein layer of the capsid is 17 kbp long and comprises 13 exons separated by large group I and spliceosomal-like introns defined by non-canonical splice sites [116]. Homology searches of the MCP gene in other faustoviruses showed the presence of six different splicing profiles that correlate with the described lineages (Unpublished under characterization).
Kaumoebavirus strain Sc was the second giant virus of amoeba isolated on Vermamoeba vermiformis [20] (Figure 4B). Although phylogenetically distant from faustoviruses and with a unique genome topology, Kaumoebavirus shares several characteristics with faustoviruses: (i) a comparable morphology with 250-nm large icosahedral particles protected by a double-layered capsid, (ii) replication cycles of similar lengths in their common amoebal host, and (iii) the spliced structure of the MCP gene, predicted to be 5 kbp long in Kaumoebavirus. Moreover, a recent study showed the presence of an AT-rich motif similar to a known promotor of ASFV in the intergenic regions of Faustoviruses and Kaumoebavirus suggesting a potential shared gene expression regulation mechanism in these viruses [117].
In the Asfarvirus-related giant viruses of amoeba, Pacmanvirus was isolated on Acanthamoeba castellanii (Figure 4C). This virus induces amoebal lysis in 6 to 8 h and produces icosahedral particles of the same size range as Faustoviruses and Kaumoebavirus (250 nm), harboring a genome of intermediate length (395 kbp). Pacmanvirus shares 31 genes with Faustovirus, Asfarvirus, and Kaumeobavirus, but is the only virus in this group that encodes for a transfer RNA (tRNA; isoleucine-tRNA) [23].
7. Pithoviridae and Related Viruses
The first member of this group of giant viruses was isolated from a >30,000-year-old Siberian permafrost sample and was named Pithovirus sibericum [53]. This virus has the largest elipsoid viral particle to date, with a mean size of 1.5 µm in length, exhibiting a single cork in one extremity, from where the genome is released [53] (Figure 5A). Despite this first observation, structural analysis permitted discovering discrete forms for Pithovirus sibericum with two corks [118]. A contemporary virus was isolated in 2016 from French sewage samples, exhibiting similar virion structure and a genetic conservation compared to its prehistoric counterpart [17]. Their genome is a circular double-stranded DNA molecule of 610–683 kb coding for up to 520 genes, with a large fraction (~21%) corresponding to multiple palindromic non-coding repeat sequences [17,53]. Despite morphological and genomic differences, phylogenomic and phyletic analysis branch these viruses among other NCLDVs [119].
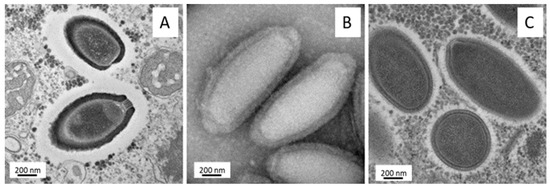
Figure 5.
Transmission electron microscopy of Pithovirus massiliensis (A), Cedratvirus A11 (B), and Orpheovirus particles (C).
New members of the putative “Pithoviridae” family were isolated from Algerian, French, and Brazilian samples and named Cedratviruses [22,120,121]. Cedratviruses have elipsoid particles of ~1.0 µm but present some differences compared to other pithoviruses. Indeed, these viruses have two striate corks, one at each extremity of the particles [22] (Figure 5B). Cedratviruses replicate in Acanthamoeba cells, entering through phagocytosis and establishing a viral factory in the host cytoplasm, wherein a complex process of morphogenesis occurs, generating a viral progeny that is released mostly by cell lysis, although exocytosis is also possible [122]. Similar to pithoviruses, the cedratviruses have a large particle containing a relatively small circular genome (from 460,038 bp for Brazilian Cedratvirus to 589,068 for Cedratvirus A11), and are possible exceptions to the allometric law involving virion volume and genome length [121]. Those exceptions raised questions about DNA compaction and about macromolecules contained inside the particle particularly, where a lower density than the Mimivirus particle could be detected in Pithovirus [118]. Meanwhile, Pithoviruses and Cedratviruses reveal an extreme variation rate of their particles. Given the genomic variations we observed between Cedratvirus lineages, this particle size could be a powerful adaptation mechanism in light of evolution where the volume of the capsid does not become a constraint for the selection process and for genomic evolution. There are currently two lineages of Cedratviruses, a possible new genus of the expanding “Pithoviridae” family [121].
In 2018, Orpheovirus was isolated from a rat stool sample in V. vermiformis [21]. The Orpheovirus particle exhibited an ovoid shape with a size exceeding 1 µm (Figure 5C). It enclosed a circular genome (more than 1,47 Mb) larger than the ones of Pithovirus and Cedratvirus, despite a relative phylognetic proximity. Moreover, this genome revealed translational components and an expansion of gene contents compared to Pithovirus, Cedratvirus, Marseilleviridae, and Irido-Ascoviridae members [123].
More recently, various viruses and virophages were detected from metagenomic studies in this emerging Pithoviridae family. One of them was misannotated as a Rickettsia bacterium (Misannotatedvirus) [124]; 15 giant viruses were detected in Loki’s castle with a variation of size from 282,320 bp to 638,759 bp, and also detection of two virophages [125] and two others, Solivirus (276 kb) and Solumvirus (316 kb) from soil samples [52]. For these viruses and virophages, there are no structural data currently available and these must be determined by future isolates. With them, particles variations would be explained and may serve to measure gigantism impact on particle variation and genome evolution in this emerging Pithoviridae familly [123].
8. Pandoraviruses
The first two strains of pandoraviruses, Pandoravirus salinus and Pandoravirus dulcis, were described in 2013, isolated from samples collected on the coast of Chile, and from a freshwater pond near Melbourne, Australia [54]. Pandoraviruses are very different from other previously described giant viruses of amoebas, both by their morphological and genomic features. They have micrometer-sized ovoid-shaped particles, encompassing genomes ranging from 1.9 to 2.5 megabases [16,54,126,127]. Consequently, to the description of these two strains, Pandoravirus inopinatum, described few years earlier as an endosymbiont, was recognized as another strain of pandoravirus [128,129]. Three new strains of pandoraviruses, named Pandoravirus massiliensis BZ81c, Pandoravirus pampulha 8.8, and Pandoravirus braziliensis SL2, were isolated from soil samples collected from Pampulha lagoon and Belo Horizonte city, and from a soda lake (Soda Lake 2), Brazil [16]. Then, the group of pandoraviruses expanded rapidly with the isolation of Pandoravirus quercus, Pandoravirus neocaledonia, and Pandoravirus macleodensis, collected from ground soil in Marseille, France; from water of a mangrove in New Caledonia; and from a freshwater pond near Melbourne, Australia, respectively [130]. All the pandoravirus strains were isolated by co-culture on Acanthamoeba castellanii.
Furthermore, metagenomic studies showed that pandoraviruses are ubiquitous, as sequences related to these viruses were detected in environmental metagenomes collected worldwide, and even from human plasma [131,132,133,134,135,136,137].
Pandoravirus virion morphology stands apart from other giant viruses, by their ovoid particles of 1.5–2 µm in length with an apical pore via which the content of particles is emptied in the amoebal cytoplasm. The study of the replication cycle showed that pandoraviruses can recruit mitochondria and membranes, inducing a modification of the amoebal cytoplasm, especially into and around the viral factories. Interestingly, the authors also showed that pandoravirus infection induces a complete degradation of the host nucleus, after which viral factories start forming [138].
Pandoraviruses harbor a linear double-stranded DNA genome, with a high GC content ranging from 59 to 64% [16,126]. They encode a huge number of genes, from 1414 (P. massiliensis) to 2693 (P. braziliensis) open reading frames (ORFs), and some tRNAs [54]. A remarkable property is their high number of ORFans, reaching up to 84% for P. salinus. Among the other predicted proteins, more than half encode MORN, F Box, and Ankyrin repeats. The proportion of duplicated genes is very high, varying from 16% to more than half of the gene content (55%), according to the parameters used [16,130].
Genomes of pandoraviruses do not encompass any gene encoding for a known capsid, another interesting and unique characteristic distinguishing further away giant viruses from canonical viruses [54]. As determined by electron microscopy, the particles harbor a three-layered tegument-like envelope. P. salinus genome also encompasses a high number of transposable elements, named MITE for miniature inverted repeat transposable elements [139]. Two pangenome analyses were performed. The first included the genomes of P. salinus, P. dulcis, P. inopinatum, P. massiliensis, P. braziliensis, and P. pampulha, and revealed a very small core genome consisting of 4.7% of the total pangenome [16]. The second included P. salinus, P. dulcis, P. inopinatum, P. quercus, P. neocaledonia, and P. macleodensis [130]. It revealed 54–88% pairwise similarity between the different pandoravirus isolates, with 80% of orthologous genes being collinear. Thus, the group of pandoraviruses harbors a big open pangenome. It mainly consists of ORFans and hypothetical proteins, for some of which transcriptomics and proteomics experiments showed that they were transcribed and translated in proteins.
Transcriptomics experiments of P. massiliensis showed that at least 25% of the predicted genes are transcribed in the conditions used in the experiment. Two-thirds of the total reads provided by the sequencing were found 6 h post infection [16]. The transcripts included some ORFans and hypothetical proteins, i.e., predicted proteins for which no function is assigned. Proteomics studies revealed 424 viral gene products for P. salinus, 357 for P. quercus, 387 for P. dulcis, and 337 for P. neocaledonia [130]. In addition, it was reported that 25% of the genetic content of P. massiliensis was detected by transcriptomics, and 11.4% of viral gene products were found by proteomic analysis in virions, more than half of which belong to the core genome [16]. On the four strains of pandoraviruses simultaneously analyzed, the core proteome was estimated at 53% of the total protein clusters globally identified in all pandoravirions, whereas the core genome is only composed of 42% of the overall number of pandoravirus-encoded protein clusters [130].
