Abstract
Varroa destructor is an ectoparasitic mite of Asian or Eastern honeybees Apis cerana (A. cerana) which has become a serious threat to European subspecies of Western honeybees Apis mellifera (A. mellifera) within the last century. V. destructor and its vectored honeybee viruses became serious threats for colony survival. This is a short period for pathogen- and host-populations to adapt. To look for possible variation in the composition of viral populations we performed RNA metagenomic analysis of the Western honeybee subspecies A. m. ligustica, A. m. syriaca, A. m. intermissa, and A. cerana and their respective V. destructor mites. The analysis revealed two novel viruses: Varroa orthomyxovirus-1 (VOV-1) in A. mellifera and V. destructor and a Hubei like-virga virus-14 homolog in V. destructor. VOV-1 was more prevalent in V. destructor than in A. mellifera and we found evidence for viral replication in both hosts. Interestingly, we found differences in viral loads of A. cerana and their V. destructor, A. m. intermissa, and its V. destructor showed partial similarity, while A. m. ligustica and A. m. syriaca and their varroa where very similar. Deformed wing virus exhibited 82.20%, 99.20%, 97.90%, and 0.76% of total viral reads in A. m. ligustica, A. m. syriaca, A. m. intermissa, and A. cerana, respectively. This is the first report of a complete segmented-single-stranded negative-sense RNA virus genome in honeybees and V. destructor mites.
1. Introduction
The mite Varroa destructor is an obligatory ectoparasite of the Eastern honeybee Apis cerana [1]. V. destructor mites spend most of their life cycle inside the colony, reproducing on the honeybee brood and feeding on the pupa. Mites transfer from one host pupa to another on nurse bees that take care of pupa [1]. During the feeding on its host this mite may transfer populations of microorganisms that it bears, and viruses in particular, as well as acquire those that belong to the host [2,3,4,5]. V. destructor made a host shift to the Western honeybee A. mellifera at the beginning of the 20th century and spread to Europe, USA, New Zealand, Africa, and the Middle East from southern and southeastern Asia during the last century [6].
Viruses are important pathogens of the honeybee. Different studies reported that Acute bee paralysis virus (ABPV), Deformed wing virus (DWV), Israel acute paralysis virus (IAPV), Kashmir bee virus (KBV), Sacbrood virus (SBV), Slow bee paralysis virus (SBPV), Chronic bee paralysis virus (CBPV), Lake Sinai virus (LSV), Apis mellifera filamentous virus (AmFV), and Black queen cell virus (BQCV) are pathogenic to honeybees [4,7,8,9,10,11].
Some of the above viruses DWV, SBPV, ABPV, IAPV, and KBV—are transmitted by V. destructor, which has become a growing threat to the existence of A. mellifera [2,3,12,13,14,15]. V. destructor and viral pathogens play a key role in the loss of A. mellifera colonies, especially because both the mite and the viruses produce a double effect in colony infestation: V. destructor being tightly linked to honeybee life cycle causes a considerable impact on the brood by feeding on it and transmitting viruses [4,16,17,18]. Viruses being vectored by mites and thus directly introduced into the pupa’s body during feeding might reach noticeably higher titers in developing honeybees [17,19,20]. Together they cause a higher impact on honeybee colony health and even induce its collapse [12,21,22].
From an evolutionary point of view, the shift of V. destructor from A. cerana to A. mellifera is a relatively short period of time for the populations of the pathogen and the host to adapt and thus, A. mellifera is a new host for V. destructor. This is valid for its viral content as well. To look for possible variations in virus composition we investigated the viral populations in samples of three A. mellifera subspecies from the Middle East and North Africa (MENA) regions and their parasitic V. destructor mites as well as from A. cerana and their V. destructor mites. For this purpose we used next-generation sequencing (NGS) and performed metagenomics analysis of viral populations from samples of A. m. ligustica colonies from Israel [23] and their V. destructor mites, from Western honeybee subspecies from the MENA region—A. m. intermissa and A. m. syriaca [24]—and their V. destructor mites, and A. cerana and their V. destructor mites from Thailand. In this study, the viral composition and loads from samples of each bee species and its mite were determined, viral loads and viruses of interest were validated and characterized using molecular biology tools. This analysis indicated differences in viral composition and load in the samples of the above honeybee subspecies and their parasitizing varroa mites. We found that in these samples A. cerana and their parasite V. destructor differed considerably in their viral load, A. m. intermissa and its varroa mites showed partial similarity in their viral load, while A. m. ligustica and A. m. syriaca and their corresponding varroa mites’ viral loads were more similar. Moreover, besides identifying known and recently discovered viruses, we found two novel viruses—an Orthomyxovirus common to A. mellifera and V. destructor—that we named Varroa orthomyxovirus-1 (VOV-1) and a homolog of the Hubei like-Virga virus 14 that we named VDV-4. VOV-1 showed 23–58% homology to the Orhomyxoviruses Thogoto and Dhori (THOV and DHOV), which bear negative-sense single stranded RNA genomes of six segments [25,26,27,28]. Replication of single-stranded sense RNA viruses requires the synthesis of the complementary positive-sense RNA, which can be detected by using strand-specific RT-PCR [29]. Using this approach we were able to demonstrate replication of VOV-1. This is the first report of the presence of a segmented negative-sense ssRNA virus in honeybees and varroa mites.
2. Materials and Methods
2.1. Sample Collection
The experimental colonies of A. m. ligustica (worker bees N = 48, from hives 1, 3, 5, and 23, (4 bees per hive), 14 (6 bees), 81 (9 bees), and 401 (1 bee); mites N = 606, from the same hives (85 of them from emerging bees and the rest from free falling mites), collected between October to February 2016, were described before [23]; A. m. syriaca (workers N = 15, from a subcollection of 500 workers from colonies from several apiaries; mites = 20, from tens of mites that were collected in 2013) and A. m. intermissa (workers N = 15, from a subcollection of 500 workers; V. destructor mites N = 27, from tens of mites that were collected in 2013) were described previously [24]. A. cerana drones (N = 6) and V. destructor mites (N = 20) were sampled during spring 2016 (December) in Phrae, Thailand from two colonies 3c and 4c untreated against mites (3 drones and 20 mites from each colony, respectively). The honeybees and corresponding mites were transported in RNA later™ and stored immediately at −80 °C until RNA extraction.
