Abstract
Replacements of animal models by advanced in vitro systems in biomedical research, despite exceptions, are currently still not satisfactory in reproducing the whole complexity of pathophysiological mechanisms that finally lead to disease. Therefore, preclinical models are additionally required to reflect analogous in vivo situations as found in humans. Despite proven limitations of both approaches, only a combined experimental arrangement guarantees generalizability of results and their transfer to the clinics. Although the laboratory mouse still stands as a paradigm for many scientific discoveries and breakthroughs, it is mandatory to broaden our view by also using nontraditional animal models. The present review will first reflect the value of experimental systems in life science and subsequently describes the preclinical rodent model Mastomys coucha that—although still not well known in the scientific community—has a long history in research of parasites, bacteria, papillomaviruses and cancer. Using Mastomys, we could recently show for the first time that cutaneous papillomaviruses—in conjunction with UV as an environmental risk factor—induce squamous cell carcinomas of the skin via a “hit-and-run” mechanism. Moreover, Mastomys coucha was also used as a proof-of-principle model for the successful vaccination against non-melanoma skin cancer even under immunosuppressive conditions.
1. Introduction
Model systems are indispensable in science, not only to visualize corresponding features of the invisible (Figure 1), but also to provide—in biomedical research—an empirical bona fide equivalent for pathophysiological processes of human diseases, such as cancer. Models are only needed as long as we do not yet have full knowledge of what they stand for. When enough certainty is obtained in form of reproducible data sets, the exploited models are obsolete and become substituted by others or by the genuine respective organism for further questions [1]. For instance, Gregor Mendel’s cross-breeding of pea plants represented a model to understand dominant and recessive inheritance, but Thomas Hunt Morgan’s work with Drosophila melanogaster became more suitable, since phenotypes could be mapped to a defined region within the chromosomes. Moreover, although the spinning top watched by the two Nobel Laureates Wolfgang Pauli and Niels Bohr (Figure 1) just represents an amusing metaphor of a scientific model for a component of the inanimate matter, namely the electron [2], it nonetheless implicates that some observations in research cannot be represented in any other way than in the form of models.

Figure 1.
Wolfgang Pauli and Niels Bohr are watching a spinning top as a model for the spinning electron. Photograph by Erik Gustafson, courtesy of AIP Emilio Segré Visual Archives. Courtesy of the Margrethe Bohr collection, Kopenhagen.
This becomes even more important when living systems like cells, three-dimensional tumors or even whole organisms with their emergent properties are considered [3,4]. A model also stands as a substitute for an inevitable reductionist approach to comprehending the complexity of an entity (e.g., primary tumors or metastases) on the basis of studying and knowing its parts (e.g., dysregulated signal transduction pathways, driver mutations) [5]. Accordingly, despite current initiatives to replace laboratory animals by sophisticated in vitro systems [6,7], biomedical research without animals as holistic models may fail to fulfill criteria and social demands of translatability of laboratory results into the clinic [8,9]. Conversely, although the “bench to bedside concept”, combined with personalized oncological treatment sounds attractive [10,11], preclinical models are further worthwhile to be funded in order to comprehend fundamental principles of cancer development without current quite obvious and ultimate clinical applications [12,13]. Clonal evolutions within tumors, for instance squamous cell carcinomas, result in tumor heterogeneity which represents an enormous problem for the treatment of cancer patients. Such evolutionary processes starting from initiation to metastasis, as recently shown in the “Confetti” mouse model, can only be obtained in vivo, but not in tissue culture [14].
Certainly, every model that represents a particular in vivo phenotype has its inherent limitations [15,16] and the choice of an animal species can even be decisive for conclusions or consequences to support future research strategies and/or programs [17,18]. A prominent historical example is the treatment of mice with penicillin, to show the therapeutic effect on staphylococcus infections. If hamsters or guinea pigs were utilized during these times, the proof-of-principle would have failed and the launch of antibiotics would have been delayed, since penicillin is highly toxic for both species [19,20]. Especially animal models for studying infectious diseases must be carefully selected. They should have similar routes of infection, should develop analogous symptoms and have to display comparable pathological changes as seen in humans [21].
