Prion and Prion-Like Protein Strains: Deciphering the Molecular Basis of Heterogeneity in Neurodegeneration
Abstract
1. Introduction
2. Prion Protein
2.1. The Cellular Prion Protein
2.2. Molecular Basis of Prion Strains
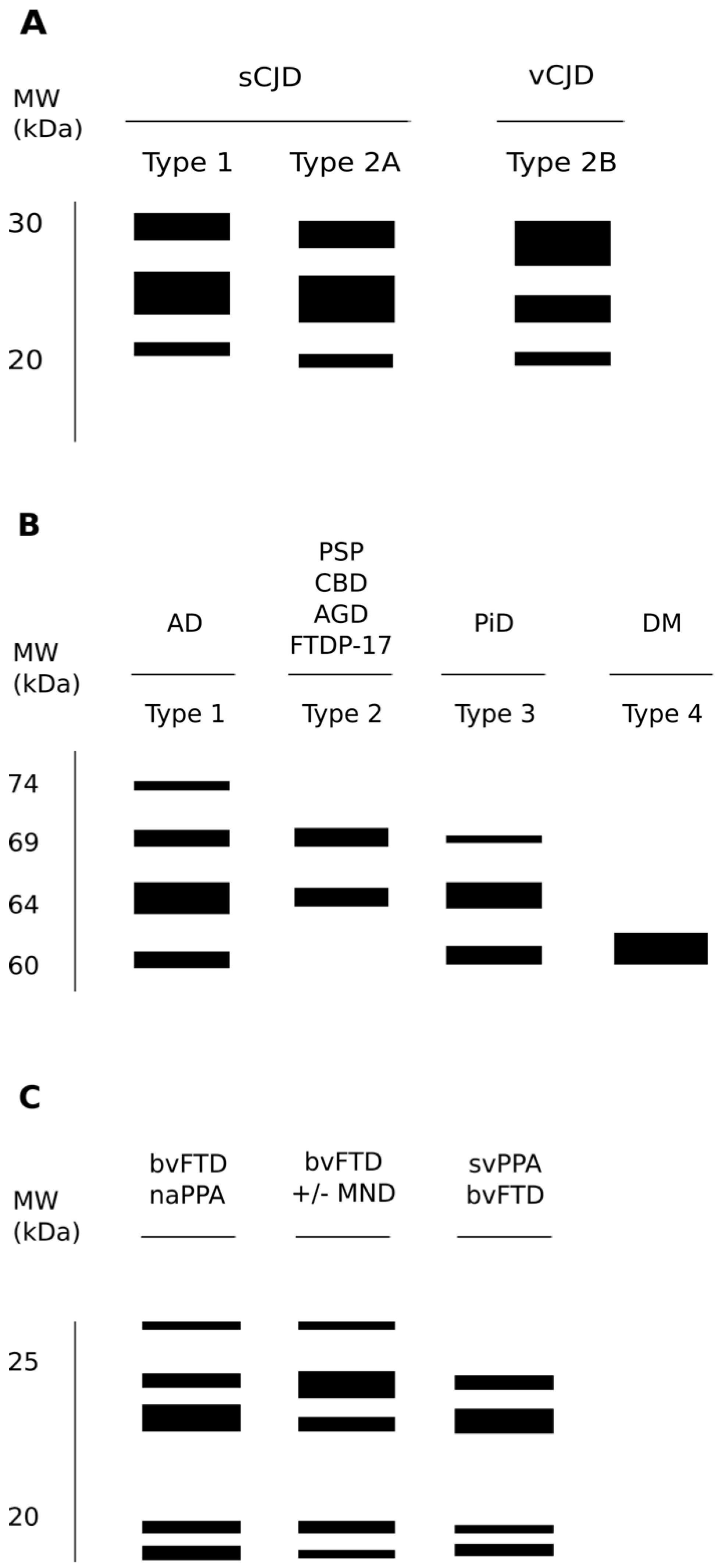
2.3. Strain Diversity in Human TSEs
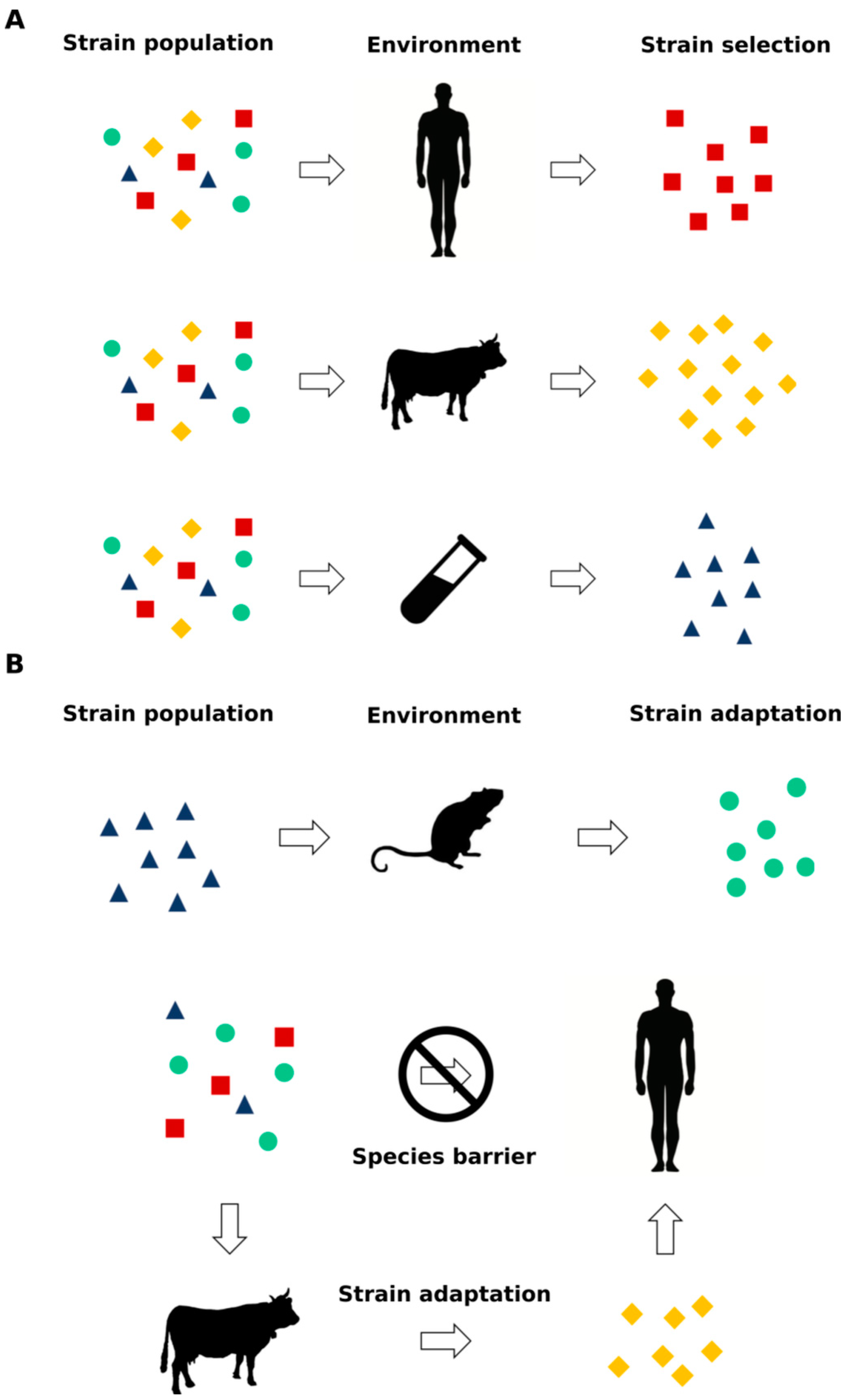
2.4. Species Barrier, Prion Strains Transmission, Selection and Adaptation
2.5. Synthetic Prion Strains
3. Amyloid-β
3.1. Amyloid-β Peptide Formation, Alzheimer’s Disease and Its Atypical Variants
3.2. Prion-Like Properties of Aβ
3.3. AD Heterogeneity: Aβ Strains
4. Tau
4.1. Tau Protein and Tauopathies
4.2. Prion-Like Properties of Tau
4.3. Tau Strains
5. Alpha-Synuclein
5.1. Alpha-Synuclein and Alpha-Synucleinopathies
5.2. Alpha-Synuclein Prion-Like Properties
5.3. Alpha-Synuclein Strains
6. TDP-43
6.1. TDP-43 in ALS and FTLD
6.2. The Prion-Like Properties of TDP-43
6.3. TDP-43 Strains
7. Conclusions
- (i)
- if prion and prion-like proteins are subjected to evolution, should we also expect an increase/evolution of misfolding-related disorders? In a recent work, it has been shown that vCJD infectious agent(s) contained in soluble or insoluble fractions of human vCJD blood donors was/were able to replicate in macaques generating typical and nonconventional vCJD phenotypes, the latter characterized by the presence of PK-sensitive PrPSc deposition and atypical clinical features mainly involving the spinal cord [93]. These results clearly suggest that prions show the ability to modify not only their structure but also their clinical manifestations after the infectious passage between human donors and nonhuman primates and that this “evolution” is dependent on the infectious source utilized.
- (ii)
- How accurate are the diagnostic tools that we are currently using to discriminate between different NDDs entities? In fact, if diagnostic molecules are not able to bind all the possible misfolded conformers, as it was shown for the PiB ligand for Aβ [24,25,148], it is not possible to reliably classify the disease. Such a lack of reliable diagnostic tests could reduce our accuracy in studying the neurodegenerative process and also the sensibility and specificity in selecting possible candidates for clinical studies and pharmacological trials.
- (iii)
- The same concept applies for therapeutic molecules. As indicated by Rasmussen et al., the discovery of multiple Aβ conformers associated with different AD clinical entities suggests the need for the development of drugs with multiple targets or for the use of a pool of polyclonal antibodies directed against the wider range of amyloid pathological structures [20].