2.2. Samples Preparation
RNA extraction from all A. mellifera and mite samples was carried out using TRI Reagent® (Sigma-Aldrich, Israel) according to the manufacturer’s instructions as published before [24,30]. RNA from A. cerana drones and the corresponding V. destructor mites was individually extracted using a GenJet RNA purification Kit (Thermo Scientific, Burlington, Canada) according to the manufacturer’s instructions.
2.3. Transcriptome and Virome Analysis
Construction and paired-end sequencing of the libraries from A. cerana and its corresponding V. destructor mites-RNA samples was performed at the Technion Genome Center on a HiSEq 2000 platform (Illumina, Haifa, Israel). Paired-end reads were assembled de novo using Trinity [31]. The obtained contigs were translated and aligned to the GenBank nonredundant (NR) database by Blastx [32]. Next-generation sequencing (NGS) of the RNA from A. m. ligustica, A. m. intermissa, and A. m. syriaca honeybees and corresponding V. destructor mites was described previously [23,24]. Metagenomic analysis of A. m. ligustica, A. m. intermissa, and A. m. syriaca subspecies and A. cerana bees, and of V. destructor mites samples were carried out as described previously and in Section 2.8 [30].
2.4. RT-PCR
cDNA was prepared using RevertAid Reverse Transcriptase (Thermo Scientific) with oligo-dT and random primers according to the manufacturer’s instructions. One-hundred nanogram and 2000 ng RNA templates were used from V. destructor and honeybee samples, respectively. RT-conditions: incubation of RNA and primers at 65 °C for 5 min., followed by addition of buffer containing 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 2 mM MgCl2, 5 mM DTT, 4 units of RNase inhibitor Ribolock® (Thermo Scientific), and the RT enzyme (200 units) in a 25 μL volume, and further incubation at 55 °C for 30 min. The reaction was terminated by heating at 85 °C for 5 min. PCR-validations were performed with GoTaq® (Promega Corporation, Madison, WI, USA) using 1 μL cDNA template and 0.2 μM of each forward and reverse primer in a 20 μL reaction with the following conditions; 95 °C for 4 min, 32 cycles at 94 °C for 30 s, then 56 °C (VOV-1 segments 1,2,4,5) or 57 °C (VOV-1 segments 3,6) for 50 s, 72 °C for 2 min. (VOV-1 segments 1,2,4,5) or 1 min. (VOV-1 segments 3,6), and a final extension step of 72 °C for 10 min. For VDV-4 the PCR conditions were identical to those used in segments 1, 2, 4, 5 of VOV-1 with the corresponding specific primers. Specific primers used for validations are described in the Tables S1 and S2 in the Supplementary material.
2.5. VOV-1 Prevalence
VOV-1 prevalence was determined by using RT-PCR to detect the presence of the segment 6 of the virus genome in samples of V. destructor mites and honeybees in apiaries located at the North (Haifa, Kibbutz Lehavot HaBashan, Kibbutz Dan), the Center (ARO, Nitzanei Oz, Herut, Kfar Ruth) and the South (Kibbutz Yad Mordechai) of Israel.
2.6. qRT-PCR
Viral genome copy number was quantified on a PikoReal 96 machine (Thermo Scientific) using a standard protocol (95 °C 2 min; 40 cycles of 95 °C 10 s, 60 °C 20 s, 72 °C 20 s). Each quantitative PCR analysis was performed in triplicate. Nontemplate controls (water) were included in triplicates in each assay. The KAPA SYBR FAST qPCR Master Mix (2×) Universal (Kapa Bio-systems) was used, in a 10 μL final volume. For each analysis 2 μL of the diluted cDNA was used (dilution factor of 4) and specific primers VOV-1-qRT-F1 and VOV-1-qRT-R1 at a concentration of 0.25 μM each (Table S1). The specificity of the amplicons synthesized during the PCR run was ascertained by performing a dissociation curve protocol from 60 °C to 95 °C. Specific primers used for quantification are provided in Table S1 in Supplementary material.
2.7. Replication Assay
Testing for viral replication (presence of the positive strand-sense RNA) was performed by synthetizing the negative-strand cDNA of fragment 6 from the RNA samples using the tagged primer VOV6-46F-TAG, as we described before to analyze replication of BRV-1 [30]. Subsequently the residual VOV6-46F-TAG primer was inactivated by adding to the mixture exonuclease-I and incubating it for another 15 min. at 37 °C (method described in de Miranda et al, 2013 [33]). Finally the exonuclease I was inactivated by heating the mixture at 80 °C for 15 min. Subsequently, PCR was performed with primers VOV6-870R and TAG (Supplementary material 1 Table S1). cDNA produced without any primer was used as control in the same reactions followed by PCR with the same primers as above. PCR was performed at 95 °C for 4 min., 30 cycles at 94 °C for 30 s, then 58 °C for 50 s, 72 °C for 1 min. and a final extension step of 72 °C for 10 min. The identity of the amplified fragment was confirmed by Sanger sequencing (performed at the Biological Services Unit of the Weizmann Institute of Science, Israel).
2.8. Bioinformatic Identification of Contigs
Each RNAseq library was de novo-assembled using Trinity assembler version 2.2.0 [31]. The assembled contigs were then searched with BLASTX [33] against the NCBI nonredundant protein database (NR) [34]. After the assembled viruses were identified in each library, each library’s raw data reads were mapped using bowtie2 [35] to evaluate the virus quantity in the transcriptome. We reanalyzed the data obtained before in A. m. ligustica [23] and realized that contigs of a new Orthomyxovirus were present. Then we analyzed data that we downloaded from recently published transcriptomes of viruses of A. m. intermissa and A. m. syriaca and their varroa from the MENA region [24] and were able to assemble the complete genome of the virus (Table 1 and Table 2).

Table 1.
Libraries used in this study.

Table 2.
Accession numbers for the new viruses.
2.9. Molecular Phylogenetic Analysis
Phylogenetic analysis was done using MEGA 6 [34]. Alignment of the proteins was done using MAFFT [36], and then Maximum likelihood Phyml 3.0 was used for creating the tree [37] with a 100 bootstrap.