These findings show that further analyses and investigations will be necessary to improve our knowledge on these very intriguing giant viruses.
9. Mobilome: From Isolation and Characterization of Mobile Genetic Elements to the Discovery of a System of Defense in Giant Viruses
In many features, giant viruses have similar characteristics to the three major branches of the tree of life. The mobilome is no exception and, in our laboratory, genetic elements were highlighted as independent or integrated genomic sequences.
9.1. The Virophage Discovery
The concept of a virophage as a parasitic agent that depends on and predates the replicative cycle of a host mimivirus dates back to 2008, when La Scola et al. observed the presence of small virions of approximately 50 nm, infiltrating the viral factory of Acanthamoeba castellanii mamavirus, a mimivirus belonging to the lineage A of the Mimiviridae family cultivated from waters of cooling towers in Paris, France [34]. This small viral entity was named Sputnik (Figure 6B). Sputnik founded a new class of microbial agents that are typical viruses characterized by their distinct reproduction strategy, known as virophages due to their functional analogy with bacteriophages [140,141]. Sputnik was produced from the same viral factory of the giant Mamavirus at a specific location and earlier stage of the co-infection cycle. Sputnik reproduction impaired the infectivity of its virus host, leading to a decrease in amoebae lysis. In addition, the morphogenesis of the mimivirus was significantly impacted resulting in a high abnormal particle production [34]. The replication of virophages relies on the presence of replication machinery of their viral host; therefore, they cannot replicate themselves in their host cell. Consequently, an isolation strategy which is based on the use of a cultivable helper giant virus was developed by our group. Our aim was to screen diverse clinical and environmental samples for the presence of virophages, to isolate them and analyze their structural, genomic, and biological features [25]. So far, nine virophage strains were successfully isolated from different origins including France, United States of America (USA), Brazil, Tunisia, and Germany using distinct sample sources including water, soil, contact lens rinse fluid, and plane tree [25,34,35,36,61,142,143]. Six of them were isolated by our team including three Sputnik strains [25,34,35], Zamilon [36], Guarani [37], and Sissivirophage (Unpublished under characterization). On the other hand, 57 uncultivated virophage population genomes were completely or partially assembled from diverse environmental metagenomes [73,144,145,146,147,148,149].
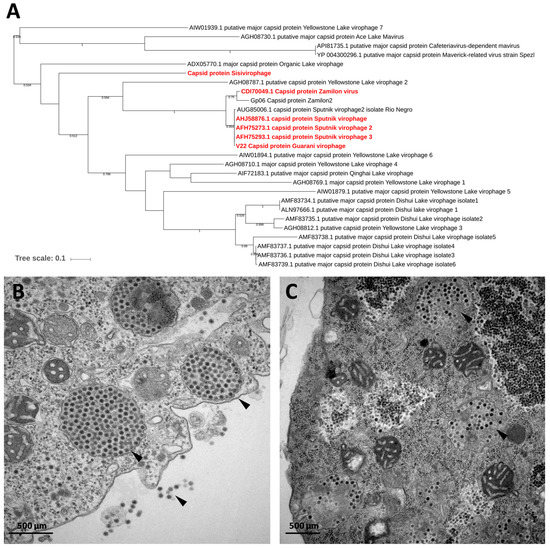
Figure 6.
(A) Phylogenetic reconstruction based on the major capsid proteins of virophages. The virophages isolated in IHU are highlighted in red. The analysis was performed using MEGA version 7.0, applying the maximum-likelihood method and WAG model of evolution with 500 bootstrap replicates (cutoff ≥ 50). (B–C) Transmission electronic microscopy of virophage particles replicating in Acanthamoeba castellanii co-infected with a giant virus host (arrows). (B) Sputnik; (C) Zamilon.
9.2. The Defense System MIMIVIRE
According to the Red Queen hypothesis [150], microorganisms are in a perpetual race to defend themselves against foreign genetic elements, and an unremitting arms race is engaged between the different hosts and invading entities. This evolutionary battle occurred between virophages and giant viruses. Based on this theory, we searched for sequences in the Mimivirus genome that could have been cannibalized and that would confer a selective advantage in the defense of Mimivirus against virophages [38]. As described above, several lineages (A, B, and C) were described in Mimiviridae and, among them, lineage A had the unique ability to resist against infection by a specific virophage, Zamilon [36]. According to our computational studies, we detected integrated and short repeated sequences of Zamilon in a gene (R349) present in the lineage A of Mimivirus. In addition, we localized, in the vicinity of these repeated sequences, genes that we assigned as a putative helicase and putative endonuclease (R350 and R354). Although this system is distinct from the structure described in the well-known prokaryotic CRISPR/Cas system, a functional analogy was proposed, and we defined this system as a defense system in giant viruses, named MIMIVIRE (Mimivirus virophage resistance element) [38]. Based on these in silico predictions, experimental assays were subsequently conducted to validate our hypothesis. By targeting the three genes of the MIMIVIRE system, RNA silencing experiments confirmed their role and that of the virophage genomic insertion sequence as key elements of the MIMIVIRE system. Recently, transformation of a lineage A Mimivirus was successfully achieved and the deletion by knockout of the R349 gene containing the repeated sequences of Zamilon restored the susceptibility of the infection by Zamilon [31]. These results confirmed our silencing experiments and definitely confirmed the R349 gene as an essential element of MIMIVIRE. An alternative hypothesis based on protein-based interaction was also proposed to explain the resistance of Mimivirus against Zamilon [151]. Finally, recent experimental results described the structure of the nuclease R354. Structural analyses and mechanistic studies confirmed the critical role of this protein as a functional Cas4-like protein as initially proposed [152].
10. Giant Virus Genes in Metagenomes and Other Microorganisms
10.1. Giant Virus-Like Sequences in Eukaryotic Genomes
Genes from giant viruses were identified in the genomes of eukaryotic organisms, including amoebae and plants [153,154,155,156]. For instance, giant virus genes encoding major capsid proteins of giant viruses were detected in the genome sequences of various Acanthamoeba species, and other giant virus genes encoding family B DNA polymerase, DNA-dependent RNA polymerase subunits, or D5 helicase-primase in the genome of Phycomitrella patens and Selaginella moellendorffii. In the case of genomes of Acanthamoeba spp., results suggested that the sequence flow between these amoebae and giant viruses could be in both directions [156]. Moreover, using sequences encoding DNA-dependent RNA polymerase subunits as baits in BLAST searches, we found that sequences previously identified as Hydra magnipapillata or Phytophtora parasitica corresponded to giant viral sequences integrated in these eukaryotes’ DNA, or alternatively were possibly previously overlooked [157].
10.2. Giant Virus-Like Sequences in Metagenomes
The onset of metagenomics for the study of microorganims and its expansion coincided with the Mimivirus discovery and the extension of giant virus diversity [158]. The power of metagenomics was tremendously enhanced by next-generation sequencing technologies. We detected, on several occasions, giant-virus-like sequences in metagenomes generated from various environmental, animal, and human samples. These results helped highlight that giant viruses are common in our biosphere. We were able to assemble ~40% of the genome of a Zamilon virophage from sequences generated from a bioreactor metagenome, in which sequences best matching with mimiviruses were concurrently detected [144]. We also implemented a tool, MG-Digger, to explore metagenomes and detect giant-virus-like sequences [131]. It homogenizes the format of metagenomes and then performs automated reciprocal BLAST hits, including comparison with BLAST searches conducted in the NCBI GenBank nucleotide or protein sequence databases. We notably tested MG-Digger on metagenomes generated from marine water and sewage, but also human blood samples. We detected sequences most related to various giant viruses including Mimiviruses and Marseilleviruses, but also more recently discovered representative members such as Faustoviruses, Pithoviruses, and Pandoraviruses, as well as virophage-like sequences. In addition, we detected sequences from putative giant virus relatives in the NCBI GenBank environmental protein sequence database (env nr), using giant virus sequences encoding a DNA-dependent RNA polymerase subunit, as well as reconstructed putative ancestral sequences for this protein as baits [157]. Metagenome analyses further provided evidence of the presence of giant viruses in humans. Thus, the fortuitous detection of Marseillevirus-like sequences in a metagenome from the stools of a young and healthy Senegalese man led to the isolation, for the first time, a giant virus named Senegalvirus from this human sample [42,159]. Then, metagenomics of blood samples from 10 blood donors from southern France led to detecting that 2.5% of the viral metagenome best matched with Marseillevirus-like sequences and could be assembled in two 10–13 kbp-large contigs closely related to the genome of the Marseillevirus prototype strain [43]. Thus, giant virus sequences, which contain large proportions of ORFans, contributed to enlighten the “dark matter” of environmental and human metagenomes since their discovery. This recently prompted other teams to assert that mimiviruses may be more abundant than prokaryotic organisms in marine waters [13] and could represent substantial fractions of the DNA virome from human specimens [160,161].
11. Giant Viruses in Humans
The presence of giant viruses in humans was documented especially for Mimiviruses and Marseilleviruses, the two families of giant viruses of amoebas first described.