Researchers introduced a plethora of animal models, such as zebrafish, rabbits, rats, dogs, pigs, goats, cattle and monkeys [22,23,24,25,26]. To be accepted as a valuable preclinical model, however, a scoring system should guarantee their careful selection, by reflecting face validity, complexity and predictability of a disease [27,28]. Nonetheless, the house mouse Mus musculus is still the best characterized organism used in biomedical research [29,30]. To create a homogeneous genetic background inbred mouse strains are used [31], a condition that may affect the experimental read-out of these model systems [27,32]. Continuous inbreeding may also result in so-called “spontaneous” models like the immunocompromised nude mouse, routinely used for xenotransplantation for several decades [33].
Nevertheless, research of infectious diseases requires models with a proper immune system to understand the spread and virulence of bacterial and viral infections, as well as the link between innate and the humoral immune responses. This aspect becomes even more important when immunosuppressive treatment is considered, that may lead to reactivation of persistent infections [34,35]. Only animals with a functioning immune system allow the development and testing of vaccination strategies, since prophylaxis is always superior to therapy. While transgenic, knock-in and knock-out mice are also valuable tools to investigate the complexity of physiological processes, natural animal models better reflect the reality with respect to the immunological surveillance of infections [36]. The holistic concept of an organism to understand the impact on the immune system, on cancer development or the efficiency of therapeutic drugs also becomes evident in current microbiome studies [37,38]. The generation of germ-free mice and their subsequent recolonization with defined microorganisms will show the impact of the microbiome or virome on the outcome of diseases [39,40]. Specific research fields and new trends in biomedical research indeed require and develop suitable nontraditional in vivo systems [41,42] as shown recently in the case of the crayfish as a novel model in epigenetics [43] and the naked mole rat as a model for chromosomal stability and senescence [44,45].
In the present overview, we describe the animal model Mastomys coucha. Although not yet very common in the scientific community, it has been known for several decades in various research fields and has recently attracted great attention in functional studies of cutaneous papillomaviruses and non-melanoma skin cancer (NMSC) formation.
2. Characteristics of Mastomys Species
The genus Mastomys belongs to the family Muridae (subfamily Murinae) and is a phylogenetic relative of mouse and rat [46]. It contains eight species referred to as multimammate mice or multimammate rats (also known as African soft-furred rats or African common rats) that can be found all over sub-Saharan Africa [47,48,49]. The animals have brown eyes and are usually covered by a dense agouti coat that is of lighter grey at the belly, although also other color variants, e.g., chamois-colored coat with pink eyes, exist in laboratory Mastomys strains (Figure 2A–D) [50]. The strain used at the German Cancer Research Center (DKFZ) for papillomavirus research and in other laboratories for parasitological studies is a variant with a chamois-colored coat and light red eyes, previously known as the GRA-Giessen strain (Figure 2E,F) [50]. With a typical head-and-body length of six to 17 cm and a tail length of six to 15 cm, Mastomys weight ranges between 20 and 100 g [51]. In animal housing M. coucha—especially the males—can reach weights of more than 160 g.
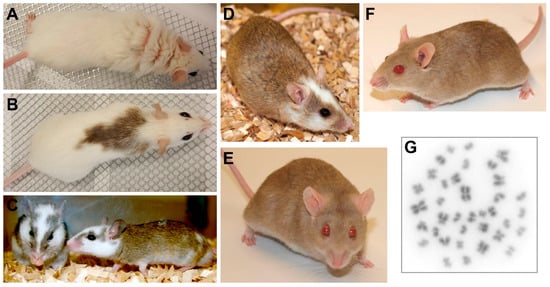
Figure 2.
The multimammate mouse Mastomys coucha. (A–D) Black-eyed Mastomys coucha with different coat colors. (E,F) Chamois-colored red-eyed Mastomys coucha used at the DKFZ (derived from the GRA-Giessen strain). (G) Metaphase chromosome spread obtained from a Mastomys coucha splenocyte (2n = 36; 630× magnification).