3. Results
3.1. Metagenomic Analysis of Viruses in A. mellifera, A. cerana and Their V. destructor Mites
In the analysis we included libraries from A. m. ligustica, A. m. syriaca, A. m. intermissa, A. cerana, and their V. destructor counterparts (see Materials and Methods). This revealed variation in composition of the viral loads of A. m. ligustica, A. m. syriaca, A. m. intermissa, A. cerana, and their V. destructor mite counterparts (libraries IB1, SB2, AB3, BCER, IV4, SV5, AV6, and VCER, respectively). Mapping of the libraries’ reads to viral contigs of honeybees and V. destructor mites resulted in
- identification of the most common honeybee viruses: Acute bee paralysis virus (ABPV), Israeli acute paralysis virus (IAPV) in IB1, IV4, and SB2; Apis mellifera filamentous virus (AmFV) in AB3; Bee Macula-like virus/Varroa Macula-like virus (BeeMLV/VdMLV) and Black queen cell virus (BQCV) in all the libraries except for BCER and VCER; Deformed wing virus (DWV) in all the libraries; Lake Sinai virus (LSV) in IB1; and Sacbrood virus (SBV) in IB1, IV4, SV5, and AV6 (details are provided in Materials and Methods and in Table 3).
 Table 3. Presence of viruses in honeybees and V. destructor mites by library.
Table 3. Presence of viruses in honeybees and V. destructor mites by library. - identification of recently described viruses Apis rhabdovirus-1/Bee rhabdovirus-1(ARV-1/BRV-1) [30], Varroa destructor virus-2 (VDV-2), and Varroa destructor virus-3 (VDV-3) [23].
- discovery of two new viruses that we designed Varroa orthomyxovirus-1 (VOV-1), an orthomyxovirus with low homology to other viruses from the Orthomyxoviridae family, and Varroa destructor virus-4 VDV-4 (Table 3 and see below).
Interestingly, ARV-1/BRV-1 and VOV-1 showed differences in their distribution across bee and V. destructor libraries; while ARV-1/BRV-1 was present in all V. destructor libraries and in two of the honeybee libraries, VOV-1 was limited to A. m. ligustica and A. m. syriaca and their corresponding varroa (Table 3).
The presence of viruses varied across A. mellifera and A. cerana bees and their corresponding V. destructor libraries. From the A. mellifera libraries analyzed, IB1 and IV4 (A. m. ligustica) showed a similar percentage of viral reads of DWV, as did SB2 and SV5 (A. m. syriaca) and A. m. intermissa, and their V. destructor libraries—AB3 and AV6—showed large differences (Figure 1). Also, A. cerana libraries—BCER and VCER—were distinct (Figure 1 and see below). For instance, AV6 showed a smaller percentage of DWV reads compared to AB3 and to the other A. mellifera—IB1 and SB2—and their corresponding V. destructor libraries—IV4 and SV5, respectively (Figure 1). The cDNA libraries of A. cerana (BCER) and its varroa (VCER) displayed extremely low percentage of viral reads for DWV (0.7601% and 0.6402%, accordingly). As can be seen, our samples BCER and VCER differed in their load of other viruses as well; while the BCER main viral component was ARV-2 [38], the main virus present in VCER was VDV-2 (Figure 1). In addition, VCER displayed two viruses present in varroa parasites of A. cerana only with 38% homology to Hubei picorna-like virus 29 [39] and 36% homology to Hubei virga-like virus 14 [40] (Table 3 and see below).
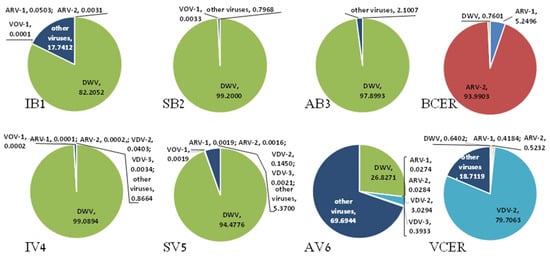
Figure 1.
Presence of viruses in honeybee and V. destructor mite by library. Percentage of total viral reads* mapping to viral contigs of the honeybees and V. destructor mites libraries. *Including: DWV and genetic variants, ARV-1, ARV-2, VOV-1, VDV-2, VDV-3, and other viruses (11 more, see Table 1), cutoff at 0.0001%.
3.2. Varroa Orthomyxovirus-1 and the Hubei Virga-like 14 Homolog Virus
We reanalyzed the data obtained before in A. m. ligustica [23] and realized that contigs of a new Orthomyxovirus were present in the data. To complete the picture we downloaded and analyzed data from the transcriptome of viruses of A. m. intermissa and A. m. syriaca and their varroa mites from the Middle East and North African (MENA) honeybees and varroa mites that were published recently but did not focus on finding new viruses [24]. This additional sequence data facilitated complete genome assembly of this new virus. According to BLASTX analysis we identified VOV-1 contigs in two cDNA libraries of bees (IB1 and SB2) and two cDNA libraries of their corresponding varroa mites (IV4 and SV5). VOV-1 showed 23–58% homology to the Orthomyxoviruses Thogoto and Dhori (THOV and DHOV), which bear negative-sense single stranded RNA genomes of six segments [25,26,27,28]. Contigs of 2198, 1899, and 358 nucleotides in length from the SV5 library showed homology of 58%, 29%, and 46% to polymerase subunits PB2, PB1, and PA encoded in segments 1, 2, and 3 of the DHOV genome, respectively (Table 4). A contig of 232 nucleotides length from the IV4 library showed homology of 41% to the glycoprotein subunit (GP) encoded in segment 4 of THOV; a contig of 1442 nucleotides length from IB1 library showed homology of 39% to the nucleoprotein subunit (NP) encoded in segment 5 of DHOV; and a contig of 983 nucleotides from the SB2 library showed homology of 23% to matrix protein (M) encoded in segment 6 of THOV (Table 4).

Table 4.
Comparison of the Varroa orthomyxovirus-1 (VOV-1) contigs to the Orthomyxoviruses Thogoto and Dhori (THOV and DHOV).
Phylogenetic analysis of open reading frames (ORFs) coding for polymerase subunits PB2, PB1, and PA showed that the polymerase was closely related to negative-sense ssRNA viruses belonging to the Orthomyxoviridae viral family: Thogoto virus (THOV); Aransas Bay virus (ABV); Upolu virus (UPOV) (Figure 2A–C); and Jos virus (JOSV) (Figure 2A,C). PB2 is phylogenetically closer to THOV, ABV, UPOV, and JOSV, and more distant from DHOV and Bourbon virus (BRBV) as well as PB1, except for JOSV, and PA, except for BRBV (Figure 2A–C).