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kovacs, G.G. Molecular Pathological Classification of Neurodegenerative Diseases: Turning towards Precision Medicine. Int. J. Mol. Sci. 2016, 17, 189. [Google Scholar] [CrossRef] [PubMed]
- Maniecka, Z.; Polymenidou, M. From nucleation to widespread propagation: A prion-like concept for ALS. Virus Res. 2015, 207, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.A.; Taubner, L.M.; Priola, S.A. Prion protein misfolding and disease. Curr. Opin. Struct. Biol. 2009, 19, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Smethurst, P.; Sidle, K.C.; Hardy, J. Review: Prion-like mechanisms of transactive response DNA binding protein of 43 kDa (TDP-43) in amyotrophic lateral sclerosis (ALS). Neuropathol. Appl. Neurobiol. 2015, 41, 578–597. [Google Scholar] [CrossRef] [PubMed]
- Holmes, B.B.; Diamond, M.I. Amyotrophic lateral sclerosis and organ donation: Is there risk of disease transmission? Ann. Neurol. 2012, 72, 832–836. [Google Scholar] [CrossRef] [PubMed]
- Jucker, M.; Walker, L.C. Pathogenic protein seeding in Alzheimer disease and other neurodegenerative disorders. Ann. Neurol. 2011, 70, 532–540. [Google Scholar] [CrossRef] [PubMed]
- Aguzzi, A. Cell biology: Beyond the prion principle. Nature 2009, 459, 924–925. [Google Scholar] [CrossRef]
- Prusiner, S.B. Biology and genetics of prions causing neurodegeneration. Annu. Rev. Genet. 2013, 47, 601–623. [Google Scholar] [CrossRef]
- Walker, L.C.; Jucker, M. Neurodegenerative diseases: Expanding the prion concept. Annu. Rev. Neurosci. 2015, 38, 87–103. [Google Scholar] [CrossRef]
- Prusiner, S.B. Novel proteinaceous infectious particles cause scrapie. Science 1982, 216, 136–144. [Google Scholar] [CrossRef]
- Prusiner, S.B. Prions. Proc. Natl. Acad. Sci. USA 1998, 95, 13363–13383. [Google Scholar] [CrossRef]
- Bistaffa, E.; Moda, F.; Virgilio, T.; Campagnani, I.; De Luca, C.M.G.; Rossi, M.; Salzano, G.; Giaccone, G.; Tagliavini, F.; Legname, G. Synthetic Prion Selection and Adaptation. Mol. Neurobiol. 2018. [Google Scholar] [CrossRef]
- Morales, R.; Abid, K.; Soto, C. The prion strain phenomenon: Molecular basis and unprecedented features. Biochim. Biophys. Acta 2007, 1772, 681–691. [Google Scholar] [CrossRef]
- Bartz, J.C.; Bessen, R.A.; McKenzie, D.; Marsh, R.F.; Aiken, J.M. Adaptation and selection of prion protein strain conformations following interspecies transmission of transmissible mink encephalopathy. J. Virol. 2000, 74, 5542–5547. [Google Scholar] [CrossRef]
- Colby, D.W.; Giles, K.; Legname, G.; Wille, H.; Baskakov, I.V.; DeArmond, S.J.; Prusiner, S.B. Design and construction of diverse mammalian prion strains. Proc. Natl. Acad. Sci. USA 2009, 106, 20417–20422. [Google Scholar] [CrossRef]
- Legname, G.; Baskakov, I.V.; Nguyen, H.O.; Riesner, D.; Cohen, F.E.; DeArmond, S.J.; Prusiner, S.B. Synthetic mammalian prions. Science 2004, 305, 673–676. [Google Scholar] [CrossRef]
- Peelaerts, W.; Bousset, L.; Baekelandt, V.; Melki, R. α-Synuclein strains and seeding in Parkinson’s disease, incidental Lewy body disease, dementia with Lewy bodies and multiple system atrophy: Similarities and differences. Cell Tissue Res. 2018, 373, 195–212. [Google Scholar] [CrossRef]
- Sanders, D.W.; Kaufman, S.K.; DeVos, S.L.; Sharma, A.M.; Mirbaha, H.; Li, A.; Barker, S.J.; Foley, A.C.; Thorpe, J.R.; Serpell, L.C.; et al. Distinct tau prion strains propagate in cells and mice and define different tauopathies. Neuron 2014, 82, 1271–1288. [Google Scholar] [CrossRef]
- Condello, C.; Lemmin, T.; Stohr, J.; Nick, M.; Wu, Y.; Maxwell, A.M.; Watts, J.C.; Caro, C.D.; Oehler, A.; Keene, C.D.; et al. Structural heterogeneity and intersubject variability of Abeta in familial and sporadic Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2018, 115, E782–E791. [Google Scholar] [CrossRef]
- Rasmussen, J.; Mahler, J.; Beschorner, N.; Kaeser, S.A.; Hasler, L.M.; Baumann, F.; Nystrom, S.; Portelius, E.; Blennow, K.; Lashley, T.; et al. Amyloid polymorphisms constitute distinct clouds of conformational variants in different etiological subtypes of Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2017, 114, 13018–13023. [Google Scholar] [CrossRef]
- Di Fede, G.; Catania, M.; Maderna, E.; Ghidoni, R.; Benussi, L.; Tonoli, E.; Giaccone, G.; Moda, F.; Paterlini, A.; Campagnani, I.; et al. Molecular subtypes of Alzheimer’s disease. Sci. Rep. 2018, 8, 3269. [Google Scholar] [CrossRef]
- Jack, C.R., Jr.; Lowe, V.J.; Senjem, M.L.; Weigand, S.D.; Kemp, B.J.; Shiung, M.M.; Knopman, D.S.; Boeve, B.F.; Klunk, W.E.; Mathis, C.A.; et al. 11C PiB and structural MRI provide complementary information in imaging of Alzheimer’s disease and amnestic mild cognitive impairment. Brain J. Neurol. 2008, 131, 665–680. [Google Scholar] [CrossRef]
- Kalimo, H.; Lalowski, M.; Bogdanovic, N.; Philipson, O.; Bird, T.D.; Nochlin, D.; Schellenberg, G.D.; Brundin, R.; Olofsson, T.; Soliymani, R.; et al. The Arctic AbetaPP mutation leads to Alzheimer’s disease pathology with highly variable topographic deposition of differentially truncated Abeta. Acta Neuropathol. Commun. 2013, 1, 60. [Google Scholar] [CrossRef]
- Scholl, M.; Wall, A.; Thordardottir, S.; Ferreira, D.; Bogdanovic, N.; Langstrom, B.; Almkvist, O.; Graff, C.; Nordberg, A. Low PiB PET retention in presence of pathologic CSF biomarkers in Arctic APP mutation carriers. Neurology 2012, 79, 229–236. [Google Scholar] [CrossRef]
- Tomiyama, T.; Nagata, T.; Shimada, H.; Teraoka, R.; Fukushima, A.; Kanemitsu, H.; Takuma, H.; Kuwano, R.; Imagawa, M.; Ataka, S.; et al. A new amyloid beta variant favoring oligomerization in Alzheimer’s-type dementia. Ann. Neurol. 2008, 63, 377–387. [Google Scholar] [CrossRef]
- Guo, J.L.; Covell, D.J.; Daniels, J.P.; Iba, M.; Stieber, A.; Zhang, B.; Riddle, D.M.; Kwong, L.K.; Xu, Y.; Trojanowski, J.Q.; et al. Distinct alpha-synuclein strains differentially promote tau inclusions in neurons. Cell 2013, 154, 103–117. [Google Scholar] [CrossRef]
- Peelaerts, W.; Bousset, L.; Van der Perren, A.; Moskalyuk, A.; Pulizzi, R.; Giugliano, M.; Van den Haute, C.; Melki, R.; Baekelandt, V. alpha-Synuclein strains cause distinct synucleinopathies after local and systemic administration. Nature 2015, 522, 340–344. [Google Scholar] [CrossRef]
- Prusiner, S.B.; Woerman, A.L.; Mordes, D.A.; Watts, J.C.; Rampersaud, R.; Berry, D.B.; Patel, S.; Oehler, A.; Lowe, J.K.; Kravitz, S.N.; et al. Evidence for alpha-synuclein prions causing multiple system atrophy in humans with parkinsonism. Proc. Natl. Acad. Sci. USA 2015, 112, E5308–E5317. [Google Scholar] [CrossRef]
- Holmqvist, S.; Chutna, O.; Bousset, L.; Aldrin-Kirk, P.; Li, W.; Bjorklund, T.; Wang, Z.Y.; Roybon, L.; Melki, R.; Li, J.Y. Direct evidence of Parkinson pathology spread from the gastrointestinal tract to the brain in rats. Acta Neuropathol. 2014, 128, 805–820. [Google Scholar] [CrossRef]
- Rey, N.L.; Petit, G.H.; Bousset, L.; Melki, R.; Brundin, P. Transfer of human alpha-synuclein from the olfactory bulb to interconnected brain regions in mice. Acta Neuropathol. 2013, 126, 555–573. [Google Scholar] [CrossRef]
- Sacino, A.N.; Brooks, M.; Thomas, M.A.; McKinney, A.B.; Lee, S.; Regenhardt, R.W.; McGarvey, N.H.; Ayers, J.I.; Notterpek, L.; Borchelt, D.R.; et al. Intramuscular injection of alpha-synuclein induces CNS alpha-synuclein pathology and a rapid-onset motor phenotype in transgenic mice. Proc. Natl. Acad. Sci. USA 2014, 111, 10732–10737. [Google Scholar] [CrossRef]
- Mao, X.; Ou, M.T.; Karuppagounder, S.S.; Kam, T.I.; Yin, X.; Xiong, Y.; Ge, P.; Umanah, G.E.; Brahmachari, S.; Shin, J.H.; et al. Pathological alpha-synuclein transmission initiated by binding lymphocyte-activation gene 3. Science 2016, 353, aah3374. [Google Scholar] [CrossRef]
- Aulic, S.; Masperone, L.; Narkiewicz, J.; Isopi, E.; Bistaffa, E.; Ambrosetti, E.; Pastore, B.; De Cecco, E.; Scaini, D.; Zago, P.; et al. alpha-Synuclein Amyloids Hijack Prion Protein to Gain Cell Entry, Facilitate Cell-to-Cell Spreading and Block Prion Replication. Sci. Rep. 2017, 7, 10050. [Google Scholar] [CrossRef]
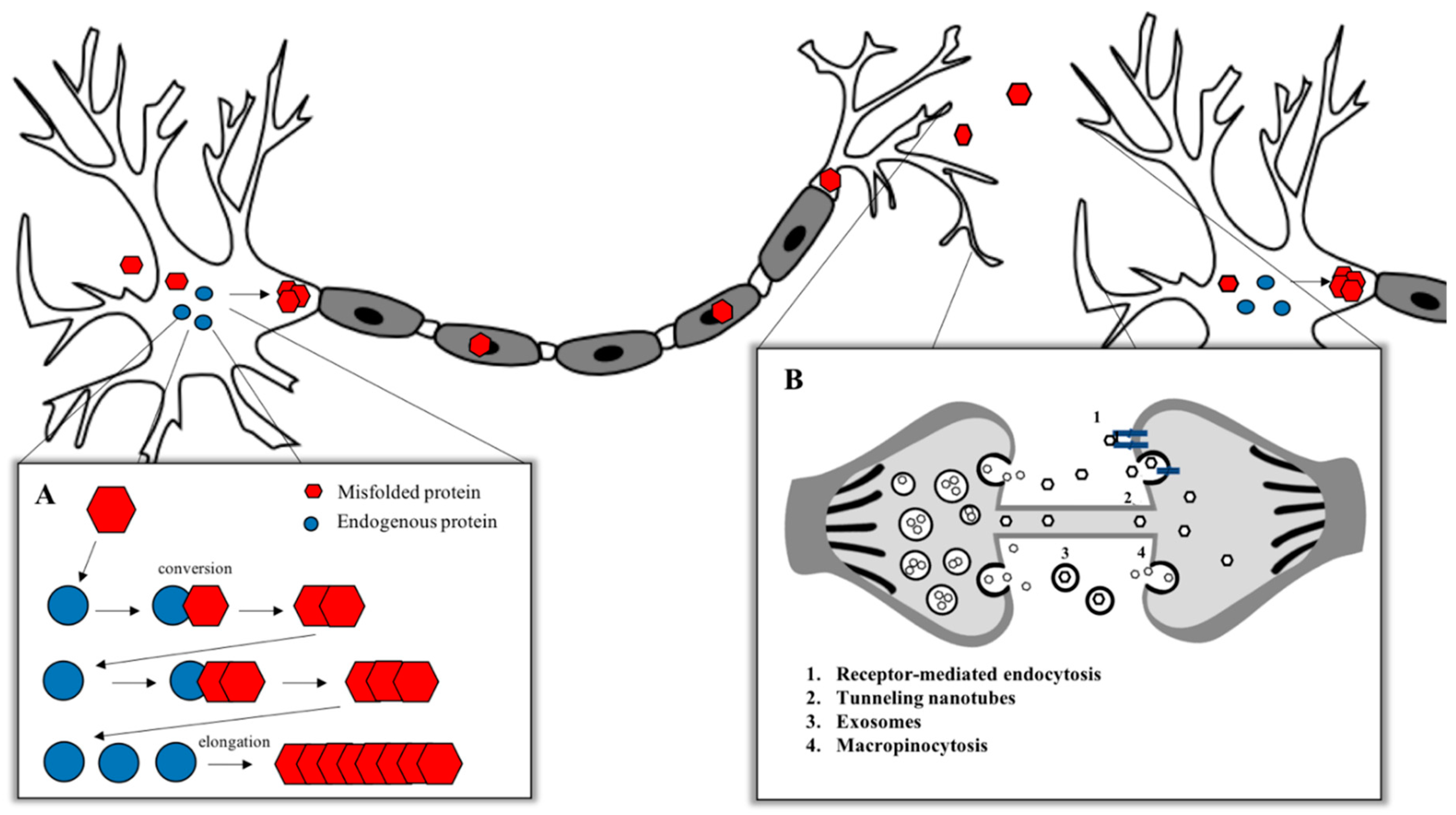
- Victoria, G.S.; Zurzolo, C. The spread of prion-like proteins by lysosomes and tunneling nanotubes: Implications for neurodegenerative diseases. J. Cell Biol. 2017, 216, 2633–2644. [Google Scholar] [CrossRef]
- Malm, T.; Loppi, S.; Kanninen, K.M. Exosomes in Alzheimer’s disease. Neurochem. Int. 2016, 97, 193–199. [Google Scholar] [CrossRef]
- Holmes, B.B.; DeVos, S.L.; Kfoury, N.; Li, M.; Jacks, R.; Yanamandra, K.; Ouidja, M.O.; Brodsky, F.M.; Marasa, J.; Bagchi, D.P.; et al. Heparan sulfate proteoglycans mediate internalization and propagation of specific proteopathic seeds. Proc. Natl. Acad. Sci. USA 2013, 110, E3138–E3147. [Google Scholar] [CrossRef]
- Aguzzi, A.; Miele, G. Recent advances in prion biology. Curr. Opin. Neurol. 2004, 17, 337–342. [Google Scholar] [CrossRef]
- Del Rio, J.A.; Ferrer, I.; Gavin, R. Role of cellular prion protein in interneuronal amyloid transmission. Prog. Neurobiol. 2018, 165–167, 87–102. [Google Scholar] [CrossRef]
- Del Rio, J.A.; Gavin, R. Functions of the cellular prion protein, the end of Moore’s law, and Ockham’s razor theory. Prion 2016, 10, 25–40. [Google Scholar] [CrossRef]