11.1. Marseilleviruses
The first giant virus isolated from a human sample was Senegalvirus, a close relative of Marseillevirus, from a stool of a healthy young Senegalese man [42]. It was serendipitously found during a metagenomic and culturomic study. Then, sequences closed to Marseillevirus were detected in the metagenome generated from the blood of blood donors from France [43]. In the same study, Marseillevirus was detected by PCR in 10% of the 20 blood samples tested. However, these results were controversial, with other studies showing negative results for all blood samples tested by PCR, with the same primers used [162]. Seroprevalence studies showed that the exposition of humans to Marseilleviruses was unexpectedly high with rates varying between 1.7 and 15% [43,44,104], and reaching up to 23% in multitransfused individuals [44]. These data suggested then that the contacts of Marseilleviruses with humans are frequent.
Marseilleviruses were also detected in lymphoid organs in three reports. A Marseillevirus-like organism was detected in a lymph node of an 11-month-old boy who presented an adenitis of undetermined etiology [45], in the lymph node from a 30-year-old woman with Hodgkin lymphoma, and in the tonsils of a 20-year-old patient with neurologic disorders [46,47]. In addition, it was described, in this latter case, the first evidence of a prolonged carriage of Marseillevirus in humans. These reports may raise questions about the pathogenicity of these Marseilleviruses, particularly in the case of lymphomas. The prolonged carriage, the presence in the blood, the detection in pathologic lymph nodes are arguments for a possible causal link between the virus and the disease. Many other infectious agents, as the Epstein–Barr virus, are linked with lymphomas, and the question on the link between Marseillevirus and lymphomas could be open; however, further investigations are required. Furthermore, a murine model confirmed the prolonged detection of the virus in a mammalian organism, after an experimental inoculation by pulmonary or intravenous routes [105].
Moreover, Marseillevirus sequences were also detected in metagenomic studies in buccal mucosa, retro-auricular crease, vagina, and stools from healthy individuals [159,160].
11.2. Mimiviruses
Mimiviruses were first isolated in an amoebal co-culture of water samples from cooling towers during the investigation of an outbreak of pneumonia [11]. Then, the association between Mimiviruses and pneumonia in humans was explored. Firstly, seroprevalence studies showed that the rates found in community-acquired pneumonia and nosocomial pneumonia were 9.7% and 10%, respectively, versus 2.3% and 3% in control patients [87,163]. Moreover, seroconversions were observed in up to 8% and 22% of nosocomial pneumonia and community-acquired pneumonia patients, while any seroconversion was observed in control patients. In addition, a study also showed that Mimivirus could be the fourth most common etiology of acquired nosocomial and community pneumonia [164].
The seroconversion of a laboratory technician who handled huge quantities of Acanthamoeba polyphaga mimivirus and developed pneumonia, for which all other pathogens were negative, was a strong argument for the association between Mimivirus and pneumonia [165]. Indeed, an experimental murine model confirmed these data through the development of pneumonia in mice inoculated by the giant virus [166]. Moreover, two strains of mimiviruses were isolated from two pneumonia patients and three mimiviruses were detected by PCR in three cases of pneumonia [39,40,167]. Recently, a mimivirus was also isolated in the urines of a kidney-transplant recipient [41]. However, these associations between giant viruses and a disease were controversial, as other studies failed to detect mimivirus by PCR in all nasopharyngeal aspirates and bronchoalveolar liquid samples [88,168]. The rarity of these findings may be due to the high genetic variability of the genomes of mimiviruses [169]. Furthermore, it was demonstrated that Mimivirus was able to inhibit the regulation of IFN-stimulated genes, thus evading the IFN system. These results suggest that Mimivirus and humans have host–pathogen interactions [170]. Another team showed that 22% of rheumatoid arthritis patients had antibodies against mimivirus collagen versus 6% in control patients, showing that Mimivirus exposure is a risk factor for the development of auto immunity against collagen [171].
Recent metagenomic studies also reported the presence of Mimivirus sequences in human stools, nasopharyngeal aspirates from patients with respiratory tract infections, buccal mucosa, saliva and retroauricular crease, vagina, blood from healthy people, and even in blood from patients with liver diseases [159,160]. Rampelli et al. also found that Mimivirus sequences were the most represented viral sequences together with Poxviridae sequences in the human gut.
12. Conclusion
This 10th-year anniversary special issue of the journal Viruses gave us the opportunity to overview 15 years of research and discoveries on giant viruses. The most recent metagenomic works [51,52] or the renewed interpretation of old observations [172] suggest that these early years were only the beginning of discoveries in a vast field of life that probably holds great surprises in the years to come.
Funding
This work was supported by the French Government under the “Investissements d’avenir” (Investments for the Future) program managed by the Agence Nationale de la Recherche (ANR, French National Agency for Research), (reference: Méditerranée Infection 10-IAHU-03), by Région Provence-Alpes-Côte d’Azur and European funding FEDER PRIMI.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Drancourt, M.; Bollet, C.; Carlioz, A.; Martelin, R.; Gayral, J.-P.; Raoult, D. 16S Ribosomal DNA Sequence Analysis of a Large Collection of Environmental and Clinical Unidentifiable Bacterial Isolates. J. Clin. Microbiol. 2000, 38, 3623. [Google Scholar] [PubMed]
- La Scola, B.; Barrassi, L.; Raoult, D. Isolation of new fastidious α Proteobacteria and Afipia felis from hospital water supplies by direct plating and amoebal co-culture procedures. FEMS Microbiol. Ecol. 2000, 34, 129–137. [Google Scholar] [CrossRef]
- La Scola, B.; Mezi, L.; Weiller, P.J.; Raoult, D. Isolation of Legionella anisa Using an Amoebic Coculture Procedure. J. Clin. Microbiol. 2001, 39, 365. [Google Scholar] [CrossRef]
- La Scola, B.; Mallet, M.-N.; Grimont, P.A.D.; Raoult, D. Description of Afipia birgiae sp. nov. and Afipia massiliensis sp. nov. and recognition of Afipia felis genospecies A. Int. J. Syst. Evol. Microbiol. 2002, 52, 1773–1782. [Google Scholar]
- Greub, G.; La Scola, B.; Raoult, D. Parachlamydia acanthamoeba Is Endosymbiotic or Lytic for Acanthamoeba polyphaga Depending on the Incubation Temperature. Ann. N. Y. Acad. Sci. 2003, 990, 628–634. [Google Scholar] [CrossRef] [PubMed]
- La Scola, B.; Mallet, M.-N.; Grimont, P.A.D.; Raoult, D. Bosea eneae sp. nov., Bosea massiliensis sp. nov. and Bosea vestrisii sp. nov., isolated from hospital water supplies, and emendation of the genus Bosea (Das et al. 1996). Int. J. Syst. Evol. Microbiol. 2003, 53, 15–20. [Google Scholar] [CrossRef]
- La Scola, B.; Barrassi, L.; Raoult, D. A novel alpha-Proteobacterium, Nordella oligomobilis gen. nov., sp. nov., isolated by using amoebal co-cultures. Res. Microbiol. 2004, 155, 47–51. [Google Scholar] [CrossRef]
- La Scola, B.; Birtles, R.J.; Greub, G.; Harrison, T.J.; Ratcliff, R.M.; Raoult, D. Legionella drancourtii sp. nov., a strictly intracellular amoebal pathogen. Int. J. Syst. Evol. Microbiol. 2004, 54, 699–703. [Google Scholar] [CrossRef] [PubMed]