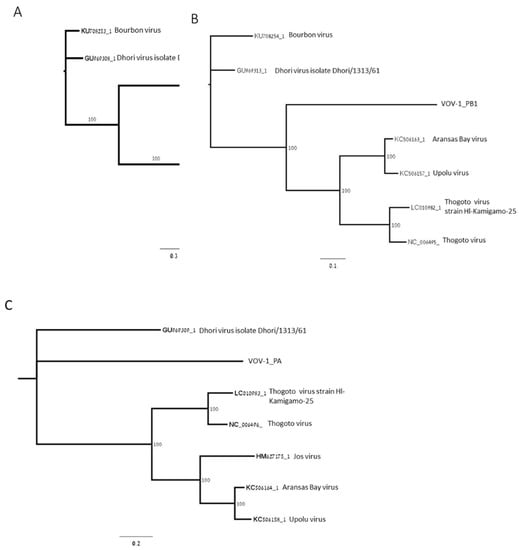
Figure 2.
Maximum-likelihood phylogenetic tree of VOV-1 contigs for segments 1, 2, and 3. The trees were constructed based on the ORF of VOV-1 polymerase subunits PB2 (A), PB1 (B), and PA (C). GeneBank accession numbers provided.
Based on the above contigs’ sequences we designed specific primers to validate the presence of each segment of VOV-1 in the viromes of Israeli A. mellifera ligustica and their counterpart V. destructor parasites (Figure 3 and see Materials and Methods). All six segments were identified in the V. destructor virome (Figure 3, lanes 1, 4, 7, 10, 13, and 16) but they were absent in the virome of honeybees (Figure 3, lanes 2, 5, 8, 11, 14, and 17).
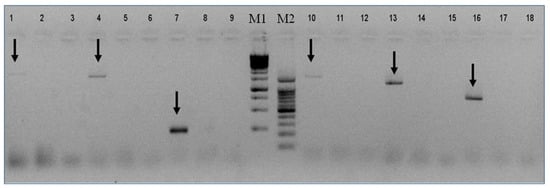
Figure 3.
Presence of VOV-1 in virome of V. destructor mites and its absence in honeybees. V. destructor mite virome: lanes 1, 4, 7, 10, 13, and 16; Honeybee virome: lanes 2, 5, 8, 11, 14, and 17; NTC (nontemplate control): lanes 3, 6, 9, 12, 15, and 18; M1 and M2: GeneRuler MarkerTM 1 kb, 100 bp DNA ladders, respectively. Arrows: VOV-1 segments (1–6).
Furthermore, we tested the presence of segment 6 of the viral genome to estimate the prevalence of VOV-1 by RT-PCR in individual mites and honeybees. We detected VOV-1 in 35.56% of V. destructor mites from Israeli colonies located in ARO, Beit Dagan (16 of 45), and in none of 32 individual honeybees sampled from the same colonies. We also analyzed its prevalence in colonies located in the Center, North, and South of Israel by testing pools of the honeybees and V. destructor mites with the same PCR method. The virus was identified in 78.57% of V. destructor pools (11 of 14 pools) and only in 8.33% of honeybee pools (5 of 60 pools).
The number of genomic copies of VOV-1 estimated by qRT-PCR was similar in individual mites collected from honeybee colonies in ARO, Beit Dagan, in pools of mites, and in honeybee pools sampled from colonies located in the Center, North, and South of Israel: 5.11×102–1.22×106, 2.33×103–4.88×105, and 4.91×102–1.38×105, respectively (Table 5).

Table 5.
Genomic copies of VOV-1 in individual mites and in pools of mites and nurse honeybees.
To investigate if VOV-1 replicates in our samples, we screened for presence of the positive-sense RNA strand of the fragment 6 of the virus using RNA-strand sense-specific primer-tagged RT-PCR (see Materials and Methods). A predicted size fragment of ~840 nucleotides corresponding to the VOV-1 positive-sense-strand RNA between nucleotides 46 and 870 was found in tested samples from individual V. destructor mites (Figure 4, panel A, lanes 3, 9, 11, and 13), or from V. destructor mite and nurse honeybee pools (Figure 4, panels B and C, lanes 15, 17, 19, 21, 23, and 27 and 33, 37, and 39, respectively). No amplification was observed in some varroa individuals, varroa and nurse honeybee pools (Figure 4, panels A, B and C, lanes 1, 5, and 7 and 25, 29, 31, and 35, respectively). Control samples obtained when PCR was performed with cDNA prepared from the same RNA without the corresponding oligonucleotide primer in the RT reaction did not show any amplicon (Figure 4, panels A, B, and C, lanes 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, and 38, respectively).
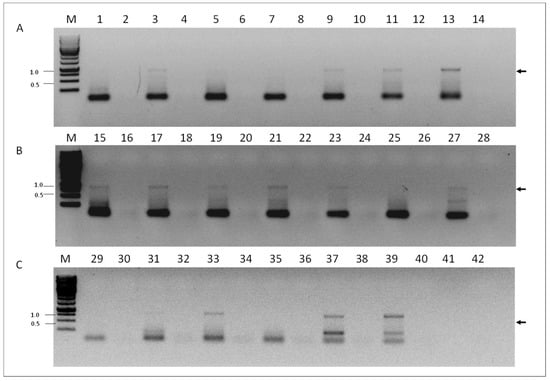
Figure 4.
Detection of the VOV-1-segment 6 positive-sense RNA strand in V. destructor and A. mellifera. (A) V. destructor individuals, odd sample numbers and their respective controls, even sample numbers. (B) V. destructor (pools) odd samples, and their respective controls, even samples. (C) Lanes 29 and 30, V. destructor (pool) and its respective control; A. mellifera (pools), odd lanes 31–39 and respective controls, even lanes 32–40. PCR control reaction of the same individual RNA performed on cDNA produced without any primer (see the section “Materials and Methods”. PCR primers: VOV6-870R and TAG. M, GeneRuler Marker 1 kb DNA Ladder (Thermo Scientific Inc.); 41 and 42: nontemplate control; Arrow, VOV-1 amplicon that was confirmed by sequencing (see “Materials and Methods”).
We confirmed by Sanger DNA sequencing that the above specific-primer-tagged amplicons were identical to the VOV-1 sequence comprising nucleotides 46 and 870 of segment 6 of the viral genome.