- Linden, R.; Martins, V.R.; Prado, M.A.; Cammarota, M.; Izquierdo, I.; Brentani, R.R. Physiology of the prion protein. Physiol. Rev. 2008, 88, 673–728. [Google Scholar] [CrossRef]
- Prusiner, S.B.; Scott, M.R.; DeArmond, S.J.; Cohen, F.E. Prion protein biology. Cell 1998, 93, 337–348. [Google Scholar] [CrossRef]
- Stahl, N.; Borchelt, D.R.; Hsiao, K.; Prusiner, S.B. Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell 1987, 51, 229–240. [Google Scholar] [CrossRef]
- Ford, M.J.; Burton, L.J.; Morris, R.J.; Hall, S.M. Selective expression of prion protein in peripheral tissues of the adult mouse. Neuroscience 2002, 113, 177–192. [Google Scholar] [CrossRef]
- Miele, G.; Alejo Blanco, A.R.; Baybutt, H.; Horvat, S.; Manson, J.; Clinton, M. Embryonic activation and developmental expression of the murine prion protein gene. Gene Expr. 2003, 11, 1–12. [Google Scholar] [CrossRef]
- Tichopad, A.; Pfaffl, M.W.; Didier, A. Tissue-specific expression pattern of bovine prion gene: Quantification using real-time RT-PCR. Mol. Cell Probes 2003, 17, 5–10. [Google Scholar] [CrossRef]
- Legname, G.; Moda, F. The Prion Concept and Synthetic Prions. Prog. Mol. Biol. Transl. Sci. 2017, 150, 147–156. [Google Scholar]
- Head, M.W.; Ironside, J.W. Review: Creutzfeldt-Jakob disease: Prion protein type, disease phenotype and agent strain. Neuropathol. Appl. Neurobiol. 2012, 38, 296–310. [Google Scholar] [CrossRef]
- Imran, M.; Mahmood, S. An overview of animal prion diseases. Virol. J. 2011, 8, 493. [Google Scholar] [CrossRef]
- Pattison, I.H.; Millson, G.C. Scrapie produced experimentally in goats with special reference to the clinical syndrome. J. Comp. Pathol. 1961, 71, 101–109. [Google Scholar] [CrossRef]
- Tenover, F.C.; Arbeit, R.D.; Goering, R.V.; Mickelsen, P.A.; Murray, B.E.; Persing, D.H.; Swaminathan, B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: Criteria for bacterial strain typing. J. Clin. Microbiol. 1995, 33, 2233–2239. [Google Scholar]
- Caughey, B.; Raymond, G.J.; Bessen, R.A. Strain-dependent differences in beta-sheet conformations of abnormal prion protein. J. Biol. Chem. 1998, 273, 32230–32235. [Google Scholar] [CrossRef]
- Safar, J.; Wille, H.; Itri, V.; Groth, D.; Serban, H.; Torchia, M.; Cohen, F.E.; Prusiner, S.B. Eight prion strains have PrP(Sc) molecules with different conformations. Nat. Med. 1998, 4, 1157–1165. [Google Scholar] [CrossRef]
- Wadsworth, J.D.; Hill, A.F.; Joiner, S.; Jackson, G.S.; Clarke, A.R.; Collinge, J. Strain-specific prion-protein conformation determined by metal ions. Nat. Cell Biol. 1999, 1, 55–59. [Google Scholar] [CrossRef]
- Aucouturier, P.; Kascsak, R.J.; Frangione, B.; Wisniewski, T. Biochemical and conformational variability of human prion strains in sporadic Creutzfeldt-Jakob disease. Neurosci. Lett. 1999, 274, 33–36. [Google Scholar] [CrossRef]
- Bellon, A.; Seyfert-Brandt, W.; Lang, W.; Baron, H.; Groner, A.; Vey, M. Improved conformation-dependent immunoassay: Suitability for human prion detection with enhanced sensitivity. J. Gen. Virol. 2003, 84, 1921–1925. [Google Scholar] [CrossRef]
- Jones, E.M.; Surewicz, W.K. Fibril conformation as the basis of species- and strain-dependent seeding specificity of mammalian prion amyloids. Cell 2005, 121, 63–72. [Google Scholar] [CrossRef]
- Bessen, R.A.; Marsh, R.F. Identification of two biologically distinct strains of transmissible mink encephalopathy in hamsters. J. Gen. Virol. 1992, 73 Pt 2, 329–334. [Google Scholar] [CrossRef]
- Bruce, M.E. Scrapie strain variation and mutation. Br. Med. Bull. 1993, 49, 822–838. [Google Scholar] [CrossRef]
- Fraser, H. Diversity in the neuropathology of scrapie-like diseases in animals. Br. Med. Bull. 1993, 49, 792–809. [Google Scholar] [CrossRef]
- Bessen, R.A.; Marsh, R.F. Biochemical and physical properties of the prion protein from two strains of the transmissible mink encephalopathy agent. J. Virol. 1992, 66, 2096–2101. [Google Scholar]
- Collinge, J.; Sidle, K.C.; Meads, J.; Ironside, J.; Hill, A.F. Molecular analysis of prion strain variation and the aetiology of ‘new variant’ CJD. Nature 1996, 383, 685–690. [Google Scholar] [CrossRef]
- Parchi, P.; Castellani, R.; Capellari, S.; Ghetti, B.; Young, K.; Chen, S.G.; Farlow, M.; Dickson, D.W.; Sima, A.A.; Trojanowski, J.Q.; et al. Molecular basis of phenotypic variability in sporadic Creutzfeldt-Jakob disease. Ann. Neurol. 1996, 39, 767–778. [Google Scholar] [CrossRef]
- Khalili-Shirazi, A.; Summers, L.; Linehan, J.; Mallinson, G.; Anstee, D.; Hawke, S.; Jackson, G.S.; Collinge, J. PrP glycoforms are associated in a strain-specific ratio in native PrPSc. J. Gen. Virol. 2005, 86, 2635–2644. [Google Scholar] [CrossRef]
- Bruce, M.E.; McBride, P.A.; Farquhar, C.F. Precise targeting of the pathology of the sialoglycoprotein, PrP, and vacuolar degeneration in mouse scrapie. Neurosci. Lett. 1989, 102, 1–6. [Google Scholar] [CrossRef]
- Bruce, M.E.; Boyle, A.; Cousens, S.; McConnell, I.; Foster, J.; Goldmann, W.; Fraser, H. Strain characterization of natural sheep scrapie and comparison with BSE. J. Gen. Virol. 2002, 83, 695–704. [Google Scholar] [CrossRef]
- Lasmezas, C.I.; Deslys, J.P.; Demaimay, R.; Adjou, K.T.; Hauw, J.J.; Dormont, D. Strain specific and common pathogenic events in murine models of scrapie and bovine spongiform encephalopathy. J. Gen. Virol. 1996, 77 Pt 7, 1601–1609. [Google Scholar] [CrossRef]
- Fraser, H.; Dickinson, A.G. Scrapie in mice. Agent-strain differences in the distribution and intensity of grey matter vacuolation. J. Comp. Pathol. 1973, 83, 29–40. [Google Scholar] [CrossRef]
- Brown, P.; Gibbs, C.J., Jr.; Rodgers-Johnson, P.; Asher, D.M.; Sulima, M.P.; Bacote, A.; Goldfarb, L.G.; Gajdusek, D.C. Human spongiform encephalopathy: The National Institutes of Health series of 300 cases of experimentally transmitted disease. Ann. Neurol. 1994, 35, 513–529. [Google Scholar] [CrossRef]
- Parchi, P.; Giese, A.; Capellari, S.; Brown, P.; Schulz-Schaeffer, W.; Windl, O.; Zerr, I.; Budka, H.; Kopp, N.; Piccardo, P.; et al. Classification of sporadic Creutzfeldt-Jakob disease based on molecular and phenotypic analysis of 300 subjects. Ann. Neurol. 1999, 46, 224–233. [Google Scholar] [CrossRef]
- Meissner, B.; Kallenberg, K.; Sanchez-Juan, P.; Collie, D.; Summers, D.M.; Almonti, S.; Collins, S.J.; Smith, P.; Cras, P.; Jansen, G.H.; et al. MRI lesion profiles in sporadic Creutzfeldt-Jakob disease. Neurology 2009, 72, 1994–2001. [Google Scholar] [CrossRef]
- Zerr, I.; Pocchiari, M.; Collins, S.; Brandel, J.P.; de Pedro Cuesta, J.; Knight, R.S.; Bernheimer, H.; Cardone, F.; Delasnerie-Laupretre, N.; Cuadrado Corrales, N.; et al. Analysis of EEG and CSF 14-3-3 proteins as aids to the diagnosis of Creutzfeldt-Jakob disease. Neurology 2000, 55, 811–815. [Google Scholar] [CrossRef]
- Mead, S.; Gandhi, S.; Beck, J.; Caine, D.; Gallujipali, D.; Carswell, C.; Hyare, H.; Joiner, S.; Ayling, H.; Lashley, T.; et al. A novel prion disease associated with diarrhea and autonomic neuropathy. N. Engl. J. Med. 2013, 369, 1904–1914. [Google Scholar] [CrossRef]
- Goldfarb, L.G.; Petersen, R.B.; Tabaton, M.; Brown, P.; LeBlanc, A.C.; Montagna, P.; Cortelli, P.; Julien, J.; Vital, C.; Pendelbury, W.W.; et al. Fatal familial insomnia and familial Creutzfeldt-Jakob disease: Disease phenotype determined by a DNA polymorphism. Science 1992, 258, 806–808. [Google Scholar] [CrossRef]
- Monari, L.; Chen, S.G.; Brown, P.; Parchi, P.; Petersen, R.B.; Mikol, J.; Gray, F.; Cortelli, P.; Montagna, P.; Ghetti, B.; et al. Fatal familial insomnia and familial Creutzfeldt-Jakob disease: Different prion proteins determined by a DNA polymorphism. Proc. Natl. Acad. Sci. USA 1994, 91, 2839–2842. [Google Scholar] [CrossRef]
- Lasmezas, C.I.; Deslys, J.P.; Robain, O.; Jaegly, A.; Beringue, V.; Peyrin, J.M.; Fournier, J.G.; Hauw, J.J.; Rossier, J.; Dormont, D. Transmission of the BSE agent to mice in the absence of detectable abnormal prion protein. Science 1997, 275, 402–405. [Google Scholar] [CrossRef]
- Manuelidis, E.E.; Gorgacz, E.J.; Manuelidis, L. Transmission of Creutzfeldt-Jakob disease with scrapie-like syndromes to mice. Nature 1978, 271, 778–779. [Google Scholar] [CrossRef]
- Muramoto, T.; Kitamoto, T.; Tateishi, J.; Goto, I. Successful transmission of Creutzfeldt-Jakob disease from human to mouse verified by prion protein accumulation in mouse brains. Brain Res. 1992, 599, 309–316. [Google Scholar] [CrossRef]
- Tateishi, J.; Sato, Y.; Nagara, H.; Boellaard, J.W. Experimental transmission of human subacute spongiform encephalopathy to small rodents. IV. Positive transmission from a typical case of Gerstmann-Straussler-Scheinker’s disease. Acta Neuropathol. 1984, 64, 85–88. [Google Scholar] [CrossRef]
- Bruce, M.; Chree, A.; McConnell, I.; Foster, J.; Pearson, G.; Fraser, H. Transmission of bovine spongiform encephalopathy and scrapie to mice: Strain variation and the species barrier. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1994, 343, 405–411. [Google Scholar]
- Fraser, H.; Bruce, M.E.; Chree, A.; McConnell, I.; Wells, G.A. Transmission of bovine spongiform encephalopathy and scrapie to mice. J. Gen. Virol. 1992, 73 Pt 8, 1891–1897. [Google Scholar] [CrossRef]