- Rowbotham, T.J. Isolation of Legionella pneumophila from clinical specimens via amoebae, and the interaction of those and other isolates with amoebae. J. Clin. Pathol. 1983, 36, 978–986. [Google Scholar] [CrossRef]
- Birtles, R.J.; Rowbotham, T.J.; Raoult, D.; Harrison, T.G. Phylogenetic diversity of intra-amoebal legionellae as revealed by 16S rRNA gene sequence comparison. Microbiology 1996, 142, 3525–3530. [Google Scholar] [CrossRef] [PubMed]
- La Scola, B.; Audic, S.; Robert, C.; Jungang, L.; De Lamballerie, X.; Drancourt, M.; Birtles, R.; Claverie, J.M.; Raoult, D. A giant virus in amoebae. Science 2003, 299, 2033. [Google Scholar] [CrossRef] [PubMed]
- Raoult, D.; Audic, S.; Robert, C.; Abergel, C.; Renesto, P.; Ogata, H.; La Scola, B.; Suzan, M.; Claverie, J.-M. The 1.2-Megabase Genome Sequence of Mimivirus. Science 2004, 306, 1344. [Google Scholar] [CrossRef]
- Mihara, T.; Koyano, H.; Hingamp, P.; Grimsley, N.; Goto, S.; Ogata, H. Taxon Richness of “Megaviridae” Exceeds those of Bacteria and Archaea in the Ocean. Microbes Environ. 2018, 33, 162–171. [Google Scholar] [CrossRef]
- La Scola, B.; Campocasso, A.; N’Dong, R.; Fournous, G.; Barrassi, L.; Flaudrops, C.; Raoult, D. Tentative Characterization of New Environmental Giant Viruses by MALDI-TOF Mass Spectrometry. Intervirology 2010, 53, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Boyer, M.; Yutin, N.; Pagnier, I.; Barrassi, L.; Fournous, G.; Espinosa, L.; Robert, C.; Azza, S.; Sun, S.; Rossmann, M.G.; et al. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc. Natl. Acad. Sci. USA 2009, 106, 21848. [Google Scholar] [CrossRef] [PubMed]
- Aherfi, S.; Andreani, J.; Baptiste, E.; Oumessoum, A.; Dornas, F.P.; dos, S.P.; Andrade, A.C.; Chabriere, E.; Abrahao, J.; Levasseur, A.; Raoult, D.; et al. A Large Open Pangenome and a Small Core Genome for Giant Pandoraviruses. Front. Microbiol. 2018, 9, 1486. [Google Scholar] [CrossRef]
- Levasseur, A.; Andreani, J.; Delerce, J.; Bou Khalil, J.Y.; Robert, C.; La Scola, B.; Raoult, D. Comparison of a Modern and Fossil Pithovirus Reveals Its Genetic Conservation and Evolution. Genome Biol. Evol. 2016, 8, 2333–2339. [Google Scholar] [CrossRef]
- Reteno, D.G.; Benamar, S.; Bou Khalil, J.Y.; Andreani, J.; Armstrong, N.; Klose, T.; Rossmann, M.; Colson, P.; Raoult, D.; La Scola, B. Faustovirus, an Asfarvirus-Related New Lineage of Giant Viruses Infecting Amoebae. J. Virol. 2015, 89, 6585–6594. [Google Scholar] [CrossRef]
- Abrahão, J.; Silva, L.; Silva, L.S.; Bou Khalil, J.Y.; Rodrigues, R.; Arantes, T.; Assis, F.; Boratto, P.; Andrade, M.; Kroon, E.G.; et al. Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere. Nat. Commun. 2018, 9, 749. [Google Scholar] [CrossRef] [PubMed]
- Bajrai, L.H.; Benamar, S.; Azhar, E.I.; Robert, C.; Levasseur, A.; Raoult, D.; La Scola, B. Kaumoebavirus, a New Virus That Clusters with Faustoviruses and Asfarviridae. Viruses 2016, 8, 278. [Google Scholar] [CrossRef]
- Andreani, J.; Bou Khalil, J.Y.; Baptiste, E.; Hasni, I.; Michelle, C.; Raoult, D.; Levasseur, A.; La Scola, B. Orpheovirus IHUMI-LCC2: A New Virus among the Giant Viruses. Front. Microbiol. 2018, 8, 2643. [Google Scholar] [CrossRef] [PubMed]
- Andreani, J.; Aherfi, S.; Bou Khalil, J.Y.; Di Pinto, F.; Bitam, I.; Raoult, D.; Colson, P.; La Scola, B. Cedratvirus, a Double-Cork Structured Giant Virus, is a Distant Relative of Pithoviruses. Viruses 2016, 8, 300. [Google Scholar] [CrossRef] [PubMed]
- Andreani, J.; Boo Khalil, J.Y.; Sevvana, M.; Benamar, S.; Di Pinto, F.; Bitam, I.; Colson, P.; Klose, T.; Rossmann, M.G.; Raoult, D.; et al. Pacmanvirus, a New Giant Icosahedral Virus at the Crossroads between Asfarviridae and Faustoviruses. J. Virol. 2017, 91, e00212-17. [Google Scholar] [CrossRef] [PubMed]
- Boughalmi, M.; Saadi, H.; Pagnier, I.; Colson, P.; Fournous, G.; Raoult, D.; La Scola, B. High-throughput isolation of giant viruses of the Mimiviridae and Marseilleviridae families in the Tunisian environment. Environ. Microbiol. 2013, 15, 2000–2007. [Google Scholar] [CrossRef]
- Gaia, M.; Pagnier, I.; Campocasso, A.; Fournous, G.; Raoult, D.; La Scola, B. Broad Spectrum of Mimiviridae Virophage Allows Its Isolation Using a Mimivirus Reporter. PLoS ONE 2013, 8, e61912. [Google Scholar] [CrossRef]
- Bou Khalil, J.Y.B.; Robert, S.; Reteno, D.G.; Andreani, J.; Raoult, D.; La Scola, B. High-throughput isolation of giant viruses in liquid medium using automated flow cytometry and fluorescence staining. Front. Microbiol. 2016, 7, 26. [Google Scholar] [CrossRef]
- Bou Khalil, J.Y.; Langlois, T.; Andreani, J.; Sorraing, J.-M.; Raoult, D.; Camoin, L.; La Scola, B. Flow Cytometry Sorting to Separate Viable Giant Viruses from Amoeba Co-culture Supernatants. Front. Cell. Infect. Microbiol. 2017, 6, 202. [Google Scholar] [CrossRef]
- Boyer, M.; Azza, S.; Barrassi, L.; Klose, T.; Campocasso, A.; Pagneir, I.; Fournous, G.; Borg, A.; Roebrt, C.; Zhang, X.; Desnues, C.; et al. Mimivirus shows dramatic genome reduction after intraamoebal culture. Proc. Natl. Acad. Sci. USA 2011, 108, 10296–10301. [Google Scholar] [CrossRef]
- Bekliz, M.; Azza, S.; Seligmann, H.; decloquement, P.; Raoult, D.; La Scola, B. Experimental analysis of Mimivirus translation initiation factor 4a reveals its importance in viral protein translation during infection of Acanthamoeba polyphaga. J. Virol. 2018, 92, 00337-18. [Google Scholar] [CrossRef]
- Sobhy, H.; Gotthard, G.; Chabriere, E.; Raoult, D.; Colson, P. Recombinant expression of mimivirus L725 ORFan gene product. Acta Virol. 2017, 61, 123–126. [Google Scholar] [CrossRef]
- Mougari, S.; Abrahao, J.S.; Pereira Oliveira, G.; Bou Khalil, J.Y.; la Scola, B. Role of the R349 gene and its repeats in the MIMIVIRE defense system. Front. Microbiol. 2019. submitted. [Google Scholar]
- Boughalmi, M.; Pagnier, I.; Aherfi, S.; Colson, P.; Raoult, D.; La Scola, B. First Isolation of a Marseillevirus in the Diptera Syrphidae Eristalis tenax. Intervirology 2013, 56, 386–394. [Google Scholar] [CrossRef]
- Temmam, S.; Monteil-Bouchard, S.; Sambou, M.; Aubadie-Ladrix, M.; Azza, S.; Decloquement, P.; Bou Khalil, J.Y.; Baudoin, J.-P.; Jardot, P.; Robert, C.; et al. Faustovirus-Like Asfarvirus in Hematophagous Biting Midges and Their Vertebrate Hosts. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- La Scola, B.; Desnues, C.; Pagnier, I.; Robert, C.; Barrassi, L.; Fournous, G.; Merchat, M.; Suzan-Monti, M.; Forterre, P.; Koonin, E.; et al. The virophage as a unique parasite of the giant mimivirus. Nature 2008, 455, 100. [Google Scholar] [CrossRef] [PubMed]
- Desnues, C.; La Scola, B.; Yutin, N.; Fournous, G.; Robert, C.; Azza, S.; Jardot, P.; Monteil, S.; Campocasso, A.; Koonin, E.V.; et al. Provirophages and transpovirons as the diverse mobilome of giant viruses. Proc. Natl. Acad. Sci. USA 2012, 109, 18078. [Google Scholar] [CrossRef]
- Gaia, M.; Benamar, S.; Boughalmi, M.; Pagnier, I.; Croce, O.; Colson, P.; Raoult, D.; La Scola, B. Zamilon, a Novel Virophage with Mimiviridae Host Specificity. PLoS ONE 2014, 9, e94923. [Google Scholar] [CrossRef] [PubMed]
- Mougari, S.; Bekliz, M.; Abrahao, J.S.; Di Pinto, F.; Levasseur, A.; La Scola, B. Guarani virophage, a new sputnik-like isolate from Brazilian lake. Front. Microbiol. 2019. submitted. [Google Scholar]
- Levasseur, A.; Bekliz, M.; Chabrière, E.; Pontarotti, P.; La Scola, B.; Raoult, D. MIMIVIRE is a defence system in mimivirus that confers resistance to virophage. Nature 2016, 531, 249–252. [Google Scholar] [CrossRef]
- Saadi, H.; Pagnier, I.; Colson, P.; Cherif, J.K.; Beji, M.; Boughalmi, M.; Azza, S.; Armstrong, N.; Robert, C.; Fournous, G.; et al. First isolation of Mimivirus in a patient with pneumonia. Clin. Infect. Dis. 2013, 57, 127–134. [Google Scholar] [CrossRef]