From the two undescribed viruses that we found in VCER we further investigated VDV-4. Phylogenetic analysis using the putative large ORF protein of the virus showed that it is 36% homologous to the hypothetical protein gene of spider viruses Hubei virga- like virus 14 and Hubei virga-like virus 13 as well as to the spider putative protein of the virus Nephila clavipes virus 4 (Figure 5).
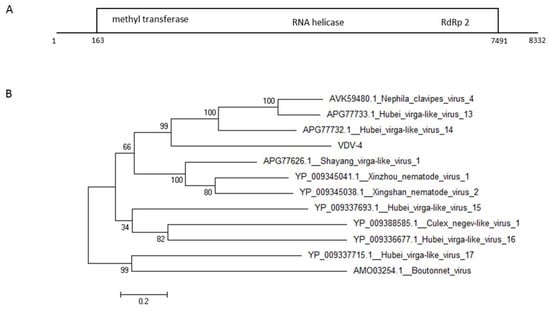
Figure 5.
Genomic organization (A) and maximum-likelihood phylogenetic tree (B) of VDV-4. The tree was constructed based on the ORF of VDV-4. GeneBank accession numbers provided.
RT-PCR validation of its presence in a small sample of A. cerana drones and V. destructor individuals suggested that this virus was predominant in the latter (Figure 6). Therefore, we decided to name it VDV-4.
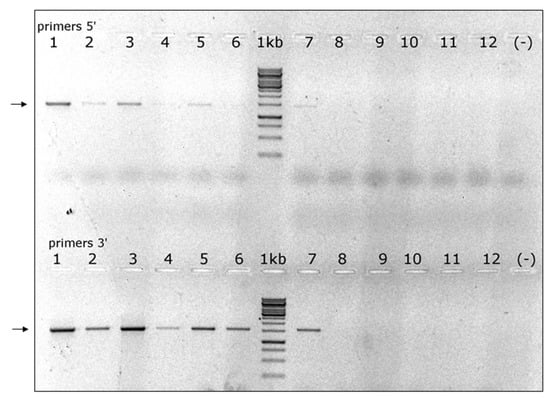
Figure 6.
Presence of VDV-4 in V. destructor mites and A. cerana drones: lanes 1–3 and 4–6 pools of 5 V. destructor cDNAs each from colonies 3c and 4c, respectively; lanes 7–9 and 10–12 individual A. cerana drones, 3 per colony; lanes (−) nontemplate control; lanes 1 kb, GeneRuler MarkerTM 1 kb DNA ladder. Arrows: VDV-4, Upper panel VDV-4 genome 5’-end primers VDV4-1451F and VDV4-2889R, expected fragment 1438 bp; lower panel VDV-4 genome 3’-end primers VDV4-6242F and VDV4-7818R, expected fragment 1576 bp.
4. Discussion
We presumed that since Apis mellifera is a new host to Varroa destructor they might show differences both in the composition and distribution of their viral load. Our analysis of the data illustrates changes in viral composition and load among samples from A. mellifera subspecies and their V. destructor. Interestingly, a small-scale sample of A. cerana and its V. destructor showed variation in viral composition and load as well. The variation in virus composition was based on a n = 1 repetition per bee species and V. destructor transcriptomes and, consequently, the results do not necessarily reflect variations at the level of species/subspecies and could be due to other factors such as sampling region, season and/or diverse time of sampling of the different bee species and there mites, etc. A higher number of samples in a coordinated effort will be required for species/subspecies comparative purposes.
We found that in the libraries studied the viral reads of Deformed wing virus, one of the most important factors affecting honeybee colony health and survival, were 82.20%, 99.20%, and 97.90% in A. m. ligustica, A. m. syriaca, and A. m. intermissa, respectively, and only 0.76% in A. cerana. In addition, we observed that a few viruses of importance were present in A. mellifera libraries but not in A. cerana’s. Namely, ABPV + IAPV in A. m. ligustica and its V. destructor, and in A. m. syriaca; BeeMLV/VdMLV in all A. mellifera libraries and corresponding V. destructor mites, as well as in varroa from A. cerana; BQCV in all A. mellifera and corresponding V. destructor and SBV in A. m. ligustica and all three libraries of varroa mites parasitizing A. mellifera. ABPV seems to be uncommon to A. cerana in China and South Korea [41,42], or to show low prevalence in wild colonies [43], as was in A. cerana in Northern Thailand [44]. SBV was reported with high prevalence in Southeast Asia [41,45,46,47] as well as BQCV, that showed relatively high prevalence in viral populations of A. cerana in China, South Korea, and Vietnam [42,43,48]. These two viruses were absent in our libraries and that could be due also to the small sample size of them.
Furthermore, we identified recently characterized viruses in A. mellifera and/or in varroa mites, namely Apis rhabdovirus-1/Bee rahbdovirus-1 (ARV-1/BRV-1) [30,38], Varroa destructor virus-2 and -3 (VDV-2 and VDV-3) [23], and a new Varroa orthomyxovirus-1 (VOV-1). Some of these viruses were identified as common to honeybees and V. destructor like ARV-1/BRV-1 and VOV-1, while others appeared to be restricted to mites, such as VDV-2 and VDV-3. Interestingly, VOV-1’s presence was limited to A. m. ligustica and A. m. syriaca as well as to V. destructor mites parasitizing them. VDV-2 was detected in all V. destructor libraries and VDV-3 was absent in Varroa destructor mites parasitizing A. cerana. ARV-1/BRV-1 was found previously in A. mellifera, V. destructor, and in Bombus impatiens, but our finding that it is present in A. cerana suggests that it may have a broader host range. V. destructor mite parasites of A. cerana appear to bear two novel viruses with low homology to Hubei picorna-like virus-29 [39] and to Hubei virga-like virus-14 [40], which were absent in V. destructor mites from A. mellifera. We validated the presence of a Hubei virga-like virus-14 and designed it VDV-4.
Data suggest that following V. destructor invasion there is high selection on DWV strains such that only a single strain seems to dominate, though which strain dominates varies across colonies and studies [20,49,50,51]. This process is accompanied with increase in the collapse of V. destructor-infested colonies [15,51,52]. Moreover, laboratory experiments showed that DWV undergoes rapid selection following its injection in the honeybee hemolymph, similarly to what happens during parasitization of Varroa destructor on A. mellifera [20]. In our study, we measured differences in DWV loads between samples of A. mellifera subspecies and their V. destructor counterparts. V. destructor from A. m. intermissa showed lower DWV levels compared to its parasitized host (26.83% and 97.90%, respectively). Interestingly, it was reported that A. m. intermissa was more resistant to V. destructor parasitization than other A. mellifera subspecies [53,54]. Again, the results are subjected to the above-mentioned limitations of the analysis including the n = 1 repetitions of the transcriptomic data per subspecies.