- Tateishi, J.; Ohta, M.; Koga, M.; Sato, Y.; Kuroiwa, Y. Transmission of chronic spongiform encephalopathy with kuru plaques from humans to small rodents. Ann. Neurol. 1979, 5, 581–584. [Google Scholar] [CrossRef]
- Moore, R.A.; Vorberg, I.; Priola, S.A. Species barriers in prion diseases—Brief review. In Infectious Diseases from Nature: Mechanisms of Viral Emergence and Persistence; Springer: Vienna, Austria, 2005; pp. 187–202. [Google Scholar]
- Peretz, D.; Williamson, R.A.; Legname, G.; Matsunaga, Y.; Vergara, J.; Burton, D.R.; DeArmond, S.J.; Prusiner, S.B.; Scott, M.R. A change in the conformation of prions accompanies the emergence of a new prion strain. Neuron 2002, 34, 921–932. [Google Scholar] [CrossRef]
- Chianini, F.; Fernandez-Borges, N.; Vidal, E.; Gibbard, L.; Pintado, B.; de Castro, J.; Priola, S.A.; Hamilton, S.; Eaton, S.L.; Finlayson, J.; et al. Rabbits are not resistant to prion infection. Proc. Natl. Acad. Sci. USA 2012, 109, 5080–5085. [Google Scholar] [CrossRef]
- Bruce, M.E.; Will, R.G.; Ironside, J.W.; McConnell, I.; Drummond, D.; Suttie, A.; McCardle, L.; Chree, A.; Hope, J.; Birkett, C.; et al. Transmissions to mice indicate that ‘new variant’ CJD is caused by the BSE agent. Nature 1997, 389, 498–501. [Google Scholar] [CrossRef]
- Hill, A.F.; Desbruslais, M.; Joiner, S.; Sidle, K.C.; Gowland, I.; Collinge, J.; Doey, L.J.; Lantos, P. The same prion strain causes vCJD and BSE. Nature 1997, 389, 448–450. [Google Scholar] [CrossRef]
- Ritchie, D.L.; Boyle, A.; McConnell, I.; Head, M.W.; Ironside, J.W.; Bruce, M.E. Transmissions of variant Creutzfeldt-Jakob disease from brain and lymphoreticular tissue show uniform and conserved bovine spongiform encephalopathy-related phenotypic properties on primary and secondary passage in wild-type mice. J. Gen. Virol. 2009, 90, 3075–3082. [Google Scholar] [CrossRef]
- Head, M.W.; Bunn, T.J.; Bishop, M.T.; McLoughlin, V.; Lowrie, S.; McKimmie, C.S.; Williams, M.C.; McCardle, L.; MacKenzie, J.; Knight, R.; et al. Prion protein heterogeneity in sporadic but not variant Creutzfeldt-Jakob disease: UK cases 1991–2002. Ann. Neurol. 2004, 55, 851–859. [Google Scholar] [CrossRef]
- Zeidler, M.; Johnstone, E.C.; Bamber, R.W.; Dickens, C.M.; Fisher, C.J.; Francis, A.F.; Goldbeck, R.; Higgo, R.; Johnson-Sabine, E.C.; Lodge, G.J.; et al. New variant Creutzfeldt-Jakob disease: Psychiatric features. Lancet 1997, 350, 908–910. [Google Scholar] [CrossRef]
- Mok, T.; Jaunmuktane, Z.; Joiner, S.; Campbell, T.; Morgan, C.; Wakerley, B.; Golestani, F.; Rudge, P.; Mead, S.; Jager, H.R.; et al. Variant Creutzfeldt-Jakob Disease in a Patient with Heterozygosity at PRNP Codon 129. N. Engl. J. Med. 2017, 376, 292–294. [Google Scholar] [CrossRef]
- Bishop, M.T.; Diack, A.B.; Ritchie, D.L.; Ironside, J.W.; Will, R.G.; Manson, J.C. Prion infectivity in the spleen of a PRNP heterozygous individual with subclinical variant Creutzfeldt-Jakob disease. Brain J. Neurol. 2013, 136, 1139–1145. [Google Scholar] [CrossRef]
- Peden, A.; McCardle, L.; Head, M.W.; Love, S.; Ward, H.J.; Cousens, S.N.; Keeling, D.M.; Millar, C.M.; Hill, F.G.; Ironside, J.W. Variant CJD infection in the spleen of a neurologically asymptomatic UK adult patient with haemophilia. Haemophilia 2010, 16, 296–304. [Google Scholar] [CrossRef]
- Comoy, E.E.; Mikol, J.; Jaffre, N.; Lebon, V.; Levavasseur, E.; Streichenberger, N.; Sumian, C.; Perret-Liaudet, A.; Eloit, M.; Andreoletti, O.; et al. Experimental transfusion of variant CJD-infected blood reveals previously uncharacterised prion disorder in mice and macaque. Nat. Commun. 2017, 8, 1268. [Google Scholar] [CrossRef]
- Bessen, R.A.; Marsh, R.F. Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J. Virol. 1994, 68, 7859–7868. [Google Scholar]
- Li, J.; Browning, S.; Mahal, S.P.; Oelschlegel, A.M.; Weissmann, C. Darwinian evolution of prions in cell culture. Science 2010, 327, 869–872. [Google Scholar] [CrossRef]
- Berry, D.B.; Lu, D.; Geva, M.; Watts, J.C.; Bhardwaj, S.; Oehler, A.; Renslo, A.R.; DeArmond, S.J.; Prusiner, S.B.; Giles, K. Drug resistance confounding prion therapeutics. Proc. Natl. Acad. Sci. USA 2013, 110, E4160–E4169. [Google Scholar] [CrossRef]
- Baskakov, I.V.; Aagaard, C.; Mehlhorn, I.; Wille, H.; Groth, D.; Baldwin, M.A.; Prusiner, S.B.; Cohen, F.E. Self-assembly of recombinant prion protein of 106 residues. Biochemistry 2000, 39, 2792–2804. [Google Scholar] [CrossRef]
- Baskakov, I.V.; Legname, G.; Baldwin, M.A.; Prusiner, S.B.; Cohen, F.E. Pathway complexity of prion protein assembly into amyloid. J. Biol. Chem. 2002, 277, 21140–21148. [Google Scholar] [CrossRef]
- Baskakov, I.V.; Legname, G.; Prusiner, S.B.; Cohen, F.E. Folding of prion protein to its native alpha-helical conformation is under kinetic control. J. Biol. Chem. 2001, 276, 19687–19690. [Google Scholar] [CrossRef]
- Hill, A.F.; Antoniou, M.; Collinge, J. Protease-resistant prion protein produced in vitro lacks detectable infectivity. J. Gen. Virol. 1999, 80 Pt 1, 11–14. [Google Scholar] [CrossRef]
- Hsiao, K.K.; Groth, D.; Scott, M.; Yang, S.L.; Serban, H.; Rapp, D.; Foster, D.; Torchia, M.; Dearmond, S.J.; Prusiner, S.B. Serial transmission in rodents of neurodegeneration from transgenic mice expressing mutant prion protein. Proc. Natl. Acad. Sci. USA 1994, 91, 9126–9130. [Google Scholar] [CrossRef]
- Kaneko, K.; Ball, H.L.; Wille, H.; Zhang, H.; Groth, D.; Torchia, M.; Tremblay, P.; Safar, J.; Prusiner, S.B.; DeArmond, S.J.; et al. A synthetic peptide initiates Gerstmann-Straussler-Scheinker (GSS) disease in transgenic mice. J. Mol. Biol. 2000, 295, 997–1007. [Google Scholar] [CrossRef]
- Tremblay, P.; Ball, H.L.; Kaneko, K.; Groth, D.; Hegde, R.S.; Cohen, F.E.; DeArmond, S.J.; Prusiner, S.B.; Safar, J.G. Mutant PrPSc conformers induced by a synthetic peptide and several prion strains. J. Virol. 2004, 78, 2088–2099. [Google Scholar] [CrossRef]
- Peretz, D.; Scott, M.R.; Groth, D.; Williamson, R.A.; Burton, D.R.; Cohen, F.E.; Prusiner, S.B. Strain-specified relative conformational stability of the scrapie prion protein. Protein Sci. 2001, 10, 854–863. [Google Scholar] [CrossRef]
- Kim, C.; Xiao, X.; Chen, S.; Haldiman, T.; Smirnovas, V.; Kofskey, D.; Warren, M.; Surewicz, K.; Maurer, N.R.; Kong, Q.; et al. Artificial strain of human prions created in vitro. Nat. Commun. 2018, 9, 2166. [Google Scholar] [CrossRef]
- Legname, G.; Nguyen, H.O.; Baskakov, I.V.; Cohen, F.E.; Dearmond, S.J.; Prusiner, S.B. Strain-specified characteristics of mouse synthetic prions. Proc. Natl. Acad. Sci. USA 2005, 102, 2168–2173. [Google Scholar] [CrossRef]
- Legname, G.; Nguyen, H.O.; Peretz, D.; Cohen, F.E.; DeArmond, S.J.; Prusiner, S.B. Continuum of prion protein structures enciphers a multitude of prion isolate-specified phenotypes. Proc. Natl. Acad. Sci. USA 2006, 103, 19105–19110. [Google Scholar] [CrossRef]
- Ghaemmaghami, S.; Watts, J.C.; Nguyen, H.O.; Hayashi, S.; DeArmond, S.J.; Prusiner, S.B. Conformational transformation and selection of synthetic prion strains. J. Mol. Biol. 2011, 413, 527–542. [Google Scholar] [CrossRef]
- Saborio, G.P.; Permanne, B.; Soto, C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature 2001, 411, 810–813. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, Y.; Wang, F.; Wang, X.; Xu, Y.; Yang, H.; Yu, G.; Yuan, C.; Ma, J. De novo generation of infectious prions with bacterially expressed recombinant prion protein. FASEB J. 2013, 27, 4768–4775. [Google Scholar] [CrossRef]
- Barria, M.A.; Telling, G.C.; Gambetti, P.; Mastrianni, J.A.; Soto, C. Generation of a new form of human PrP(Sc) in vitro by interspecies transmission from cervid prions. J. Biol. Chem. 2011, 286, 7490–7495. [Google Scholar] [CrossRef]
- Castilla, J.; Gonzalez-Romero, D.; Saa, P.; Morales, R.; De Castro, J.; Soto, C. Crossing the species barrier by PrP(Sc) replication in vitro generates unique infectious prions. Cell 2008, 134, 757–768. [Google Scholar] [CrossRef]
- Gonzalez-Montalban, N.; Lee, Y.J.; Makarava, N.; Savtchenko, R.; Baskakov, I.V. Changes in prion replication environment cause prion strain mutation. FASEB J. 2013, 27, 3702–3710. [Google Scholar] [CrossRef]
- Moda, F.; Le, T.N.; Aulic, S.; Bistaffa, E.; Campagnani, I.; Virgilio, T.; Indaco, A.; Palamara, L.; Andreoletti, O.; Tagliavini, F.; et al. Synthetic prions with novel strain-specified properties. PLoS Pathog. 2015, 11, e1005354. [Google Scholar] [CrossRef]
- Goedert, M.; Spillantini, M.G. A century of Alzheimer’s disease. Science 2006, 314, 777–781. [Google Scholar] [CrossRef]
- O’Brien, R.J.; Wong, P.C. Amyloid precursor protein processing and Alzheimer’s disease. Annu. Rev. Neurosci. 2011, 34, 185–204. [Google Scholar] [CrossRef]
- Bergmans, B.A.; De Strooper, B. gamma-secretases: From cell biology to therapeutic strategies. Lancet Neurol. 2010, 9, 215–226. [Google Scholar] [CrossRef]
- Shen, J.; Kelleher, R.J., 3rd. The presenilin hypothesis of Alzheimer’s disease: Evidence for a loss-of-function pathogenic mechanism. Proc. Natl. Acad. Sci. USA 2007, 104, 403–409. [Google Scholar] [CrossRef]