Like rats the animals do not have a gall bladder, but the females carry the characteristic eight to 18 pairs of mammary glands giving these animals the name multimammate mouse. The different Mastomys species can be roughly assigned to certain geographic locations, e.g., Southern Africa for M. coucha and M. natalensis, whereby the latter is also found in other regions [49]. The distinction of both species by superficial appearance is impossible and needs analysis of molecular markers e.g., by isoelectric focusing of serum proteins [52,53,54] or karyotyping (e.g., M. natalensis 2n = 32 vs. M. coucha 2n = 36; Figure 2G) [51,55]. This is why it was originally thought that both are the same species, but actually their mating only produces sterile offspring [56]. Of the several Mastomys species, M. natalensis and M. coucha are the ones that are used the most in biomedical research.
3. Housing of Mastomys in the Laboratory
The housing of M. natalensis and M. coucha is comparable to mice (Mus musculus) and rats (Rattus norvegicus). They develop well under typical animal facility conditions (20–25 °C, 55–65% humidity) and standard diets and have intermediate space requirements. They are very curious and explore wood shavings or paper towels newly introduced in the cage, which they also use for nest-building (Figure 3). Their gestation period is approximately 23 days and the litter size can be in maximum up to 19 [51], although it is usually around 10 newborns with a birth weight of two to three grams (Figure 3A–C). The young are weaned after approximately four weeks with a weight of approximately 12 g, become sexually mature after one to three months and can reach an age of up to 2.5 years [51].

Figure 3.
Young Mastomys coucha. (A) Young Mastomys usually huddle together. (B,C) Parental Mastomys, especially the mother, have a strong protective instinct and always stay close to their offspring.
4. Mastomys as Model Systems in Biomedical Research—Historical Flashback
Moving back in science history and inspecting different research fields, Mastomys is definitely an interesting animal species to study many infectious diseases and cancer. As already noticed very early, Mastomys is highly susceptible to plague without any resistance to Yersinia pestis [57], which 1939 initially led to the breeding of Mastomys for plague research at the Medical Ecology Center in Johannesburg, South-Africa. In 1974, a plague outbreak in Zimbabwe (the former Rhodesia) [58] emerged with Mastomys as its primary reservoir host which were in turn used to test attenuated Y. pestis strains for vaccination [59]. Although previously suggested that there may be different sibling species of Mastomys [60], the discrimination between M. natalensis and M. coucha via chromosomal G-banding, was not successful before 1977 [61] and in 1983 it turned out that the latter is actually sensitive to Y. pestis [62,63]. Interestingly, the geographical distribution of M. coucha also correlates with human plague, while M. natalensis is resistant and predominates in areas without recorded human plague [62,64].
In 1969, a new virus was isolated from three American missionary nurses who worked in a town called Lassa and incurred a severe and previously undescribed hemorrhagic fever [65]. The identified so called Lassa virus (LASV), a member of the Arenaviridae, emerged to be a major public health problem in Western Africa during the following three years [66]. Here, M. natalensis was found to be the reservoir of this virus [66,67] and is still used as an animal model [68,69].
Mastomys are also highly susceptible to several helminths (nematodes, cestodes, trematodes) [70] and experiments with chemotherapeutics on Schistosoma mansoni-infected animals were performed already decades ago [71]. Filarial parasites like Wuchereria bancrofti, Brugia malayi and Brugia timori occupy the lymphatic system and cause the mosquito-borne disease lymphatic filariasis [72], characterized by long-term disability and severe immunopathology, e.g., elephantiasis [73]. Since available drugs only have low macrofilaricidal activity and have to be applied in long-term regimens, there is a need for new treatment and prevention strategies. In this context, M. coucha served as a permissive system for B. malayi infection to study immune responses [74,75,76,77], as well as a preclinical model for novel vaccines, including the use of recombinant filarial proteins [78,79,80,81] and DNA vaccination approaches [82].
Mastomys have also been reported to develop spontaneous adenocarcinomas and gastric carcinoids in high frequencies and were in turn used to study these stomach cancers [83,84,85,86,87,88]. In humans, gastritis and carcinoids can be triggered—amongst other reasons—by gram-negative bacteria of the genus Helicobacter. The main member H. pylori is prevalent in approximately 50% of the global human population (estimated 4.4 billion people) [89] and responsible for 75% of non-cardia gastric carcinomas worldwide [90]. Consequently, H. pylori was classified as a carcinogen for humans [91]. Mastomys have been used to study the effects of H. pylori-colonization on carcinoid formation [92]. Distributed throughout the gastrointestinal epithelium, so-called enterochromaffin (EC) cells are regarded as the predominant neuroendocrine cells of the bowel [93] and enterochromaffin-like (ECL) cells in the stomach regulate acid secretion [94]. As shown in a Mastomys-derived gastric enterochromaffin-like (ECL) cell neoplasia in vitro model, H. pylori lipopolysaccharides (LPS) have mitogenic effects on tumor ECL cells [95]. Notably, in 2005, during analyses of specimens from Mastomys, the novel Helicobacter species H. mastomyrinus has been isolated [96], which can cause severe inflammatory bowel disease in certain mouse strains [97].