- Saadi, H.; Reteno, D.-G.I.; Colson, P.; Aherfi, S.; Minodier, P.; Pagnier, I.; Raoult, D.; La Scola, B. Shan Virus: A New Mimivirus Isolated from the Stool of a Tunisian Patient with Pneumonia. Intervirology 2013, 56, 424–429. [Google Scholar] [CrossRef]
- Moal, V.; Kodangasse, W.; Aherfi, S.; Berland, Y.; Raoult, D.; La Scola, B. Mimivirus in the urine of a kidney-transplant recipient. Clin. Microbiol. Infect. 2018, 24, 561–563. [Google Scholar] [CrossRef]
- Lagier, J.-C.; Armougom, F.; Million, M.; Hugon, P.; Pagnier, I.; Robert, C.; Bittar, F.; Fournous, G.; Gimenez, G.; Maraninchi, M.; et al. Microbial culturomics: Paradigm shift in the human gut microbiome study. Clin. Microbiol. Infect. 2012, 18, 1185–1193. [Google Scholar] [CrossRef] [PubMed]
- Popgeorgiev, N.; Boyer, M.; Fancello, L.; Monteil, S.; Robert, C.; Rivet, R.; Nappez, C.; Azza, S.; Chiaroni, J.; Raoult, D.; et al. Marseillevirus-Like Virus Recovered from Blood Donated by Asymptomatic Humans. J. Infect. Dis. 2013, 208, 1042–1050. [Google Scholar] [CrossRef] [PubMed]
- Popgeorgiev, N.; Colson, P.; Thuret, I.; Chiarioni, J.; Gallian, P.; Raoult, D.; Desnues, C. Marseillevirus prevalence in multitransfused patients suggests blood transmission. J. Clin. Virol. 2013, 58, 722–725. [Google Scholar] [CrossRef] [PubMed]
- Popgeorgiev, N.; Michel, G.; Lepidi, H.; Raoult, D.; Desnues, C. Marseillevirus adenitis in an 11-month-old child. J. Clin. Microbiol. 2013, 51, 4102–4105. [Google Scholar] [CrossRef]
- Aherfi, S.; Colson, P.; Audoly, G.; Nappez, C.; Xerri, L.; Valensi, A.; Million, M.; Lepidi, H.; Costello, R.; Raoult, D. Marseillevirus in lymphoma: A giant in the lymph node. Lancet Infect. Dis. 2016, 16, e225–e234. [Google Scholar] [CrossRef]
- Aherfi, S.; Colson, P.; Raoult, D. Marseillevirus in the Pharynx of a Patient with Neurologic Disorders. Emerg. Infect. Dis. 2016, 22, 11. [Google Scholar] [CrossRef]
- Arslan, D.; Legendre, M.; Seltzer, V.; Abergel, C.; Claverie, J.-M. Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae. Proc. Natl. Acad. Sci. USA 2011, 108, 17486. [Google Scholar] [CrossRef]
- Legendre, M.; Arslan, D.; Abergel, C.; Claverie, J.-M. Genomics of Megavirus and the elusive fourth domain of Life. Commun. Integr. Biol. 2012, 5, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Grishin, N.V.; Shabalina, S.A.; Wolf, Y.I.; Koonin, E.V. A putative RNA-interference-based immune system in prokaryotes: Computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct 2006, 1, 7. [Google Scholar] [CrossRef] [PubMed]
- Schulz, F.; Yutin, N.; Ivanova, N.N.; Ortega, D.R.; Lee, T.K.; Vierheilig, J.; Daims, H.; Horn, M.; Wagner, M.; Jensen, G.J.; et al. Giant viruses with an expanded complement of translation system components. Science 2017, 356, 82–85. [Google Scholar] [CrossRef] [PubMed]
- Schulz, F.; Alteio, L.; Goudeau, D.; Ryan, E.M.; Yu, F.B.; Malmstrom, R.R.; Blanchard, J.; Woyke, T. Hidden diversity of soil giant viruses. Nat. Commun. 2018, 9, 4881. [Google Scholar] [CrossRef] [PubMed]
- Legendre, M.; Bartoli, J.; Shmakova, L.; Jeudy, S.; Labadie, K.; Adrait, A.; Lescot, M.; Poirot, O.; Bertaux, L.; Bruley, C.; et al. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc. Natl. Acad. Sci. USA 2014, 111, 4274. [Google Scholar] [CrossRef] [PubMed]
- Philippe, N.; Legendre, M.; Doutre, G.; Couté, Y.; Poirot, O.; Lescot, M.; Arslan, D.; Seltzer, V.; Bertaux, L.; Bruley, C.; et al. Pandoraviruses: Amoeba Viruses with Genomes Up to 2.5 Mb Reaching That of Parasitic Eukaryotes. Science 2013, 341, 281. [Google Scholar] [CrossRef] [PubMed]
- Hingamp, P.; Grimsley, N.; Acinas, S.G.; Clerissi, C.; Subirana, L.; Poulain, J.; Ferrera, I.; Sarmento, H.; Villar, E.; Lima-Mendez, G.; et al. Exploring nucleo-cytoplasmic large DNA viruses in Tara Oceans microbial metagenomes. ISME J. 2013, 7, 1678–1695. [Google Scholar] [CrossRef] [PubMed]
- Martínez, J.M.; Swan, B.K.; Wilson, W.H. Marine viruses, a genetic reservoir revealed by targeted viromics. ISME J. 2014, 8, 1079–1088. [Google Scholar] [CrossRef]
- Monier, A.; Claverie, J.-M.; Ogata, H. Taxonomic distribution of large DNA viruses in the sea. Genome Biol. 2008, 9, R106. [Google Scholar] [CrossRef]
- Ghedin, E.; Claverie, J.-M. Mimivirus relatives in the Sargasso sea. Virol. J. 2005, 2, 62. [Google Scholar] [CrossRef] [PubMed]
- Boughalmi, M.; Pagnier, I.; Aherfi, S.; Colson, P.; Raoult, D.; La Scola, B. First isolation of a giant virus from wild Hirudo medicinalis leech: Mimiviridae isolation in Hirudo medicinalis. Viruses 2013, 5, 2920–2930. [Google Scholar] [CrossRef]
- Legendre, M.; Lartigue, A.; Bertaux, L.; Jeudy, S.; Bartoli, J.; Lescot, M.; Alempic, J.-M.; Ramus, C.; Bruley, C.; Labadie, K.; et al. In-depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba. Proc. Natl. Acad. Sci. USA 2015, 112, E5327–E5335. [Google Scholar] [CrossRef]
- Campos, R.K.; Boratto, P.V.; Assis, F.L.; Aguiar, E.R.G.R.; Silva, L.C.F.; Albarnaz, J.D.; Dornas, F.P.; Trindade, G.S.; Ferreira, P.P.; Marques, J.T.; et al. Samba virus: A novel mimivirus from a giant rain forest, the Brazilian Amazon. Virol. J. 2014, 11. [Google Scholar] [CrossRef] [PubMed]
- Bou Khalil, J.Y.; Andreani, J.; La Scola, B. Updating strategies for isolating and discovering giant viruses. Curr. Opin. Microbiol. 2016, 31, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Dornas, F.P.; Bou Khalil, J.Y.; Pagnier, I.; Raoult, D.; Abrahão, J.; La Scola, B. Isolation of new Brazilian giant viruses from environmental samples using a panel of protozoa. Front. Microbiol. 2015, 6, 1086. [Google Scholar] [CrossRef] [PubMed]
- Pagnier, I.; Reteno, D.G.I.; Saadi, H.; Boughalmi, M.; Gaia, M.; Slimani, M.; Ngounga, T.; Bekliz, M.; Colson, P.; Raoult, D.; et al. A decade of improvements in mimiviridae and marseilleviridae isolation from amoeba. Intervirology 2013, 56, 354–363. [Google Scholar] [CrossRef]
- Francis, R.; Ominami, Y.; Bou Khalid, J.Y.; La Scola, B. High throughput strategies for the isolation of giant viruses using high content screening. Comm. Biol. 2019. submitted. [Google Scholar]
- Colson, P.; La Scola, B.; Levasseur, A.; Caetano-Anollés, G.; Raoult, D. Mimivirus: Leading the way in the discovery of giant viruses of amoebae. Nat. Rev. Microbiol. 2017, 15, 243–254. [Google Scholar] [CrossRef]
- Colson, P.; Yutin, N.; Shabalina, S.A.; Robert, C.; Fournous, G.; La Scola, B.; Raoult, D.; Koonin, E.V. Viruses with More Than 1000 Genes: Mamavirus, a New Acanthamoeba polyphagamimivirus Strain, and Reannotation of Mimivirus Genes. Genome Biol. Evol. 2011, 3, 737–742. [Google Scholar] [CrossRef]
- Yoosuf, N.; Yutin, N.; Colson, P.; Shabalina, S.A.; Pagnier, I.; Robert, C.; Azza, S.; Klose, T.; Wong, J.; Rossmann, M.G.; et al. Related Giant Viruses in Distant Locations and Different Habitats: Acanthamoeba polyphaga moumouvirus Represents a Third Lineage of the Mimiviridae That Is Close to the Megavirus Lineage. Genome Biol. Evol. 2012, 4, 1324–1330. [Google Scholar] [CrossRef]
- dos, S.P.; Andrade, A.C.; Arantes, T.S.; Rodrigues, R.A.L.; Machado, T.B.; Dornas, F.P.; Landell, M.F.; Furst, C.; Borges, L.G.A.; Dutra, L.A.L.; Almeida, G.; et al. Ubiquitous giants: A plethora of giant viruses found in Brazil and Antarctica. Virol. J. 2018, 15, 22. [Google Scholar]
- Fischer, M.G.; Allen, M.J.; Wilson, W.H.; Suttle, C.A. Giant virus with a remarkable complement of genes infects marine zooplankton. Proc. Natl. Acad. Sci. USA 2010, 107, 19508–19513. [Google Scholar] [CrossRef]
- Fischer, M.G.; Hackl, T. Host genome integration and giant virus-induced reactivation of the virophage mavirus. Nature 2016, 540, 288–291. [Google Scholar] [CrossRef] [PubMed]