We characterized VOV-1, a novel virus common to A. mellifera and V. destructor and VDV-4, a novel virus of V. destructor from A. cerana. VOV-1 possess a single-stranded negative-sense RNA genome and belongs to the Orthomyxoviridae family that among others includes the genus Thogotovirus. Most of the Thogotoviruses have been associated with ticks [55] and relatively few of them have been described in Acari or other types of hematophagous arthropods [56,57]. The VOV-1 genome has six segments and this is the first report of the complete genome of a single-stranded negative-sense RNA segmented virus seen in honeybees and varroa mites. We provide evidence that VOV-1 replicates in individual varroa mites, and we found positive sense-virus RNA in pools collected from A. m. ligustica. Interestingly, it showed greater prevalence in V. destructor mites compared with honeybees of 78.57% and 8.33%, respectively, in samples from apiaries located at the North (Haifa, Kibbutz Lehavot HaBashan, Kibbutz Dan), the Center (ARO, Nitzanei Oz, Herut, Kfar Ruth), and the South (Kibbutz Yad Mordechai) of Israel. This is why, taking together the above data, we decided to name it Varroa orthomyxovirus-1, VOV-1.
The discovery of these novel viruses in Apis mellifera and its recently acquired obligatory parasite Varroa destructor opens a new venue for investigation of viral interactions in honeybee colonies. A number of questions emerge concerning this new host–pathogen relationship that could interfere the preexisting balance. What is the pathology associated with VOV-1? Do varroa mites transmit VOV-1 and VDV-4 directly, e.g., transovarially, or via their host bee? Where do the viruses accumulate in varroa? Is VOV-1 infectious to A. mellifera?
Furthermore, there is another issue concerning those viruses crucial for colony health and survival—DWV, ABPV, IAPV, and CBPV—and their interaction with newly discovered ARV1/ BRV-1, VOV-1, and VDV-2 and -3: Do they affect one another on a mutual base? If they do, on what level and what are the factors that may be involved (e.g., colony location, kind of treatment against varroa, season, colony resistance and/or hygienic behavior, etc.).
Our findings and the tools that we have developed in this study pave the way to investigate these questions and extend our knowledge and understanding of the role played by viral pathogens in honeybee colonies.
Supplementary Materials
The following are available online at http://www.mdpi.com/1999-4915/11/2/94/s1, Figure S1: title, Table S1: Table S1. Primers used for validation and quantitation of Varroa orthomyxovirus-1 (VOV-1). Table S2. Primers used for validation and quantitation of Varroa destructor virus-4 (VDV-4). Table S3. Complete VOV-1 nucleotide sequences. Table S4. Nucleotide sequence of VDV-4.
Author Contributions
Conceptualization, S.L. and N.C.; Methodology, S.L., N.S., T.E., and D.N.; Software, N.S.; Validation, S.L and T.E.; Formal Analysis, S.L., N.S., D.N., and N.C.; Investigation, S.L., N.S., and N.C.; Resources, S.L., J.P., and T.E.; Data Curation, S.L. and N.S.; Writing—Original Draft Preparation, S.L. and P.N.; Writing—Review and Editing S.L., N.S., P.N., J.P., D.N., and N.C.; Visualization, S.L.,P.N., and N.C.; Supervision, N.C.; Project Administration and Funding Acquisition, N.C.
Funding
The research was supported by a USAID Grant number TA-MOU-11-M32-035 and a Grant of the Chief Scientist of the Ministry of Agriculture of Israel number 131-1857.
Acknowledgments
We thank Dr. Victoria Soroker, Mr. Yossef Kamer, and Mrs Nurit Eliash for their assistance with the honeybee colonies from ARO, and Rotem Zirler for her assistance with honeybee samples.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Rosenkranz, P.; Aumeier, P.; Ziegelmann, B. Biology and control of Varroa destructor. J. Invertebr. Pathol. 2010, 103, S96–S119. [Google Scholar] [CrossRef] [PubMed]
- Bowen-Walker, P.L.; Martin, S.J.; Gunn, A. The transmission of deformed wing virus between honeybees (Apis mellifera L.) by the ectoparasitic mite Varroa jacobsoni Oud. J. Invertebr. Pathol. 1999, 73, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Pettis, J.S.; Evans, J.D.; Kramer, M.; Feldlaufer, M.F. Transmission of Kashmir bee virus by the ectoparasitic mite Varroa destructor. Apidologie 2004, 35, 441–448. [Google Scholar] [CrossRef]
- Shen, M.; Cui, L.; Ostiguy, N.; Cox-Foster, D. Intricate transmission routes and interactions between picorna-like viruses (Kashmir bee virus and sacbrood virus) with the honeybee host and the parasitic varroa mite. J. Gen. Virol. 2005, 86, 2281–2289. [Google Scholar] [CrossRef] [PubMed]
- De Miranda, J.R.; Genersch, E. Deformed wing virus. J. Invertebr. Pathol. 2010, 103 (Suppl. 1), S48–S61. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.M.K.; Anderson, D.L.; Tay, W.T. Multiple host shifts by the emerging honeybee parasite, Varroa jacobsoni. Mol. Ecol. 2015, 24, 2379–2391. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.P.; Siede, R. Honey bee viruses. In Advances in Virus Research; Academic Press: Cambridge, MA, USA, 2007; pp. 33–80. ISSN 0065-3527. [Google Scholar]