Like mice and rats, Mastomys were shown to be susceptible to the Murine Sarcoma Virus-Harvey (MSV-H) and develop erythroblastic splenomegaly and large sarcomas upon infection [98]. In the past, Mastomys were also infected with autonomous parvoviruses (minute virus of mice prototype strain, MVMp and H-1) to evaluate their usability for parvovirus-based vectors. However, this was not found to be applicable, since both viruses were pathogenic for Mastomys while harmless for mice and rats, respectively [99].
Notably, in 2011 a novel polyomavirus (PyV) was identified in a wild African Mastomys that is phylogenetically closely related to the murine pneumotropic PyV [100] and was named Mastomys PyV (MasPyV). Like human polyomaviruses, e.g., BKV, JCV or MCPyV, causing hemorrhagic cystitis [101], progressive multifocal leukoencephalopathy [102] or Merkel-cell carcinoma [103], respectively, MasPyV persistently infect their host cells and do not cause pathological symptoms under immunocompetent conditions [100,104].
In the following sections, we will provide some historical and recent aspects about how the animal model Mastomys coucha contributes to the understanding of cutaneous papillomaviruses and their role in the development of non-melanoma skin cancer (NMSC).
5. Mastomys coucha as a Preclinical Model in Papillomavirus Research
5.1. History of Mastomys coucha in Papillomavirus Research
The use of Mastomys coucha in papillomavirus (PV) research is attractive, since it offers the possibility to study the function of a cutaneous PV in a natural infection model. The colony currently housed at the German Cancer Research Center (DKFZ) in Heidelberg (Germany) emerged from the chamois-colored, red-eyed GRA-Giessen strain that was held after 1966 at the Institute for Parasitology in Giessen (Germany). Their offspring were transferred to the DKFZ in 1969, because initially a high incidence of stomach cancer was noticed (Oettle, 1957). Whether these particular tumors were induced by H. mastomyrinus [96], by papillomaviruses [105] or by both is still an interesting question that remains to be clarified. Spontaneously appearing epithelial skin lesions in the GRA-Giessen strain were first described and initially classified as “so-called” keratoacanthomas [106,107]. Due to the endemic occurrence in the colony, a virus was suspected to be the etiological agent of these lesions.
Indeed, when transferring homogenized cell-free tumor tissue to scarified skin of different animals, according to the Koch postulates [108], new tumors indistinguishable from spontaneous tumors emerged [106]. Morphologically identical viral particles could be isolated from these lesions (papillomas and keratoacanthomas, KAs) [109] (Figure 4A,B), as well as from well-differentiated squamous cell carcinomas (SCCs) [110,111,112]. Serum obtained from rabbits immunized with purified virions could further prevent tumor formation in experimentally infected animals [112].
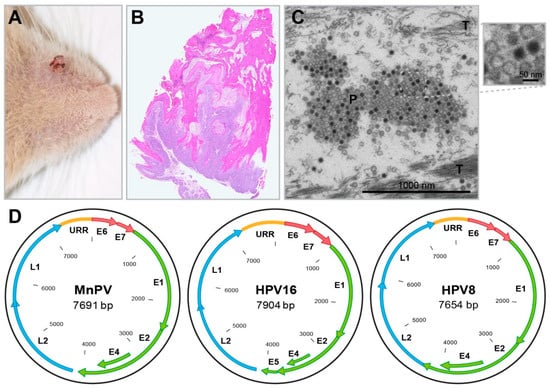
Figure 4.