- Hof, P.R.; Bouras, C.; Perl, D.P.; Sparks, D.L.; Mehta, N.; Morrison, J.H. Age-related distribution of neuropathologic changes in the cerebral cortex of patients with Down’s syndrome. Quantitative regional analysis and comparison with Alzheimer’s disease. Arch. Neurol. 1995, 52, 379–391. [Google Scholar] [CrossRef]
- Karantzoulis, S.; Galvin, J.E. Distinguishing Alzheimer’s disease from other major forms of dementia. Expert Rev. Neurother. 2011, 11, 1579–1591. [Google Scholar] [CrossRef]
- Lam, B.; Masellis, M.; Freedman, M.; Stuss, D.T.; Black, S.E. Clinical, imaging, and pathological heterogeneity of the Alzheimer’s disease syndrome. Alzheimers Res. 2013, 5, 1. [Google Scholar] [CrossRef]
- Cohen, M.L.; Kim, C.; Haldiman, T.; ElHag, M.; Mehndiratta, P.; Pichet, T.; Lissemore, F.; Shea, M.; Cohen, Y.; Chen, W.; et al. Rapidly progressive Alzheimer’s disease features distinct structures of amyloid-beta. Brain J. Neurol. 2015, 138, 1009–1022. [Google Scholar] [CrossRef]
- Cohen, M.; Appleby, B.; Safar, J.G. Distinct prion-like strains of amyloid beta implicated in phenotypic diversity of Alzheimer’s disease. Prion 2016, 10, 9–17. [Google Scholar] [CrossRef]
- Goudsmit, J.; Morrow, C.H.; Asher, D.M.; Yanagihara, R.T.; Masters, C.L.; Gibbs, C.J., Jr.; Gajdusek, D.C. Evidence for and against the transmissibility of Alzheimer disease. Neurology 1980, 30, 945–950. [Google Scholar] [CrossRef]
- Baker, H.F.; Ridley, R.M.; Duchen, L.W.; Crow, T.J.; Bruton, C.J. Evidence for the experimental transmission of cerebral beta-amyloidosis to primates. Int. J. Exp. Pathol. 1993, 74, 441–454. [Google Scholar]
- Duran-Aniotz, C.; Morales, R.; Moreno-Gonzalez, I.; Hu, P.P.; Fedynyshyn, J.; Soto, C. Aggregate-depleted brain fails to induce Abeta deposition in a mouse model of Alzheimer’s disease. PLoS ONE 2014, 9, e89014. [Google Scholar] [CrossRef]
- Duran-Aniotz, C.; Morales, R.; Moreno-Gonzalez, I.; Hu, P.P.; Soto, C. Brains from non-Alzheimer’s individuals containing amyloid deposits accelerate Abeta deposition in vivo. Acta Neuropathol. Commun. 2013, 1, 76. [Google Scholar] [CrossRef]
- Hamaguchi, T.; Eisele, Y.S.; Varvel, N.H.; Lamb, B.T.; Walker, L.C.; Jucker, M. The presence of Abeta seeds, and not age per se, is critical to the initiation of Abeta deposition in the brain. Acta Neuropathol. 2012, 123, 31–37. [Google Scholar] [CrossRef]
- Heilbronner, G.; Eisele, Y.S.; Langer, F.; Kaeser, S.A.; Novotny, R.; Nagarathinam, A.; Aslund, A.; Hammarstrom, P.; Nilsson, K.P.; Jucker, M. Seeded strain-like transmission of beta-amyloid morphotypes in APP transgenic mice. EMBO Rep. 2013, 14, 1017–1022. [Google Scholar] [CrossRef]
- Kane, M.D.; Lipinski, W.J.; Callahan, M.J.; Bian, F.; Durham, R.A.; Schwarz, R.D.; Roher, A.E.; Walker, L.C. Evidence for seeding of beta -amyloid by intracerebral infusion of Alzheimer brain extracts in beta -amyloid precursor protein-transgenic mice. J. Neurosci. 2000, 20, 3606–3611. [Google Scholar] [CrossRef]
- Langer, F.; Eisele, Y.S.; Fritschi, S.K.; Staufenbiel, M.; Walker, L.C.; Jucker, M. Soluble Abeta seeds are potent inducers of cerebral beta-amyloid deposition. J. Neurosci. 2011, 31, 14488–14495. [Google Scholar] [CrossRef]
- Meyer-Luehmann, M.; Coomaraswamy, J.; Bolmont, T.; Kaeser, S.; Schaefer, C.; Kilger, E.; Neuenschwander, A.; Abramowski, D.; Frey, P.; Jaton, A.L.; et al. Exogenous induction of cerebral beta-amyloidogenesis is governed by agent and host. Science 2006, 313, 1781–1784. [Google Scholar] [CrossRef]
- Morales, R.; Duran-Aniotz, C.; Castilla, J.; Estrada, L.D.; Soto, C. De novo induction of amyloid-beta deposition in vivo. Mol. Psychiatry 2012, 17, 1347–1353. [Google Scholar] [CrossRef]
- Rosen, R.F.; Fritz, J.J.; Dooyema, J.; Cintron, A.F.; Hamaguchi, T.; Lah, J.J.; LeVine, H., 3rd; Jucker, M.; Walker, L.C. Exogenous seeding of cerebral beta-amyloid deposition in betaAPP-transgenic rats. J. Neurochem. 2012, 120, 660–666. [Google Scholar] [CrossRef]
- Walker, L.C.; Callahan, M.J.; Bian, F.; Durham, R.A.; Roher, A.E.; Lipinski, W.J. Exogenous induction of cerebral beta-amyloidosis in betaAPP-transgenic mice. Peptides 2002, 23, 1241–1247. [Google Scholar] [CrossRef]
- Ye, L.; Hamaguchi, T.; Fritschi, S.K.; Eisele, Y.S.; Obermuller, U.; Jucker, M.; Walker, L.C. Progression of Seed-Induced Abeta Deposition within the Limbic Connectome. Brain Pathol. 2015, 25, 743–752. [Google Scholar] [CrossRef]
- Fritschi, S.K.; Langer, F.; Kaeser, S.A.; Maia, L.F.; Portelius, E.; Pinotsi, D.; Kaminski, C.F.; Winkler, D.T.; Maetzler, W.; Keyvani, K.; et al. Highly potent soluble amyloid-beta seeds in human Alzheimer brain but not cerebrospinal fluid. Brain J. Neurol. 2014, 137, 2909–2915. [Google Scholar] [CrossRef]
- Eisele, Y.S.; Obermuller, U.; Heilbronner, G.; Baumann, F.; Kaeser, S.A.; Wolburg, H.; Walker, L.C.; Staufenbiel, M.; Heikenwalder, M.; Jucker, M. Peripherally applied Abeta-containing inoculates induce cerebral beta-amyloidosis. Science 2010, 330, 980–982. [Google Scholar] [CrossRef]
- Eisele, Y.S.; Fritschi, S.K.; Hamaguchi, T.; Obermuller, U.; Fuger, P.; Skodras, A.; Schafer, C.; Odenthal, J.; Heikenwalder, M.; Staufenbiel, M.; et al. Multiple factors contribute to the peripheral induction of cerebral beta-amyloidosis. J. Neurosci. 2014, 34, 10264–10273. [Google Scholar] [CrossRef]
- Eisele, Y.S.; Bolmont, T.; Heikenwalder, M.; Langer, F.; Jacobson, L.H.; Yan, Z.X.; Roth, K.; Aguzzi, A.; Staufenbiel, M.; Walker, L.C.; et al. Induction of cerebral beta-amyloidosis: Intracerebral versus systemic Abeta inoculation. Proc. Natl. Acad. Sci. USA 2009, 106, 12926–12931. [Google Scholar] [CrossRef]
- Stohr, J.; Watts, J.C.; Mensinger, Z.L.; Oehler, A.; Grillo, S.K.; DeArmond, S.J.; Prusiner, S.B.; Giles, K. Purified and synthetic Alzheimer’s amyloid beta (Abeta) prions. Proc. Natl. Acad. Sci. USA 2012, 109, 11025–11030. [Google Scholar] [CrossRef]
- Stohr, J.; Condello, C.; Watts, J.C.; Bloch, L.; Oehler, A.; Nick, M.; DeArmond, S.J.; Giles, K.; DeGrado, W.F.; Prusiner, S.B. Distinct synthetic Abeta prion strains producing different amyloid deposits in bigenic mice. Proc. Natl. Acad. Sci. USA 2014, 111, 10329–10334. [Google Scholar] [CrossRef]
- Purro, S.A.; Farrow, M.A.; Linehan, J.; Nazari, T.; Thomas, D.X.; Chen, Z.; Mengel, D.; Saito, T.; Saido, T.; Rudge, P.; et al. Transmission of amyloid-beta protein pathology from cadaveric pituitary growth hormone. Nature 2018, 564, 415–419. [Google Scholar] [CrossRef]
- Heuer, E.; Rosen, R.F.; Cintron, A.; Walker, L.C. Nonhuman primate models of Alzheimer-like cerebral proteopathy. Curr. Pharm. Des. 2012, 18, 1159–1169. [Google Scholar] [CrossRef]
- Jucker, M. The benefits and limitations of animal models for translational research in neurodegenerative diseases. Nat. Med. 2010, 16, 1210–1214. [Google Scholar] [CrossRef]
- Petkova, A.T.; Leapman, R.D.; Guo, Z.; Yau, W.M.; Mattson, M.P.; Tycko, R. Self-propagating, molecular-level polymorphism in Alzheimer’s beta-amyloid fibrils. Science 2005, 307, 262–265. [Google Scholar] [CrossRef]
- Qiang, W.; Yau, W.M.; Lu, J.X.; Collinge, J.; Tycko, R. Structural variation in amyloid-beta fibrils from Alzheimer’s disease clinical subtypes. Nature 2017, 541, 217–221. [Google Scholar] [CrossRef]
- Rosen, R.F.; Ciliax, B.J.; Wingo, T.S.; Gearing, M.; Dooyema, J.; Lah, J.J.; Ghiso, J.A.; LeVine, H., 3rd; Walker, L.C. Deficient high-affinity binding of Pittsburgh compound B in a case of Alzheimer’s disease. Acta Neuropathol. 2010, 119, 221–233. [Google Scholar] [CrossRef]
- Binder, L.I.; Frankfurter, A.; Rebhun, L.I. The distribution of tau in the mammalian central nervous system. J. Cell Biol. 1985, 101, 1371–1378. [Google Scholar] [CrossRef]
- Dugger, B.N.; Whiteside, C.M.; Maarouf, C.L.; Walker, D.G.; Beach, T.G.; Sue, L.I.; Garcia, A.; Dunckley, T.; Meechoovet, B.; Reiman, E.M.; et al. The Presence of Select Tau Species in Human Peripheral Tissues and Their Relation to Alzheimer’s Disease. J. Alzheimers Dis. 2016, 51, 345–356. [Google Scholar] [CrossRef]
- Kahlson, M.A.; Colodner, K.J. Glial Tau Pathology in Tauopathies: Functional Consequences. J. Exp. Neurosci. 2015, 9, 43–50. [Google Scholar] [CrossRef]
- Buee, L.; Bussiere, T.; Buee-Scherrer, V.; Delacourte, A.; Hof, P.R. Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain Res. Brain Res. Rev. 2000, 33, 95–130. [Google Scholar] [CrossRef]
- Andreadis, A.; Brown, W.M.; Kosik, K.S. Structure and novel exons of the human tau gene. Biochemistry 1992, 31, 10626–10633. [Google Scholar] [CrossRef]
- Goedert, M.; Spillantini, M.G.; Jakes, R.; Rutherford, D.; Crowther, R.A. Multiple isoforms of human microtubule-associated protein tau: Sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron 1989, 3, 519–526. [Google Scholar] [CrossRef]
- Liu, F.; Gong, C.X. Tau exon 10 alternative splicing and tauopathies. Mol. Neurodegener. 2008, 3, 8. [Google Scholar] [CrossRef]
- Avila, J.; Jimenez, J.S.; Sayas, C.L.; Bolos, M.; Zabala, J.C.; Rivas, G.; Hernandez, F. Tau Structures. Front. Aging Neurosci. 2016, 8, 262. [Google Scholar] [CrossRef]
- Leterrier, J.F.; Liem, R.K.; Shelanski, M.L. Interactions between neurofilaments and microtubule-associated proteins: A possible mechanism for intraorganellar bridging. J. Cell Biol. 1982, 95, 982–986. [Google Scholar] [CrossRef]