- Nielsen, S.L.; Nicolaisen, M.; Kryger, P. Incidence of acute bee paralysis virus, black queen cell virus, chronic bee paralysis virus, deformed wing virus, Kashmir bee virus and sacbrood virus in honey bees Apis mellifera in Denmark. Apidologie 2008, 39, 310–314. [Google Scholar] [CrossRef]
- Gauthier, L.; Cornman, S.; Hartmann, U.; Cousserans, F.; Evans, J.D.; de Miranda, J.R.; Neumann, P. The Apis mellifera filamentous virus genome. Viruses 2015, 7, 3798–3815. [Google Scholar] [CrossRef]
- Amakpe, F.; Smet, L.D.; Brunain, M.; Ravoet, J.; Jacobs, F.J.; Reybroeck, W.; Sinsin, B.; de Graaf, D.C. Discovery of Lake Sinai virus and an unusual strain of acute bee paralysis virus in West African apiaries. Apidologie 2016, 47, 35–47. [Google Scholar] [CrossRef]
- Brutscher, L.M.; McMenamin, A.J.; Flenniken, M.L. The Buzz about honey bee viruses. PLoS Pathog. 2016, 12, e1005757. [Google Scholar] [CrossRef]
- Martin, S.J. The role of varroa and viral pathogens in the collapse of honeybee colonies: A modelling approach: Collapse of varroa-infested honeybee colonies. J. Appl. Ecol. 2002, 38, 1082–1093. [Google Scholar] [CrossRef]
- Santillán-Galicia, M.T.; Ball, B.V.; Clark, S.J.; Alderson, P.G. Transmission of Deformed wing virus and Slow paralysis virus to adult bees (Apis mellifera L.) by Varroa destructor. J. Apic. Res. 2010, 49, 141–148. [Google Scholar]
- Di Prisco, G.; Pennacchio, F.; Caprio, E.; Boncristiani, H.F.; Evans, J.D.; Chen, Y. Varroa destructor is an effective vector of Israeli acute paralysis virus in the honeybee, Apis mellifera. J. Gen. Virol. 2011, 92, 151–155. [Google Scholar] [CrossRef] [PubMed]
- Francis, R.M.; Nielsen, S.L.; Kryger, P. Varroa-virus Interaction in collapsing honey bee colonies. PLoS ONE 2013, 8, e57540. [Google Scholar] [CrossRef] [PubMed]
- Gisder, S.; Aumeier, P.; Genersch, E. Deformed wing virus: Replication and viral load in mites (Varroa destructor). J. Gen. Virol. 2009, 90, 463–467. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.; Jironkin, A.; Chandler, D.; Burroughs, N.; Evans, D.J.; Ryabov, E.V. Recombinants between Deformed wing virus and Varroa destructor virus-1 may prevail in Varroa destructor-infested honeybee colonies. J. Gen. Virol. 2011, 92, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Mondet, F.; de Miranda, J.R.; Kretzschmar, A.; Le Conte, Y.; Mercer, A.R. On the front line: Quantitative virus dynamics in honeybee (Apis mellifera L.) colonies along a new expansion front of the parasite Varroa destructor. PLoS Pathog. 2014, 10, e1004323. [Google Scholar] [CrossRef] [PubMed]
- Nazzi, F.; Brown, S.P.; Annoscia, D.; Del Piccolo, F.; Di Prisco, G.; Varricchio, P.; Della Vedova, G.; Cattonaro, F.; Caprio, E.; Pennacchio, F. Synergistic parasite-pathogen interactions mediated by host immunity can drive the collapse of honeybee colonies. PLoS Pathog. 2012, 8, e1002735. [Google Scholar] [CrossRef]
- Ryabov, E.V.; Wood, G.R.; Fannon, J.M.; Moore, J.D.; Bull, J.C.; Chandler, D.; Mead, A.; Burroughs, N.; Evans, D.J. A virulent strain of Deformed wing virus (DWV) of honeybees (Apis mellifera) prevails after Varroa destructor-mediated, or in vitro, transmission. PLoS Pathog. 2014, 10, e1004230. [Google Scholar] [CrossRef]
- Carreck, N.L.; Ball, B.V.; Martin, S.J. Honey bee colony collapse and changes in viral prevalence associated with Varroa destructor. J. Apic. Res. 2010, 49, 93–94. [Google Scholar] [CrossRef]
- Dainat, B.; Evans, J.D.; Chen, Y.P.; Gauthier, L.; Neumann, P. Predictive markers of honey bee colony collapse. PLoS ONE 2012, 7, e32151. [Google Scholar] [CrossRef] [PubMed]
- Levin, S.; Sela, N.; Chejanovsky, N. Two novel viruses associated with the Apis mellifera pathogenic mite Varroa destructor. Sci. Rep. 2016, 6, 37710. [Google Scholar] [CrossRef] [PubMed]
- Haddad, N.J.; Horth, L.; Al-Shagour, B.; Adjlane, N.; Loucif-Ayad, W. Next-generation sequence data demonstrate several pathogenic bee viruses in Middle East and African honey bee subspecies (Apis mellifera syriaca, Apis mellifera intermissa) as well as their cohabiting pathogenic mites (Varroa destructor). Virus Genes 2018, 54, 694–705. [Google Scholar] [CrossRef] [PubMed]
- Haig, D.A.; Woodall, J.P.; Danskin, D. Thogoto Virus: A hitherto undescribed agent isolated from ticks in Kenya. Microbiology 1965, 38, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Calisher, C.H.; Karabatsos, N.; Filipe, A.R. Antigenic uniformity of topotype strains of Thogoto virus from Africa, Europe, and Asia. Am. J. Trop. Med. Hyg. 1987, 37, 670–673. [Google Scholar] [CrossRef] [PubMed]
- Davies, C.R.; Jones, L.D.; Green, B.M.; Nuttall, P.A. In vivo reassortment of Thogoto virus (a tick-borne influenza-like virus) following oral infection of Rhipicephalus appendiculatus ticks. J. Gen. Virol. 1987, 68 Pt 9, 2331–2338. [Google Scholar] [CrossRef]
- Mateo, R.I.; Xiao, S.-Y.; Lei, H.; DA ROSA, A.P.T.; Tesh, R.B. Dhori virus (Orthomyxoviridae: Thogotovirus) infection in mice: A model of the pathogenesis of severe orthomyxovirus infection. Am. J. Trop. Med. Hyg. 2007, 76, 785–790. [Google Scholar] [CrossRef] [PubMed]
- Horsington, J.; Zhang, Z. Analysis of foot-and-mouth disease virus replication using strand-specific quantitative RT-PCR. J. Virol. Methods 2007, 144, 149–155. [Google Scholar] [CrossRef]