- Yutin, N.; Colson, P.; Raoult, D.; Koonin, E.V. Mimiviridae: Clusters of orthologous genes, reconstruction of gene repertoire evolution and proposed expansion of the giant virus family. Virol. J. 2013, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Santini, S.; Jeudy, S.; Bartoli, J.; Poirot, O.; Lescot, M.; Abergel, C.; Barbe, V.; Wommack, K.E.; Noordeloos, A.A.M.; Brussaard, C.P.D.; et al. Genome of Phaeocystis globosa virus PgV-16T highlights the common ancestry of the largest known DNA viruses infecting eukaryotes. Proc. Natl. Acad. Sci. USA 2013, 110, 10800–10805. [Google Scholar] [CrossRef]
- Deeg, C.M.; Chow, C.E.T.; Suttle, C.A. The kinetoplastid-infecting bodo saltans virus (Bsv), a window into the most abundant giant viruses in the sea. eLife 2018, 7, e33014. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, R.A.L.; Arantes, T.S.; Oliveira, G.P.; dos Santos Silva, L.K.; Abrahão, J.S. The Complex Nature of Tupanviruses. In Advances in Virus Research; Academic Press: Cambridge, MA, USA, 2018. [Google Scholar]
- Rodrigues, R.A.L.; Mougari, S.; Colson, P.; La Scola, B.; Abrahão, J.S. “Tupanvirus”, a new genus in the family Mimiviridae. Arch. Virol. 2018, 164, 325–331. [Google Scholar] [CrossRef]
- dos, S.P.; Andrade, A.C.; Rodrigues, R.A.L.; Oliveira, G.P.; Andrade, K.R.; Bonjardim, C.A.; La Scola, B.; Kroon, E.G.; Abrahão, J.S. Filling Knowledge Gaps for Mimivirus Entry, Uncoating, and Morphogenesis. J. Virol. 2017, 91, e01335-17. [Google Scholar]
- Zauberman, N.; Mutsafi, Y.; Halevy, D.B.; Shimoni, E.; Klein, E.; Xiao, C.; Sun, S.; Minsky, A. Distinct DNA Exit and Packaging Portals in the Virus Acanthamoeba polyphaga mimivirus. PLoS Biol. 2008, 6, e114. [Google Scholar] [CrossRef] [PubMed]
- Suzan-Monti, M.; La Scola, B.; Barrassi, L.; Espinosa, L.; Raoult, D. Ultrastructural Characterization of the Giant Volcano-like Virus Factory of Acanthamoeba polyphaga Mimivirus. PLoS ONE 2007, 2, e328. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, Y.G.; Klose, T.; Rossmann, M.; McPherson, A. Morphogenesis of Mimivirus and Its Viral Factories: An Atomic Force Microscopy Study of Infected Cells. J. Virol. 2013, 87, 11200. [Google Scholar] [CrossRef]
- Mutsafi, Y.; Shimoni, E.; Shimon, A.; Minsky, A. Membrane Assembly during the Infection Cycle of the Giant Mimivirus. PLoS Pathog. 2013, 9, e1003367. [Google Scholar] [CrossRef]
- Boyer, M.; Madoui, M.-A.; Gimenez, G.; La Scola, B.; Raoult, D. Phylogenetic and Phyletic Studies of Informational Genes in Genomes Highlight Existence of a 4th Domain of Life Including Giant Viruses. PLoS ONE 2010, 5, e15530. [Google Scholar] [CrossRef] [PubMed]
- Abrahão, J.S.; Araújo, R.; Colson, P.; La Scola, B. The analysis of translation-related gene set boosts debates around origin and evolution of mimiviruses. PLoS Genet. 2017, 13, e1006532. [Google Scholar] [CrossRef]
- Colson, P.; Levasseur, A.; La Scola, B.; Sharma, V.; Nasir, A.; Pontarotti, P.; Caetano-Anollés, G.; Raoult, D. Ancestrality and Mosaicism of Giant Viruses Supporting the Definition of the Fourth TRUC of Microbes. Front. Microbiol. 2018, 9, 2668. [Google Scholar] [CrossRef] [PubMed]
- Yutin, N.; Wolf, Y.I.; Koonin, E.V. Origin of giant viruses from smaller DNA viruses not from a fourth domain of cellular life. Virology 2014, 466–467, 38–52. [Google Scholar] [CrossRef]
- Moreira, D.; López-García, P. Evolution of viruses and cells: Do we need a fourth domain of life to explain the origin of eukaryotes? Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140327. [Google Scholar] [CrossRef]
- La Scola, B.; Marrie, T.J.; Auffray, J.-P.; Raoult, D. Mimivirus in Pneumonia Patients. Emerg. Infect. Dis. 2005, 11, 449–452. [Google Scholar] [CrossRef]
- Dare, R.K.; Chittaganpitch, M.; Erdman, D.D. Screening Pneumonia Patients for Mimivirus. Emerg. Infect. Dis. 2008, 14, 3. [Google Scholar] [CrossRef]
- Colson, P.; Pagnier, I.; Yoosuf, N.; Fournous, G.; La Scola, B.; Raoult, D. “Marseilleviridae”, a new family of giant viruses infecting amoebae. Arch. Virol. 2013, 158, 915–920. [Google Scholar] [CrossRef] [PubMed]
- Raoult, D.; La Scola, B.; Birtles, R. The Discovery and Characterization of Mimivirus, the Largest Known Virus and Putative Pneumonia Agent. Clin. Infect. Dis. 2007, 45, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Aherfi, S.; Pagnier, I.; Fournous, G.; Raoult, D.; La Scola, B.; Colson, P. Complete genome sequence of Cannes 8 virus, a new member of the proposed family “Marseilleviridae”. Virus Genes 2013, 47, 550–555. [Google Scholar] [CrossRef]
- Doutre, G.; Philippe, N.; Abergel, C.; Claverie, J.-M. Genome Analysis of the First Marseilleviridae Representative from Australia Indicates that Most of Its Genes Contribute to Virus Fitness. J. Virol. 2014, 88, 14340–14349. [Google Scholar] [CrossRef] [PubMed]
- Takemura, M. Morphological and Taxonomic Properties of Tokyovirus, the First Marseilleviridae Member Isolated from Japan. Microbes Environ. 2016, 31, 442–448. [Google Scholar] [CrossRef] [PubMed]
- Thomas, V.; Bertelli, C.; Collyn, F.; Casson, N.; Telenti, A.; Goesmann, A.; Croxatto, A.; Greub, G. Lausannevirus, a giant amoebal virus encoding histone doublets. Environ. Microbiol. 2011, 13, 1454–1466. [Google Scholar] [CrossRef]
- Fabre, E.; Jeudy, S.; Santini, S.; Legendre, M.; Trauchessec, M.; Couté, Y.; Claverie, J.-M.; Abergel, C. Noumeavirus replication relies on a transient remote control of the host nucleus. Nat. Commun. 2017, 8, 15087. [Google Scholar] [CrossRef]
- Doutre, G.; Arfib, B.; Rochette, P.; Claverie, J.-M.; Bonin, P.; Abergel, C. Complete Genome Sequence of a New Member of the Marseilleviridae Recovered from the Brackish Submarine Spring in the Cassis Port-Miou Calanque, France. Genome Announc. 2015, 3, e01148-15. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Kondabagil, K. Complete genome sequence of Kurlavirus, a novel member of the family Marseilleviridae isolated in Mumbai, India. Arch. Virol. 2017, 162, 3243–3245. [Google Scholar] [CrossRef] [PubMed]
- Aherfi, S.; Boughalmi, M.; Pagnier, I.; Fournous, G.; La Scola, B.; Raoult, D.; Colson, P. Complete genome sequence of Tunisvirus, a new member of the proposed family Marseilleviridae. Arch. Virol. 2014, 159, 2349–2358. [Google Scholar] [CrossRef]
- Dornas, F.P.; Assis, F.L.; Aherfi, S.; Arantes, T.; Abrahão, J.S.; Colson, P.; La Scola, B. A Brazilian Marseillevirus Is the Founding Member of a Lineage in Family Marseilleviridae. Viruses 2016, 8, 76. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, R.N.; Campos, F.S.; Medeiros de Albuquerque, N.R.; Finoketti, F.; Côrrea, R.A.; Cano-Ortiz, L.; Assis, F.L.; Arantes, T.S.; Roehe, P.M.; Franco, A.C. A new marseillevirus isolated in Southern Brazil from Limnoperna fortunei. Sci. Rep. 2016, 6, 35237. [Google Scholar] [CrossRef] [PubMed]
- Arantes, T.S.; Rodrigues, R.A.L.; Dos Santos Silva, L.K.; Oliveira, G.P.; de Souza, H.L.; Bou Khalil, J.Y.; de Oliveira, D.B.; Torres, A.A.; da Silva, L.L.; Colson, P.; et al. The Large Marseillevirus Explores Different Entry Pathways by Forming Giant Infectious Vesicles. J. Virol. 2016, 90, 5246–5255. [Google Scholar] [CrossRef]
- Oliveira, G.P.; Lima, M.T.; Arantes, T.S.; Assis, F.L.; Rodrigues, R.A.L.; da Fonseca, F.G.; Bonjardim, C.A.; Kroon, E.G.; Colson, P.; La Scola, B.; et al. The Investigation of Promoter Sequences of Marseilleviruses Highlights a Remarkable Abundance of the AAATATTT Motif in Intergenic Regions. J. Virol. 2017, 91, e01088-17. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, R.A.L.; Cherif Louazani, A.; Colson, P.; La Scola, B.; Abrahao, J.S. J. Virol. 2019. submitted.