- Rendon, A.; Jung, D.; Jancsik, V. Interaction of microtubules and microtubule-associated proteins (MAPs) with rat brain mitochondria. Biochem. J. 1990, 269, 555–556. [Google Scholar] [CrossRef]
- Chen, J.; Kanai, Y.; Cowan, N.J.; Hirokawa, N. Projection domains of MAP2 and tau determine spacings between microtubules in dendrites and axons. Nature 1992, 360, 674–677. [Google Scholar] [CrossRef]
- Lee, G.; Newman, S.T.; Gard, D.L.; Band, H.; Panchamoorthy, G. Tau interacts with src-family non-receptor tyrosine kinases. J. Cell Sci. 1998, 111 Pt 21, 3167–3177. [Google Scholar]
- Hwang, S.C.; Jhon, D.Y.; Bae, Y.S.; Kim, J.H.; Rhee, S.G. Activation of phospholipase C-gamma by the concerted action of tau proteins and arachidonic acid. J. Biol. Chem. 1996, 271, 18342–18349. [Google Scholar] [CrossRef]
- Drechsel, D.N.; Hyman, A.A.; Cobb, M.H.; Kirschner, M.W. Modulation of the dynamic instability of tubulin assembly by the microtubule-associated protein tau. Mol. Biol. Cell 1992, 3, 1141–1154. [Google Scholar] [CrossRef]
- Correas, I.; Padilla, R.; Avila, J. The tubulin-binding sequence of brain microtubule-associated proteins, tau and MAP-2, is also involved in actin binding. Biochem. J. 1990, 269, 61–64. [Google Scholar] [CrossRef]
- Griffith, L.M.; Pollard, T.D. The interaction of actin filaments with microtubules and microtubule-associated proteins. J. Biol. Chem. 1982, 257, 9143–9151. [Google Scholar]
- Carlier, M.F.; Simon, C.; Cassoly, R.; Pradel, L.A. Interaction between microtubule-associated protein tau and spectrin. Biochimie 1984, 66, 305–311. [Google Scholar] [CrossRef]
- Martin, L.; Latypova, X.; Terro, F. Post-translational modifications of tau protein: Implications for Alzheimer’s disease. Neurochem. Int. 2011, 58, 458–471. [Google Scholar] [CrossRef]
- Drewes, G.; Trinczek, B.; Illenberger, S.; Biernat, J.; Schmitt-Ulms, G.; Meyer, H.E.; Mandelkow, E.M.; Mandelkow, E. Microtubule-associated protein/microtubule affinity-regulating kinase (p110mark). A novel protein kinase that regulates tau-microtubule interactions and dynamic instability by phosphorylation at the Alzheimer-specific site serine 262. J. Biol. Chem. 1995, 270, 7679–7688. [Google Scholar] [CrossRef]
- Eidenmuller, J.; Fath, T.; Maas, T.; Pool, M.; Sontag, E.; Brandt, R. Phosphorylation-mimicking glutamate clusters in the proline-rich region are sufficient to simulate the functional deficiencies of hyperphosphorylated tau protein. Biochem. J. 2001, 357, 759–767. [Google Scholar] [CrossRef]
- Liu, F.; Li, B.; Tung, E.J.; Grundke-Iqbal, I.; Iqbal, K.; Gong, C.X. Site-specific effects of tau phosphorylation on its microtubule assembly activity and self-aggregation. Eur. J. Neurosci. 2007, 26, 3429–3436. [Google Scholar] [CrossRef]
- Hoover, B.R.; Reed, M.N.; Su, J.; Penrod, R.D.; Kotilinek, L.A.; Grant, M.K.; Pitstick, R.; Carlson, G.A.; Lanier, L.M.; Yuan, L.L.; et al. Tau mislocalization to dendritic spines mediates synaptic dysfunction independently of neurodegeneration. Neuron 2010, 68, 1067–1081. [Google Scholar] [CrossRef]
- Dickey, C.A.; Kamal, A.; Lundgren, K.; Klosak, N.; Bailey, R.M.; Dunmore, J.; Ash, P.; Shoraka, S.; Zlatkovic, J.; Eckman, C.B.; et al. The high-affinity HSP90-CHIP complex recognizes and selectively degrades phosphorylated tau client proteins. J. Clin. Investig. 2007, 117, 648–658. [Google Scholar] [CrossRef]
- Sergeant, N.; Delacourte, A.; Buee, L. Tau protein as a differential biomarker of tauopathies. Biochim. Biophys. Acta 2005, 1739, 179–197. [Google Scholar] [CrossRef]
- Falcon, B.; Zhang, W.; Schweighauser, M.; Murzin, A.G.; Vidal, R.; Garringer, H.J.; Ghetti, B.; Scheres, S.H.W.; Goedert, M. Tau filaments from multiple cases of sporadic and inherited Alzheimer’s disease adopt a common fold. Acta Neuropathol. 2018, 136, 699–708. [Google Scholar] [CrossRef] [PubMed]
- Falcon, B.; Zhang, W.; Murzin, A.G.; Murshudov, G.; Garringer, H.J.; Vidal, R.; Crowther, R.A.; Ghetti, B.; Scheres, S.H.W.; Goedert, M. Structures of filaments from Pick’s disease reveal a novel tau protein fold. Nature 2018, 561, 137–140. [Google Scholar] [CrossRef] [PubMed]
- Clavaguera, F.; Bolmont, T.; Crowther, R.A.; Abramowski, D.; Frank, S.; Probst, A.; Fraser, G.; Stalder, A.K.; Beibel, M.; Staufenbiel, M.; et al. Transmission and spreading of tauopathy in transgenic mouse brain. Nat. Cell Biol. 2009, 11, 909–913. [Google Scholar] [CrossRef]
- Mirbaha, H.; Holmes, B.B.; Sanders, D.W.; Bieschke, J.; Diamond, M.I. Tau Trimers Are the Minimal Propagation Unit Spontaneously Internalized to Seed Intracellular Aggregation. J. Biol. Chem. 2015, 290, 14893–14903. [Google Scholar] [CrossRef]
- Mukrasch, M.D.; von Bergen, M.; Biernat, J.; Fischer, D.; Griesinger, C.; Mandelkow, E.; Zweckstetter, M. The “jaws” of the tau-microtubule interaction. J. Biol. Chem. 2007, 282, 12230–12239. [Google Scholar] [CrossRef]
- Clavaguera, F.; Akatsu, H.; Fraser, G.; Crowther, R.A.; Frank, S.; Hench, J.; Probst, A.; Winkler, D.T.; Reichwald, J.; Staufenbiel, M.; et al. Brain homogenates from human tauopathies induce tau inclusions in mouse brain. Proc. Natl. Acad. Sci. USA 2013, 110, 9535–9540. [Google Scholar] [CrossRef]
- Falcon, B.; Cavallini, A.; Angers, R.; Glover, S.; Murray, T.K.; Barnham, L.; Jackson, S.; O’Neill, M.J.; Isaacs, A.M.; Hutton, M.L.; et al. Conformation determines the seeding potencies of native and recombinant Tau aggregates. J. Biol. Chem. 2015, 290, 1049–1065. [Google Scholar] [CrossRef] [PubMed]
- Iba, M.; McBride, J.D.; Guo, J.L.; Zhang, B.; Trojanowski, J.Q.; Lee, V.M. Tau pathology spread in PS19 tau transgenic mice following locus coeruleus (LC) injections of synthetic tau fibrils is determined by the LC’s afferent and efferent connections. Acta Neuropathol. 2015, 130, 349–362. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, S.K.; Sanders, D.W.; Thomas, T.L.; Ruchinskas, A.J.; Vaquer-Alicea, J.; Sharma, A.M.; Miller, T.M.; Diamond, M.I. Tau Prion Strains Dictate Patterns of Cell Pathology, Progression Rate, and Regional Vulnerability In Vivo. Neuron 2016, 92, 796–812. [Google Scholar] [CrossRef] [PubMed]
- Rauch, J.N.; Chen, J.J.; Sorum, A.W.; Miller, G.M.; Sharf, T.; See, S.K.; Hsieh-Wilson, L.C.; Kampmann, M.; Kosik, K.S. Tau Internalization is Regulated by 6-O Sulfation on Heparan Sulfate Proteoglycans (HSPGs). Sci. Rep. 2018, 8, 6382. [Google Scholar] [CrossRef]
- Wu, J.W.; Herman, M.; Liu, L.; Simoes, S.; Acker, C.M.; Figueroa, H.; Steinberg, J.I.; Margittai, M.; Kayed, R.; Zurzolo, C.; et al. Small misfolded Tau species are internalized via bulk endocytosis and anterogradely and retrogradely transported in neurons. J. Biol. Chem. 2013, 288, 1856–1870. [Google Scholar] [CrossRef]
- Abounit, S.; Bousset, L.; Loria, F.; Zhu, S.; de Chaumont, F.; Pieri, L.; Olivo-Marin, J.C.; Melki, R.; Zurzolo, C. Tunneling nanotubes spread fibrillar alpha-synuclein by intercellular trafficking of lysosomes. EMBO J. 2016, 35, 2120–2138. [Google Scholar] [CrossRef]
- Tardivel, M.; Begard, S.; Bousset, L.; Dujardin, S.; Coens, A.; Melki, R.; Buee, L.; Colin, M. Tunneling nanotube (TNT)-mediated neuron-to neuron transfer of pathological Tau protein assemblies. Acta Neuropathol. Commun. 2016, 4, 117. [Google Scholar] [CrossRef]
- Ahmed, Z.; Cooper, J.; Murray, T.K.; Garn, K.; McNaughton, E.; Clarke, H.; Parhizkar, S.; Ward, M.A.; Cavallini, A.; Jackson, S.; et al. A novel in vivo model of tau propagation with rapid and progressive neurofibrillary tangle pathology: The pattern of spread is determined by connectivity, not proximity. Acta Neuropathol. 2014, 127, 667–683. [Google Scholar] [CrossRef]
- Dujardin, S.; Lecolle, K.; Caillierez, R.; Begard, S.; Zommer, N.; Lachaud, C.; Carrier, S.; Dufour, N.; Auregan, G.; Winderickx, J.; et al. Neuron-to-neuron wild-type Tau protein transfer through a trans-synaptic mechanism: Relevance to sporadic tauopathies. Acta Neuropathol. Commun. 2014, 2, 14. [Google Scholar] [CrossRef]
- Peeraer, E.; Bottelbergs, A.; Van Kolen, K.; Stancu, I.C.; Vasconcelos, B.; Mahieu, M.; Duytschaever, H.; Ver Donck, L.; Torremans, A.; Sluydts, E.; et al. Intracerebral injection of preformed synthetic tau fibrils initiates widespread tauopathy and neuronal loss in the brains of tau transgenic mice. Neurobiol. Dis. 2015, 73, 83–95. [Google Scholar] [CrossRef]
- De Calignon, A.; Polydoro, M.; Suarez-Calvet, M.; William, C.; Adamowicz, D.H.; Kopeikina, K.J.; Pitstick, R.; Sahara, N.; Ashe, K.H.; Carlson, G.A.; et al. Propagation of tau pathology in a model of early Alzheimer’s disease. Neuron 2012, 73, 685–697. [Google Scholar] [CrossRef]
- Liu, L.; Drouet, V.; Wu, J.W.; Witter, M.P.; Small, S.A.; Clelland, C.; Duff, K. Trans-synaptic spread of tau pathology in vivo. PLoS ONE 2012, 7, e31302. [Google Scholar] [CrossRef]
- Asai, H.; Ikezu, S.; Tsunoda, S.; Medalla, M.; Luebke, J.; Haydar, T.; Wolozin, B.; Butovsky, O.; Kugler, S.; Ikezu, T. Depletion of microglia and inhibition of exosome synthesis halt tau propagation. Nat. Neurosci. 2015, 18, 1584–1593. [Google Scholar] [CrossRef]
- Siddiqua, A.; Margittai, M. Three- and four-repeat Tau coassemble into heterogeneous filaments: An implication for Alzheimer disease. J. Biol. Chem. 2010, 285, 37920–37926. [Google Scholar] [CrossRef]