- Levin, S.; Galbraith, D.; Sela, N.; Erez, T.; Grozinger, C.M.; Chejanovsky, N. Presence of Apis Rhabdovirus-1 in populations of pollinators and their parasites from two continents. Front. Microbiol. 2017, 8, 2482. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- De Miranda, J.R.; Bailey, L.; Ball, B.V.; Blanchard, P.; Budge, G.E.; Chejanovsky, N.; Chen, Y.P.; Gauthier, L.; Genersch, E.; de Graaf, D.C.; et al. Standard methods for virus research in Apis mellifera. J. Apic. Res. 2013, 52, 32. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Remnant, E.J.; Shi, M.; Buchmann, G.; Blacquière, T.; Holmes, E.C.; Beekman, M.; Ashe, A. A Diverse range of novel RNA viruses in geographically distinct honey bee populations. J. Virol. 2017, 91, e00158-17. [Google Scholar] [CrossRef]
- Shi, M.; Lin, X.-D.; Tian, J.-H.; Chen, L.-J.; Chen, X.; Li, C.-X.; Qin, X.-C.; Li, J.; Cao, J.-P.; Eden, J.-S.; et al. Redefining the invertebrate RNA virosphere. Nature 2016, 540, 539–543. [Google Scholar] [CrossRef]
- Debat, H.J. An RNA virome associated to the golden orb-weaver spider Nephila clavipes. Front. Microbiol. 2017, 8, 2097. [Google Scholar] [CrossRef]
- Ai, H.; Yan, X.; Han, R. Occurrence and prevalence of seven bee viruses in Apis mellifera and Apis cerana apiaries in China. J. Invertebr. Pathol. 2012, 109, 160–164. [Google Scholar] [CrossRef]
- Choe, S.E.; Nguyen, L.T.K.; Noh, J.H.; Koh, H.B.; Jean, Y.H.; Kweon, C.H.; Kang, S.W. Prevalence and distribution of six bee viruses in Korean Apis cerana populations. J. Invertebr. Pathol. 2012, 109, 330–333. [Google Scholar] [CrossRef] [PubMed]
- Forsgren, E.; Wei, S.; Ding, G.; Liu, Z.; Tran, T.V.; Tang, P.T.; Truong, T.A.; Dinh, T.Q.; Fries, I. Preliminary observations on possible pathogen spill-over from Apis mellifera to Apis cerana. Apidologie 2015, 46, 265–275. [Google Scholar] [CrossRef]
- Chanpanitkitchote, P.; Chen, Y.; Evans, J.D.; Li, W.; Li, J.; Hamilton, M.; Chantawannakul, P. Acute bee paralysis virus occurs in the Asian honey bee Apis cerana and parasitic mite Tropilaelaps mercedesae. J. Invertebr. Pathol. 2018, 151, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Rana, B.; Garg, I.; Khurana, S.P.; Verma, L.; Agrawal, H. Thai sacbrood virus of honeybees (Apis cerana indica F) in north-west Himalayas. Indian J. Virol. 1986, 2, 127–131. [Google Scholar]
- Chantawannakul, P.; de Guzman, L.I.; Li, J.; Williams, G.R. Parasites, pathogens, and pests of honeybees in Asia. Apidologie 2016, 47, 301–324. [Google Scholar] [CrossRef]
- Li, J.; Qin, H.; Wu, J.; Sadd, B.M.; Wang, X.; Evans, J.D.; Peng, W.; Chen, Y. The prevalence of parasites and pathogens in Asian honeybees Apis cerana in China. PLoS ONE 2012, 7, e47955. [Google Scholar] [CrossRef]
- Thu, H.T.; Thi, N.; Lien, K.; Linh, M.T.; Le, T.H.; Thi, N.; Reddy, K.E.; Yoo, M.S.; Kim, Y.H.; Cho, Y.S.; et al. Prevalence of bee viruses among Apis cerana populations in Vietnam. J. Apic. Res. 2016, 55, 379–385. [Google Scholar] [CrossRef]
- Dalmon, A.; Desbiez, C.; Coulon, M.; Thomasson, M.; Le Conte, Y.; Alaux, C.; Vallon, J.; Moury, B. Evidence for positive selection and recombination hotspots in Deformed wing virus (DWV). Sci. Rep. 2017, 7, 41045. [Google Scholar] [CrossRef]
- Natsopoulou, M.E.; McMahon, D.P.; Doublet, V.; Frey, E.; Rosenkranz, P.; Paxton, R.J. The virulent, emerging genotype B of Deformed wing virus is closely linked to overwinter honeybee worker loss. Sci. Rep. 2017, 7, 5242. [Google Scholar] [CrossRef]
- Martin, S.J.; Highfield, A.C.; Brettell, L.; Villalobos, E.M.; Budge, G.E.; Powell, M.; Nikaido, S.; Schroeder, D.C. Global honey bee viral landscape altered by a parasitic mite. Science 2012, 336, 1304–1306. [Google Scholar] [CrossRef]
- Highfield, A.C.; Nagar, A.E.; Mackinder, L.C.M.; Noël, L.M.-L.J.; Hall, M.J.; Martin, S.J.; Schroeder, D.C. Deformed wing virus implicated in overwintering honeybee colony losses. Appl. Environ. Microbiol. 2009, 75, 7212–7220. [Google Scholar] [CrossRef] [PubMed]
- Boecking, O.; Ritter, W. Grooming and removal behavior of Apis mellifera intermissa in Tunisia against Varroa jacobsoni. J. Apic. Res. 1993, 32, 127–134. [Google Scholar] [CrossRef]
- Adjlane, N.; Dainat, B.; Gauthier, L.; Dietemann, V. Atypical viral and parasitic pattern in Algerian honey bee subspecies Apis mellifera intermissa and A. m. sahariensis. Apidologie 2016, 47, 631–641. [Google Scholar] [CrossRef]
- Contreras-Gutiérrez, M.A.; Nunes, M.R.T.; Guzman, H.; Uribe, S.; Suaza Vasco, J.D.; Cardoso, J.F.; Popov, V.L.; Widen, S.G.; Wood, T.G.; Vasilakis, N.; et al. Sinu virus, a novel and divergent orthomyxovirus related to members of the genus Thogotovirus isolated from mosquitoes in Colombia. Virology 2017, 501, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Temmam, S.; Monteil-Bouchard, S.; Robert, C.; Baudoin, J.-P.; Sambou, M.; Aubadie-Ladrix, M.; Labas, N.; Raoult, D.; Mediannikov, O.; Desnues, C. Characterization of viral communities of biting midges and identification of novel Thogotovirus species and Rhabdovirus genus. Viruses 2016, 8, 77. [Google Scholar] [CrossRef] [PubMed]
- Bolling, B.G.; Weaver, S.C.; Tesh, R.B.; Vasilakis, N. Insect-specific virus discovery: Significance for the Arbovirus community. Viruses 2015, 7, 4911–4928. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).