- Mueller, L.; Baud, D.; Bertelli, C.; Greub, G. Lausannevirus Seroprevalence among Asymptomatic Young Adults. Intervirology 2013, 56, 430–433. [Google Scholar] [CrossRef]
- Aherfi, S.; Nappez, C.; Lepidi, H.; Bedotto, M.; Barassi, L.; Jardot, P.; Colson, P.; La Scola, B.; Raoult, D.; Bregeon, F. Experimental Inoculation in Rats and Mice by the Giant Marseillevirus Leads to Long-Term Detection of Virus. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Pagnier, I.; Valles, C.; Raoult, D.; La Scola, B. Isolation of Vermamoeba vermiformis and associated bacteria in hospital water. Microb. Pathog. 2015, 80, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Thomas, V.; Herrera-Rimann, K.; Blanc, D.S.; Greub, G. Biodiversity of Amoebae and Amoeba-Resisting Bacteria in a Hospital Water Network. Appl. Environ. Microbiol. 2006, 72, 2428–2438. [Google Scholar] [CrossRef] [PubMed]
- Delafont, V.; Brouke, A.; Bouchon, D.; Moulin, L.; Héchard, Y. Microbiome of free-living amoebae isolated from drinking water. Microb. Ecol. Drink. Water Wastewater Treat. 2013, 47, 6958–6965. [Google Scholar] [CrossRef]
- Bradbury, R.S. Free-Living Amoebae Recovered from Human Stool Samples in Strongyloides Agar Culture. J. Clin. Microbiol. 2014, 52, 699–700. [Google Scholar] [CrossRef]
- Hajialilo, E.; Niyyati, M.; Solaymani, M.; Rezaeian, M. Pathogenic Free-Living Amoebae Isolated from Contact Lenses of Keratitis Patients. Iran. J. Parasitol. 2015, 10, 541–546. [Google Scholar]
- Abedkhojasteh, H.; Niyyati, M.; Rahimi, F.; Hei-Dari, M.; Farnia, S.; Rezaeian, M. First report of Hartmannella keratitis in a cosmetic soft contact lens wearer in Iran. Iran. J. Parasitol. 2013, 8, 481–485. [Google Scholar]
- Bou Khalil, J.Y.; Andreani, J.; Raoult, D.; La Scola, B. A Rapid Strategy for the Isolation of New Faustoviruses from Environmental Samples Using Vermamoeba vermiformis. J. Vis. Exp. 2016. [Google Scholar] [CrossRef]
- Louazani, A.C.; Andreani, J.; Ouarhache, M.; Aherfi, S.; Baptiste, E.; Levasseur, A.; La Scola, B. Genome Sequences of New Faustovirus Strains ST1 and LC9, Isolated from the South of France. Genome Announc. 2017, 5, e00613-17. [Google Scholar]
- Benamar, S.; Reteno, D.G.I.; Bandaly, V.; Labas, N.; Raoult, D.; La Scola, B. Faustoviruses: Comparative Genomics of New Megavirales Family Members. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Klose, T.; Reteno, D.G.; Benamar, S.; Hollerbach, A.; Colson, P.; La Scola, B.; Rossmann, M.G. Structure of faustovirus, a large dsDNA virus. Proc. Natl. Acad. Sci. USA 2016, 113, 6206–6211. [Google Scholar] [CrossRef] [PubMed]
- Cherif Louazani, A.; Baptiste, E.; Levasseur, A.; Colson, P.; La Scola, B. Faustovirus E12 Transcriptome Analysis Reveals Complex Splicing in Capsid Gene. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, G.P.; de Aquino, I.L.M.; Luiz, A.P.M.F.; Abrahão, J.S. Putative Promoter Motif Analyses Reinforce the Evolutionary Relationships Among Faustoviruses, Kaumoebavirus, and Asfarvirus. Front. Microbiol. 2018, 9, 1041. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, K.; Miyazaki, N.; Song, C.; Maia, F.R.N.C.; Reddy, H.K.N.; Abergel, C.; Claverie, J.-M.; Hajdu, J.; Svenda, M.; Murata, K. Structural variability and complexity of the giant Pithovirus sibericum particle revealed by high-voltage electron cryo-tomography and energy-filtered electron cryo-microscopy. Sci. Rep. 2017, 7, 13291. [Google Scholar] [CrossRef]
- Sharma, V.; Colson, P.; Chabrol, O.; Pontarotti, P.; Raoult, D. Pithovirus sibericum, a new bona fide member of the “Fourth TRUC” club. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Bertelli, C.; Mueller, L.; Thomas, V.; Pillonel, T.; Jacquier, N.; Greub, G. Cedratvirus lausannensis—Digging into Pithoviridae diversity. Environ. Microbiol. 2017, 19, 4022–4034. [Google Scholar] [CrossRef]
- Rodrigues, R.A.L.; Andreani, J.; dos, S.P.; Andrade, A.C.; Machado, T.B.; Abdi, S.; Levasseur, A.; Abrahão, J.S.; La Scola, B. Morphologic and Genomic Analyses of New Isolates Reveal a Second Lineage of Cedratviruses. J. Virol. 2018, 92, e00372-18. [Google Scholar] [CrossRef]
- Silva, L.K.; dos, S.; dos, S.P.; Andrade, A.C.; Dornas, F.P.; Rodrigues, R.A.L.; Arantes, T.; Kroon, E.G.; Bonjardim, C.A.; Abrahão, J.S. Cedratvirus getuliensis replication cycle: An in-depth morphological analysis. Sci. Rep. 2018, 8, 4000. [Google Scholar] [CrossRef]
- Koonin, E.V.; Yutin, N. Multiple evolutionary origins of giant viruses. F1000Research 2018, 7, 1840. [Google Scholar] [CrossRef] [PubMed]
- Andreani, J.; Verneau, J.; Raoult, D.; Levasseur, A.; La Scola, B. Deciphering viral presences: Two novel partial giant viruses detected in marine metagenome and in a mine drainage metagenome. Virol. J. 2018, 15, 66. [Google Scholar] [CrossRef]
- Backstrom, D.; Yutin, N.; Jorgensen, S.L.; Dharamshi, J.; Homa, F.; Zaremba-Niedwiedzka, K.; Spang, A.; Wolf, Y.I.; Koonin, E.V.; Ettema, T.J.G. Virus genomes from deep sea sediments expand the ocean megavirome and support independent origins of viral gigantism. bioRxiv 2018, 469403. [Google Scholar] [CrossRef]
- Abergel, C.; Legendre, M.; Claverie, J.-M. The rapidly expanding universe of giant viruses: Mimivirus, Pandoravirus, Pithovirus and Mollivirus. FEMS Microbiol. Rev. 2015, 39, 779–796. [Google Scholar] [CrossRef]
- Colson, P.; La Scola, B.; Raoult, D. Giant Viruses of Amoebae: A Journey through Innovative Research and Paradigm Changes. Annu. Rev. Virol. 2017, 4, 61–85. [Google Scholar] [CrossRef]
- Antwerpen, M.H.; Georgi, E.; Zoeller, L.; Woelfel, R.; Stoecker, K.; Scheid, P. Whole-Genome Sequencing of a Pandoravirus Isolated from Keratitis-Inducing Acanthamoeba. Genome Announc. 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Scheid, P. A strange endocytobiont revealed as largest virus. Curr. Opin. Microbiol. 2016, 31, 58–62. [Google Scholar] [CrossRef]
- Legendre, M.; Fabre, E.; Poirot, O.; Jeudy, S.; Lartigue, A.; Alempic, J.-M.; Beucher, L.; Philippe, N.; Bertaux, L.; Christo-Foroux, E.; et al. Diversity and evolution of the emerging Pandoraviridae family. Nat. Commun. 2018, 9, 2285. [Google Scholar] [CrossRef]
- Verneau, J.; Levasseur, A.; Raoult, D.; La Scola, B.; Colson, P. MG-Digger: An Automated Pipeline to Search for Giant Virus-Related Sequences in Metagenomes. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Atoni, E.; Wang, Y.; Karungu, S.; Waruhiu, C.; Zohaib, A.; Obanda, V.; Agwanda, B.; Mutua, M.; Xia, H.; Yuan, Z. Metagenomic Virome Analysis of Culex Mosquitoes from Kenya and China. Viruses 2018, 10, 30. [Google Scholar] [CrossRef] [PubMed]
- Brinkman, N.E.; Villegas, E.N.; Garland, J.L.; Keely, S.P. Reducing inherent biases introduced during DNA viral metagenome analyses of municipal wastewater. PLoS ONE 2018, 13, e0195350. [Google Scholar] [CrossRef] [PubMed]
- Halary, S.; Temmam, S.; Raoult, D.; Desnues, C. Viral metagenomics: Are we missing the giants? Curr. Opin. Microbiol. 2016, 31, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Szabó, A.; Korponai, K.; Kerepesi, C.; Somogyi, B.; Vörös, L.; Bartha, D.; Márialigeti, K.; Felföldi, T. Soda pans of the Pannonian steppe harbor unique bacterial communities adapted to multiple extreme conditions. Extremophiles 2017, 21, 639–649. [Google Scholar] [CrossRef]
- Temmam, S.; Davoust, B.; Chaber, A.-L.; Lignereux, Y.; Michelle, C.; Monteil-Bouchard, S.; Raoult, D.; Desnues, C. Screening for Viral Pathogens in African Simian Bushmeat Seized at A French Airport. Transbound. Emerg. Dis. 2017, 64, 1159–1167. [Google Scholar] [CrossRef]
- Temmam, S.; Monteil-Bouchard, S.; Robert, C.; Baudoin, J.-P.; Sambou, M.; Aubadie-Ladrix, M.; Labas, N.; Raoult, D.; Mediannikov, O.; Desnues, C. Characterization of Viral Communities of Biting Midges and Identification of Novel Thogotovirus Species and Rhabdovirus Genus. Viruses 2016, 8, 77. [Google Scholar] [CrossRef] [PubMed]
- dos, S.P.; Andrade, A.C.; de Miranda Boratto, P.V.; Rodrigues, R.A.L.; Bastos, T.M.; Azevedo, B.L.; Dornas, F.P.; Oliveira, D.B.; Drumond, B.P.; Kroon, E.G.; et al. New isolates of pandoraviruses: Contribution to the study of replication cycle steps. J. Virol. 2018, 93. [Google Scholar] [CrossRef]
- Sun, C.; Feschotte, C.; Wu, Z.; Mueller, R.L. DNA transposons have colonized the genome of the giant virus Pandoravirus salinus. BMC Biol. 2015, 13, 38. [Google Scholar] [CrossRef]
- Desnues, C.; Raoult, D. Virophages question the existence of satellites. Nat. Rev. Microbiol. 2012, 10, 234. [Google Scholar] [CrossRef]
- Desnues, C.; Boyer, M.; Raoult, D. Sputnik, a virophage infecting the viral domain of life. Adv. Virus Res. 2012, 82, 63–89. [Google Scholar]
- Fischer, M.G.; Suttle, C.A. A Virophage at the Origin of Large DNA Transposons. Science 2011, 332, 231–234. [Google Scholar] [CrossRef] [PubMed]
- Experimental Co-Infection of Saccamoeba lacustris with Mimivirus-Like Giant Virus and a Small Satellite Virus—JPortal. Available online: https://zs.thulb.uni-jena.de/receive/jportal_jparticle_00560514 (accessed on 3 January 2019).