- Dinkel, P.D.; Siddiqua, A.; Huynh, H.; Shah, M.; Margittai, M. Variations in filament conformation dictate seeding barrier between three- and four-repeat tau. Biochemistry 2011, 50, 4330–4336. [Google Scholar] [CrossRef]
- Weismiller, H.A.; Murphy, R.; Wei, G.; Ma, B.; Nussinov, R.; Margittai, M. Structural disorder in four-repeat Tau fibrils reveals a new mechanism for barriers to cross-seeding of Tau isoforms. J. Biol. Chem. 2018, 293, 17336–17348. [Google Scholar] [CrossRef]
- Fitzpatrick, A.W.P.; Falcon, B.; He, S.; Murzin, A.G.; Murshudov, G.; Garringer, H.J.; Crowther, R.A.; Ghetti, B.; Goedert, M.; Scheres, S.H.W. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 2017, 547, 185–190. [Google Scholar] [CrossRef]
- Goedert, M.; Jakes, R. Mutations causing neurodegenerative tauopathies. Biochim. Biophys. Acta 2005, 1739, 240–250. [Google Scholar] [CrossRef]
- Frost, B.; Ollesch, J.; Wille, H.; Diamond, M.I. Conformational diversity of wild-type Tau fibrils specified by templated conformation change. J. Biol. Chem. 2009, 284, 3546–3551. [Google Scholar] [CrossRef]
- Von Bergen, M.; Friedhoff, P.; Biernat, J.; Heberle, J.; Mandelkow, E.M.; Mandelkow, E. Assembly of tau protein into Alzheimer paired helical filaments depends on a local sequence motif ((306)VQIVYK(311)) forming beta structure. Proc. Natl. Acad. Sci. USA 2000, 97, 5129–5134. [Google Scholar] [CrossRef]
- Aoyagi, H.; Hasegawa, M.; Tamaoka, A. Fibrillogenic nuclei composed of P301L mutant tau induce elongation of P301L tau but not wild-type tau. J. Biol. Chem. 2007, 282, 20309–20318. [Google Scholar] [CrossRef]
- Guo, J.L.; Narasimhan, S.; Changolkar, L.; He, Z.; Stieber, A.; Zhang, B.; Gathagan, R.J.; Iba, M.; McBride, J.D.; Trojanowski, J.Q.; et al. Unique pathological tau conformers from Alzheimer’s brains transmit tau pathology in nontransgenic mice. J. Exp. Med. 2016, 213, 2635–2654. [Google Scholar] [CrossRef]
- Saijo, E.; Ghetti, B.; Zanusso, G.; Oblak, A.; Furman, J.L.; Diamond, M.I.; Kraus, A.; Caughey, B. Ultrasensitive and selective detection of 3-repeat tau seeding activity in Pick disease brain and cerebrospinal fluid. Acta Neuropathol. 2017, 133, 751–765. [Google Scholar] [CrossRef]
- Narasimhan, S.; Guo, J.L.; Changolkar, L.; Stieber, A.; McBride, J.D.; Silva, L.V.; He, Z.; Zhang, B.; Gathagan, R.J.; Trojanowski, J.Q.; et al. Pathological Tau Strains from Human Brains Recapitulate the Diversity of Tauopathies in Nontransgenic Mouse Brain. J. Neurosci. Off. J. Soc. Neurosci. 2017, 37, 11406–11423. [Google Scholar] [CrossRef]
- Emamzadeh, F.N. Alpha-synuclein structure, functions, and interactions. J. Res. Med. Sci. 2016, 21, 29. [Google Scholar] [CrossRef]
- Sode, K.; Ochiai, S.; Kobayashi, N.; Usuzaka, E. Effect of reparation of repeat sequences in the human alpha-synuclein on fibrillation ability. Int. J. Biol. Sci. 2006, 3, 1–7. [Google Scholar]
- Giasson, B.I.; Murray, I.V.; Trojanowski, J.Q.; Lee, V.M. A hydrophobic stretch of 12 amino acid residues in the middle of alpha-synuclein is essential for filament assembly. J. Biol. Chem. 2001, 276, 2380–2386. [Google Scholar] [CrossRef]
- Bertoncini, C.W.; Jung, Y.S.; Fernandez, C.O.; Hoyer, W.; Griesinger, C.; Jovin, T.M.; Zweckstetter, M. Release of long-range tertiary interactions potentiates aggregation of natively unstructured alpha-synuclein. Proc. Natl. Acad. Sci. USA 2005, 102, 1430–1435. [Google Scholar] [CrossRef]
- Li, W.; West, N.; Colla, E.; Pletnikova, O.; Troncoso, J.C.; Marsh, L.; Dawson, T.M.; Jakala, P.; Hartmann, T.; Price, D.L.; et al. Aggregation promoting C-terminal truncation of alpha-synuclein is a normal cellular process and is enhanced by the familial Parkinson’s disease-linked mutations. Proc. Natl. Acad. Sci. USA 2005, 102, 2162–2167. [Google Scholar] [CrossRef]
- Kim, T.D.; Choi, E.; Rhim, H.; Paik, S.R.; Yang, C.H. Alpha-synuclein has structural and functional similarities to small heat shock proteins. Biochem. Biophys. Res. Commun. 2004, 324, 1352–1359. [Google Scholar] [CrossRef]
- Baldwin, A.J.; Knowles, T.P.; Tartaglia, G.G.; Fitzpatrick, A.W.; Devlin, G.L.; Shammas, S.L.; Waudby, C.A.; Mossuto, M.F.; Meehan, S.; Gras, S.L.; et al. Metastability of native proteins and the phenomenon of amyloid formation. J. Am. Chem. Soc. 2011, 133, 14160–14163. [Google Scholar] [CrossRef]
- Savica, R.; Grossardt, B.R.; Bower, J.H.; Ahlskog, J.E.; Rocca, W.A. Incidence and pathology of synucleinopathies and tauopathies related to parkinsonism. JAMA Neurol. 2013, 70, 859–866. [Google Scholar] [CrossRef]
- Marti, M.J.; Tolosa, E.; Campdelacreu, J. Clinical overview of the synucleinopathies. Mov. Disord. 2003, 18 (Suppl. 6), 21–27. [Google Scholar] [CrossRef]
- Beitz, J.M. Parkinson’s disease: A review. Front. Biosci. (Sch. Ed.) 2014, 6, 65–74. [Google Scholar] [CrossRef]
- Dickson, D.W. Parkinson’s disease and parkinsonism: Neuropathology. Cold Spring Harb. Perspect. Med. 2012, 2, a009258. [Google Scholar] [CrossRef] [PubMed]
- Donaghy, P.C.; McKeith, I.G. The clinical characteristics of dementia with Lewy bodies and a consideration of prodromal diagnosis. Alzheimers Res. 2014, 6, 46. [Google Scholar] [CrossRef] [PubMed]
- Stefanova, N.; Bucke, P.; Duerr, S.; Wenning, G.K. Multiple system atrophy: An update. Lancet Neurol. 2009, 8, 1172–1178. [Google Scholar] [CrossRef]
- Kordower, J.H.; Chu, Y.; Hauser, R.A.; Freeman, T.B.; Olanow, C.W. Lewy body-like pathology in long-term embryonic nigral transplants in Parkinson’s disease. Nat. Med. 2008, 14, 504–506. [Google Scholar] [CrossRef]
- Li, J.Y.; Englund, E.; Holton, J.L.; Soulet, D.; Hagell, P.; Lees, A.J.; Lashley, T.; Quinn, N.P.; Rehncrona, S.; Bjorklund, A.; et al. Lewy bodies in grafted neurons in subjects with Parkinson’s disease suggest host-to-graft disease propagation. Nat. Med. 2008, 14, 501–503. [Google Scholar] [CrossRef] [PubMed]
- Desplats, P.; Lee, H.J.; Bae, E.J.; Patrick, C.; Rockenstein, E.; Crews, L.; Spencer, B.; Masliah, E.; Lee, S.J. Inclusion formation and neuronal cell death through neuron-to-neuron transmission of alpha-synuclein. Proc. Natl. Acad. Sci. USA 2009, 106, 13010–13015. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.C.; Giles, K.; Oehler, A.; Middleton, L.; Dexter, D.T.; Gentleman, S.M.; DeArmond, S.J.; Prusiner, S.B. Transmission of multiple system atrophy prions to transgenic mice. Proc. Natl. Acad. Sci. USA 2013, 110, 19555–19560. [Google Scholar] [CrossRef]
- Luk, K.C.; Kehm, V.; Carroll, J.; Zhang, B.; O’Brien, P.; Trojanowski, J.Q.; Lee, V.M. Pathological alpha-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science 2012, 338, 949–953. [Google Scholar] [CrossRef]
- Luk, K.C.; Kehm, V.M.; Zhang, B.; O’Brien, P.; Trojanowski, J.Q.; Lee, V.M. Intracerebral inoculation of pathological alpha-synuclein initiates a rapidly progressive neurodegenerative alpha-synucleinopathy in mice. J. Exp. Med. 2012, 209, 975–986. [Google Scholar] [CrossRef]
- Luk, K.C.; Song, C.; O’Brien, P.; Stieber, A.; Branch, J.R.; Brunden, K.R.; Trojanowski, J.Q.; Lee, V.M. Exogenous alpha-synuclein fibrils seed the formation of Lewy body-like intracellular inclusions in cultured cells. Proc. Natl. Acad. Sci. USA 2009, 106, 20051–20056. [Google Scholar] [CrossRef] [PubMed]
- Masuda-Suzukake, M.; Nonaka, T.; Hosokawa, M.; Oikawa, T.; Arai, T.; Akiyama, H.; Mann, D.M.; Hasegawa, M. Prion-like spreading of pathological alpha-synuclein in brain. Brain J. Neurol. 2013, 136, 1128–1138. [Google Scholar] [CrossRef] [PubMed]
- Sacino, A.N.; Brooks, M.; McGarvey, N.H.; McKinney, A.B.; Thomas, M.A.; Levites, Y.; Ran, Y.; Golde, T.E.; Giasson, B.I. Induction of CNS alpha-synuclein pathology by fibrillar and non-amyloidogenic recombinant alpha-synuclein. Acta Neuropathol. Commun. 2013, 1, 38. [Google Scholar] [CrossRef]
- Volpicelli-Daley, L.A.; Luk, K.C.; Patel, T.P.; Tanik, S.A.; Riddle, D.M.; Stieber, A.; Meaney, D.F.; Trojanowski, J.Q.; Lee, V.M. Exogenous alpha-synuclein fibrils induce Lewy body pathology leading to synaptic dysfunction and neuron death. Neuron 2011, 72, 57–71. [Google Scholar] [CrossRef]
- Aulic, S.; Le, T.T.; Moda, F.; Abounit, S.; Corvaglia, S.; Casalis, L.; Gustincich, S.; Zurzolo, C.; Tagliavini, F.; Legname, G. Defined alpha-synuclein prion-like molecular assemblies spreading in cell culture. BMC Neurosci. 2014, 15, 69. [Google Scholar] [CrossRef] [PubMed]
- Recasens, A.; Dehay, B.; Bove, J.; Carballo-Carbajal, I.; Dovero, S.; Perez-Villalba, A.; Fernagut, P.O.; Blesa, J.; Parent, A.; Perier, C.; et al. Lewy body extracts from Parkinson disease brains trigger alpha-synuclein pathology and neurodegeneration in mice and monkeys. Ann. Neurol. 2014, 75, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Brundin, P.; Melki, R. Prying into the Prion Hypothesis for Parkinson’s Disease. J. Neurosci. 2017, 37, 9808–9818. [Google Scholar] [CrossRef] [PubMed]
- Aguzzi, A.; Lakkaraju, A.K.K. Cell Biology of Prions and Prionoids: A Status Report. Trends Cell Biol. 2016, 26, 40–51. [Google Scholar] [CrossRef]
- Tamguney, G.; Korczyn, A.D. A critical review of the prion hypothesis of human synucleinopathies. Cell Tissue Res. 2018, 373, 213–220. [Google Scholar] [CrossRef]