- Bekliz, M.; Verneau, J.; Benamar, S.; Raoult, D.; La Scola, B.; Colson, P. A New Zamilon-like Virophage Partial Genome Assembled from a Bioreactor Metagenome. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Yau, S.; Lauro, F.M.; DeMaere, M.Z.; Brown, M.V.; Thomas, T.; Raftery, M.J.; Andrews-Pfannkoch, C.; Lewis, M.; Hoffman, J.M.; Gibson, J.A.; et al. Virophage control of antarctic algal host–virus dynamics. Proc. Natl. Acad. Sci. USA 2011, 108, 6163–6168. [Google Scholar] [CrossRef]
- Zhou, J.; Sun, D.; Childers, A.; McDermott, T.R.; Wang, Y.; Liles, M.R. Three Novel Virophage Genomes Discovered from Yellowstone Lake Metagenomes. J. Virol. 2015, 89, 1278–1285. [Google Scholar] [CrossRef]
- Oh, S.; Yoo, D.; Liu, W.-T. Metagenomics Reveals a Novel Virophage Population in a Tibetan Mountain Lake. Microbes Environ. 2016, 31, 173–177. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, W.; Yan, S.; Xiao, J.; Zhang, Y.; Li, B.; Pan, Y.; Wang, Y. Diversity of Virophages in Metagenomic Data Sets. J. Virol. 2013, 87, 4225–4236. [Google Scholar] [CrossRef] [PubMed]
- Yutin, N.; Kapitonov, V.V.; Koonin, E.V. A new family of hybrid virophages from an animal gut metagenome. Biol. Direct 2015, 10, 19. [Google Scholar] [CrossRef]
- Van Valen, L. A new evolutionary law. Evol. Theory 1973, 1, 1–30. [Google Scholar]
- Claverie, J.-M.; Abergel, C. CRISPR-Cas-like system in giant viruses: Why MIMIVIRE is not likely to be an adaptive immune system. Virol. Sin. 2016, 31, 193–196. [Google Scholar] [CrossRef] [PubMed]
- Dou, C.; Yu, M.; Gu, Y.; Wang, J.; Yin, K.; Nie, C.; Zhu, X.; Qi, S.; Wei, Y.; Cheng, W. Structural and Mechanistic Analyses Reveal a Unique Cas4-like Protein in the Mimivirus Virophage Resistance Element System. iScience 2018, 3, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Colson, P.; Gimenez, G.; Boyer, M.; Fournous, G.; Raoult, D. The Giant Cafeteria roenbergensis Virus That Infects a Widespread Marine Phagocytic Protist Is a New Member of the Fourth Domain of Life. PLoS ONE 2011, 6, e18935. [Google Scholar] [CrossRef] [PubMed]
- Maumus, F.; Epert, A.; Nogué, F.; Blanc, G. Plant genomes enclose footprints of past infections by giant virus relatives. Nat. Commun. 2014, 5, 4268. [Google Scholar] [CrossRef] [PubMed]
- Maumus, F.; Blanc, G. Study of Gene Trafficking between Acanthamoeba and Giant Viruses Suggests an Undiscovered Family of Amoeba-Infecting Viruses. Genome Biol. Evol. 2016, 8, 3351–3363. [Google Scholar] [CrossRef]
- Chelkha, N.; Levasseur, A.; Pontarotti, P.; Raoult, D.; La Scola, B.; Colson, P. A Phylogenomic Study of Acanthamoeba polyphaga Draft Genome Sequences Suggests Genetic Exchanges with Giant Viruses. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Colson, P.; Giorgi, R.; Pontarotti, P.; Raoult, D. DNA-Dependent RNA Polymerase Detects Hidden Giant Viruses in Published Databanks. Genome Biol. Evol. 2014, 6, 1603–1610. [Google Scholar] [CrossRef]
- Mokili, J.L.; Rohwer, F.; Dutilh, B.E. Metagenomics and future perspectives in virus discovery. Curr. Opin. Virol. 2012, 2, 63–77. [Google Scholar] [CrossRef]
- Colson, P.; Fancello, L.; Gimenez, G.; Armougom, F.; Desnues, C.; Fournous, G.; Yoosuf, N.; Million, M.; La Scola, B.; Raoult, D. Evidence of the megavirome in humans. J. Clin. Virol. 2013, 57, 191–200. [Google Scholar] [CrossRef]
- Rampelli, S.; Soverini, M.; Turroni, S.; Quercia, S.; Biagi, E.; Brigidi, P.; Candela, M. ViromeScan: A new tool for metagenomic viral community profiling. BMC Genom. 2016, 17, 165. [Google Scholar] [CrossRef] [PubMed]
- Bzhalava, Z.; Hultin, E.; Dillner, J. Extension of the viral ecology in humans using viral profile hidden Markov models. PLoS ONE 2018, 13, e0190938. [Google Scholar] [CrossRef] [PubMed]
- Sauvage, V.; Livartowski, A.; Boizeau, L.; Servant-Delmas, A.; Lionnet, F.; Lefrère, J.-J.; Laperche, S. No Evidence of Marseillevirus-like Virus Presence in Blood Donors and Recipients of Multiple Blood Transfusions. J. Infect. Dis. 2014, 210, 2017–2018. [Google Scholar] [CrossRef] [PubMed]
- Bousbia, S.; Papazian, L.; Saux, P.; Forel, J.-M.; Auffray, J.-P.; Martin, C.; Raoult, D.; La Scola, B. Serologic Prevalence of Amoeba-Associated Microorganisms in Intensive Care Unit Pneumonia Patients. PLoS ONE 2013, 8, e58111. [Google Scholar] [CrossRef]
- Berger, P.; Papazian, L.; Drancourt, M.; La Scola, B.; Auffray, J.-P.; Raoult, D. Ameba-associated Microorganisms and Diagnosis of Nosocomial Pneumonia. Emerg. Infect. Dis. 2006, 12, 248–255. [Google Scholar] [CrossRef]
- Raoult, D.; Renesto, P.; Brouqui, P. Laboratory infection of a technician by mimivirus. Ann. Intern. Med. 2006, 144, 702–703. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; La Scola, B.; Lepidi, H.; Raoult, D. Pneumonia in mice inoculated experimentally with Acanthamoeba polyphaga mimivirus. Microb. Pathog. 2007, 42, 56–61. [Google Scholar] [CrossRef]
- Zhang, X.A.; Zhu, T.; Zhang, P.H.; Li, H.; Li, Y.; Liu, E.M.; Liu, W.; Cao, W.C. Lack of Mimivirus Detection in Patients with Respiratory Disease, China. Emerg. Infect. Dis. 2016, 22, 11. [Google Scholar] [CrossRef] [PubMed]
- Larcher, C.; Jeller, V.; Fischer, H.; Huemer, H.P. Prevalence of respiratory viruses, including newly identified viruses, in hospitalised children in Austria. Eur. J. Clin. Microbiol. Infect. Dis. 2006, 25, 681–686. [Google Scholar] [CrossRef]
- Raoult, D.; Levasseur, A.; La Scola, B. PCR Detection of Mimivirus. Emerg. Infect. Dis. 2017, 23, 6. [Google Scholar] [CrossRef] [PubMed]
- de, F.; Almeida, G.M.; Silva, L.C.F.; Colson, P.; Abrahao, J.S. Mimiviruses and the Human Interferon System: Viral Evasion of Classical Antiviral Activities, But Inhibition by a Novel Interferon-β Regulated Immunomodulatory Pathway. J. Interferon Cytokine Res. 2017, 37, 1–8. [Google Scholar]
- Shah, N.; Hülsmeier, A.J.; Hochhold, N.; Neidhart, M.; Gay, S.; Hennet, T. Exposure to Mimivirus Collagen Promotes Arthritis. J. Virol. 2014, 88, 838–845. [Google Scholar] [CrossRef]
- Barthélémy, R.-M.; Faure, E.; Goto, T.; Rm, B. Serendipitous Discovery in a Marine Invertebrate (Phylum Chaetognatha) of the Longest Giant Viruses Reported till Date. Virology 2019, 3, 13. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).