- Nonaka, T.; Watanabe, S.T.; Iwatsubo, T.; Hasegawa, M. Seeded aggregation and toxicity of α-synuclein and tau: Cellular models of neurodegenerative diseases. J. Biol. Chem. 2010, 285, 34885–34898. [Google Scholar] [CrossRef]
- Bousset, L.; Pieri, L.; Ruiz-Arlandis, G.; Gath, J.; Jensen, P.H.; Habenstein, B.; Madiona, K.; Olieric, V.; Bockmann, A.; Meier, B.H.; et al. Structural and functional characterization of two alpha-synuclein strains. Nat. Commun. 2013, 4, 2575. [Google Scholar] [CrossRef]
- Fujiwara, H.; Hasegawa, M.; Dohmae, N.; Kawashima, A.; Masliah, E.; Goldberg, M.S.; Shen, J.; Takio, K.; Iwatsubo, T. alpha-Synuclein is phosphorylated in synucleinopathy lesions. Nat. Cell Biol. 2002, 4, 160–164. [Google Scholar] [CrossRef]
- Ma, M.R.; Hu, Z.W.; Zhao, Y.F.; Chen, Y.X.; Li, Y.M. Phosphorylation induces distinct alpha-synuclein strain formation. Sci. Rep. 2016, 6, 37130. [Google Scholar] [CrossRef]
- Peng, C.; Gathagan, R.J.; Covell, D.J.; Medellin, C.; Stieber, A.; Robinson, J.L.; Zhang, B.; Pitkin, R.M.; Olufemi, M.F.; Luk, K.C.; et al. Cellular milieu imparts distinct pathological alpha-synuclein strains in alpha-synucleinopathies. Nature 2018, 557, 558–563. [Google Scholar] [CrossRef]
- Arai, T.; Hasegawa, M.; Akiyama, H.; Ikeda, K.; Nonaka, T.; Mori, H.; Mann, D.; Tsuchiya, K.; Yoshida, M.; Hashizume, Y.; et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 2006, 351, 602–611. [Google Scholar] [CrossRef]
- Neumann, M.; Sampathu, D.M.; Kwong, L.K.; Truax, A.C.; Micsenyi, M.C.; Chou, T.T.; Bruce, J.; Schuck, T.; Grossman, M.; Clark, C.M.; et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006, 314, 130–133. [Google Scholar] [CrossRef]
- Hasegawa, M.; Arai, T.; Nonaka, T.; Kametani, F.; Yoshida, M.; Hashizume, Y.; Beach, T.G.; Buratti, E.; Baralle, F.; Morita, M.; et al. Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Ann. Neurol. 2008, 64, 60–70. [Google Scholar] [CrossRef]
- Gregory, R.I.; Yan, K.P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240. [Google Scholar] [CrossRef]
- Sreedharan, J.; Blair, I.P.; Tripathi, V.B.; Hu, X.; Vance, C.; Rogelj, B.; Ackerley, S.; Durnall, J.C.; Williams, K.L.; Buratti, E.; et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 2008, 319, 1668–1672. [Google Scholar] [CrossRef]
- Lagier-Tourenne, C.; Polymenidou, M.; Cleveland, D.W. TDP-43 and FUS/TLS: Emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet. 2010, 19, R46–R64. [Google Scholar] [CrossRef]
- Casafont, I.; Bengoechea, R.; Tapia, O.; Berciano, M.T.; Lafarga, M. TDP-43 localizes in mRNA transcription and processing sites in mammalian neurons. J. Struct. Biol. 2009, 167, 235–241. [Google Scholar] [CrossRef]
- Wu, L.S.; Cheng, W.C.; Hou, S.C.; Yan, Y.T.; Jiang, S.T.; Shen, C.K. TDP-43, a neuro-pathosignature factor, is essential for early mouse embryogenesis. Genesis 2010, 48, 56–62. [Google Scholar] [CrossRef]
- Kraemer, B.C.; Schuck, T.; Wheeler, J.M.; Robinson, L.C.; Trojanowski, J.Q.; Lee, V.M.; Schellenberg, G.D. Loss of murine TDP-43 disrupts motor function and plays an essential role in embryogenesis. Acta Neuropathol. 2010, 119, 409–419. [Google Scholar] [CrossRef]
- Sephton, C.F.; Good, S.K.; Atkin, S.; Dewey, C.M.; Mayer, P., 3rd; Herz, J.; Yu, G. TDP-43 is a developmentally regulated protein essential for early embryonic development. J. Biol. Chem. 2010, 285, 6826–6834. [Google Scholar] [CrossRef]
- Nonaka, T.; Hasegawa, M. TDP-43 Prions. Cold Spring Harb. Perspect. Med. 2018, 8, a024463. [Google Scholar] [CrossRef]
- Daoud, H.; Valdmanis, P.N.; Kabashi, E.; Dion, P.; Dupre, N.; Camu, W.; Meininger, V.; Rouleau, G.A. Contribution of TARDBP mutations to sporadic amyotrophic lateral sclerosis. J. Med. Genet. 2009, 46, 112–114. [Google Scholar] [CrossRef]
- Kabashi, E.; Valdmanis, P.N.; Dion, P.; Spiegelman, D.; McConkey, B.J.; Vande Velde, C.; Bouchard, J.P.; Lacomblez, L.; Pochigaeva, K.; Salachas, F.; et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet. 2008, 40, 572–574. [Google Scholar] [CrossRef]
- Rutherford, N.J.; Zhang, Y.J.; Baker, M.; Gass, J.M.; Finch, N.A.; Xu, Y.F.; Stewart, H.; Kelley, B.J.; Kuntz, K.; Crook, R.J.; et al. Novel mutations in TARDBP (TDP-43) in patients with familial amyotrophic lateral sclerosis. PLoS Genet. 2008, 4, e1000193. [Google Scholar] [CrossRef]
- Tsermentseli, S.; Leigh, P.N.; Goldstein, L.H. The anatomy of cognitive impairment in amyotrophic lateral sclerosis: More than frontal lobe dysfunction. Cortex 2012, 48, 166–182. [Google Scholar] [CrossRef]
- Mackenzie, I.R.; Neumann, M.; Baborie, A.; Sampathu, D.M.; Du Plessis, D.; Jaros, E.; Perry, R.H.; Trojanowski, J.Q.; Mann, D.M.; Lee, V.M. A harmonized classification system for FTLD-TDP pathology. Acta Neuropathol. 2011, 122, 111–113. [Google Scholar] [CrossRef]
- Lee, E.B.; Porta, S.; Michael Baer, G.; Xu, Y.; Suh, E.; Kwong, L.K.; Elman, L.; Grossman, M.; Lee, V.M.; Irwin, D.J.; et al. Expansion of the classification of FTLD-TDP: Distinct pathology associated with rapidly progressive frontotemporal degeneration. Acta Neuropathol. 2017, 134, 65–78. [Google Scholar] [CrossRef]
- Furukawa, Y.; Kaneko, K.; Watanabe, S.; Yamanaka, K.; Nukina, N. A seeding reaction recapitulates intracellular formation of Sarkosyl-insoluble transactivation response element (TAR) DNA-binding protein-43 inclusions. J. Biol. Chem. 2011, 286, 18664–18672. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.L.; Che, M.X.; Zhao, J.; Zhou, C.J.; Xie, M.Y.; Li, H.Y.; He, J.H.; Hu, H.Y. Structural transformation of the amyloidogenic core region of TDP-43 protein initiates its aggregation and cytoplasmic inclusion. J. Biol. Chem. 2013, 288, 19614–19624. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.S.; Snead, D.; Lee, J.J.; McCaffery, J.M.; Shorter, J.; Gitler, A.D. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J. Biol. Chem. 2009, 284, 20329–20339. [Google Scholar] [CrossRef] [PubMed]
- Porta, S.; Xu, Y.; Restrepo, C.R.; Kwong, L.K.; Zhang, B.; Brown, H.J.; Lee, E.B.; Trojanowski, J.Q.; Lee, V.M. Patient-derived frontotemporal lobar degeneration brain extracts induce formation and spreading of TDP-43 pathology in vivo. Nat. Commun. 2018, 9, 4220. [Google Scholar] [CrossRef] [PubMed]
- Kwong, L.K.; Uryu, K.; Trojanowski, J.Q.; Lee, V.M. TDP-43 proteinopathies: Neurodegenerative protein misfolding diseases without amyloidosis. Neurosignals 2008, 16, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Neumann, M.; Kwong, L.K.; Sampathu, D.M.; Trojanowski, J.Q.; Lee, V.M. TDP-43 proteinopathy in frontotemporal lobar degeneration and amyotrophic lateral sclerosis: Protein misfolding diseases without amyloidosis. Arch. Neurol. 2007, 64, 1388–1394. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.L.; Geser, F.; Stieber, A.; Umoh, M.; Kwong, L.K.; Van Deerlin, V.M.; Lee, V.M.; Trojanowski, J.Q. TDP-43 skeins show properties of amyloid in a subset of ALS cases. Acta Neuropathol. 2013, 125, 121–131. [Google Scholar] [CrossRef]
- Bigio, E.H.; Wu, J.Y.; Deng, H.X.; Bit-Ivan, E.N.; Mao, Q.; Ganti, R.; Peterson, M.; Siddique, N.; Geula, C.; Siddique, T.; et al. Inclusions in frontotemporal lobar degeneration with TDP-43 proteinopathy (FTLD-TDP) and amyotrophic lateral sclerosis (ALS), but not FTLD with FUS proteinopathy (FTLD-FUS), have properties of amyloid. Acta Neuropathol. 2013, 125, 463–465. [Google Scholar] [CrossRef]
- Nonaka, T.; Masuda-Suzukake, M.; Arai, T.; Hasegawa, Y.; Akatsu, H.; Obi, T.; Yoshida, M.; Murayama, S.; Mann, D.M.; Akiyama, H.; et al. Prion-like properties of pathological TDP-43 aggregates from diseased brains. Cell Rep. 2013, 4, 124–134. [Google Scholar] [CrossRef]
- Smethurst, P.; Newcombe, J.; Troakes, C.; Simone, R.; Chen, Y.R.; Patani, R.; Sidle, K. In vitro prion-like behaviour of TDP-43 in ALS. Neurobiol. Dis. 2016, 96, 236–247. [Google Scholar] [CrossRef]
- Strong, M.J.; Yang, W. The frontotemporal syndromes of ALS. Clinicopathological correlates. J. Mol. Neurosci. 2011, 45, 648–655. [Google Scholar] [CrossRef]
- Shimonaka, S.; Nonaka, T.; Suzuki, G.; Hisanaga, S.; Hasegawa, M. Templated Aggregation of TAR DNA-binding Protein of 43 kDa (TDP-43) by Seeding with TDP-43 Peptide Fibrils. J. Biol. Chem. 2016, 291, 8896–8907. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scialò, C.; De Cecco, E.; Manganotti, P.; Legname, G. Prion and Prion-Like Protein Strains: Deciphering the Molecular Basis of Heterogeneity in Neurodegeneration. Viruses 2019, 11, 261. https://doi.org/10.3390/v11030261
Scialò C, De Cecco E, Manganotti P, Legname G. Prion and Prion-Like Protein Strains: Deciphering the Molecular Basis of Heterogeneity in Neurodegeneration. Viruses. 2019; 11(3):261. https://doi.org/10.3390/v11030261
Chicago/Turabian StyleScialò, Carlo, Elena De Cecco, Paolo Manganotti, and Giuseppe Legname. 2019. "Prion and Prion-Like Protein Strains: Deciphering the Molecular Basis of Heterogeneity in Neurodegeneration" Viruses 11, no. 3: 261. https://doi.org/10.3390/v11030261
APA StyleScialò, C., De Cecco, E., Manganotti, P., & Legname, G. (2019). Prion and Prion-Like Protein Strains: Deciphering the Molecular Basis of Heterogeneity in Neurodegeneration. Viruses, 11(3), 261. https://doi.org/10.3390/v11030261