The Mastomys natalensis papillomavirus. (A) A spontaneous skin tumor (papilloma) near the nose. (B) HE staining of a spontaneous skin tumor (“so-called” keratoacanthoma) shows typical endo-exophytic growth and strong keratinization. (C) EM micrograph of MnPV particles (P) in the most upper layer of a spontaneous MnPV-induced skin lesion. While larger host cell compartments are already degraded during terminal differentiation and desquamation, tonofibrils (T) are still visible. (D) Schematic representation of the genomes of HPV16 (an alpha-type), HPV8 (a beta-type) and MnPV (an iota-type). While the genomes of all PV types harbor an upstream regulatory region (URR) and code for early genes (E1, E2, E4, E6, E7) and late genes (L1, L2), PVs from the genus beta and iota do not contain E5 open reading frames.
Viral particles can be found in large amounts in nuclei of tumor cells and keratinized masses of skin tumors [110], as well as upper parts of the stratum granulosum and stratum corneum [113] (Figure 4C). They were structurally characterized as a typical papillomavirus of 52 nm in size, referred to as Mastomys natalensis papillomavirus (MnPV) [112]. The sequence of the MnPV genome contains open reading frames for the early genes E6, E7, E1, E2 and E4, the late genes L1 and L2, as well as an untranslated URR (upstream regulatory region) (Figure 4D) [114]. The E5 gene, known from mucosal alpha-type human papillomaviruses (HPVs), is missing—a feature shared with cutaneous beta-type HPVs [115]. These, like HPV5 or HPV8, cause epithelial lesions or tumors especially in organ transplant recipients [116,117], but also in immunocompetent individuals [118].
Apparently, the so-called KAs observed in Mastomys (Figure 4B) differed from KAs known from humans regarding histology and time course. While, macroscopically, a strong keratinization and infiltration to surrounding tissue were similar, human KAs were described as emerging from hair follicles [119], while those in Mastomys seemed to arise from the surface and at best at the infundibulum of the hair follicle [109]. However, considering later data obtained by in-situ hybridization, MnPV as the etiological agent for papillomas, KAs and SCCs could be also detected in hair follicle cells, being apparently a reservoir for these viruses [105]. Up to date, a spontaneous regression described for human KAs was never reported for Mastomys where the tumor continues growing and infiltrating even down to the muscle tissue [110].
Human skin is an open ecosystem [120] and becomes colonized already shortly after birth by a wide range of cutaneous HPV types [121]. A homologous validity is true for Mastomys coucha, since MnPV DNA in normal skin, as well as seroconversion, can be detected at 4–5 weeks of age, probably, due to nursing between the mother and their offspring [122]. Due to a natural infection route within the colony and the occurrence of MnPV-positive skin lesions, Mastomys were found to be an ideal model system to investigate the contribution of MnPV to skin carcinogenesis. As shown in previous studies, persisting MnPV genomes in the skin become activated after administration of 7,12-dimethyl-benz(a)anthracene (DMBA) and/or 12-O-Tetradecanoylphorbol-13-acetate (TPA) resulting in the formation of both benign and malignant skin tumors [123,124]. The oncogenic potential of MnPV was also demonstrated in MnPV-E6 transgenic mice (E7 transgenic animals died in utero for unknown reasons), showing nearly a 100% formation of SCCs upon DMBA/TPA treatment compared to only 10% of their non-transgenic littermates [125]. Interestingly, while chemically induced SCCs in wildtype mice usually harbor specific DMBA-induced activating Hras mutations that favor cell growth upon TPA treatment [126], skin tumors obtained in MnPV-E6 transgenic mice consistently contained wild-type Ras at all three hot-spot positions [125]. Intriguingly, the same mutual exclusion of papillomavirus positivity and Hras mutations could be observed in SCCs from melanoma patients after treatment with BRAF inhibitors, such as Vemurafenib [127], indicating that cutaneous HPVs (similar to MnPV) may substitute or circumvent activating Hras mutations [128].
Chemical DMBA/TPA-induced skin carcinogenesis—although providing interesting insight into molecular mechanisms—do not really reflect physiological skin tumor development in humans [129]. More equivalent to tumor physiological promoting mechanisms was the observation that latent MnPV genomes in the skin can be activated by wounding or chronic mechanical irritation (repeated scratching of the skin with glass paper), finally leading to SCC formation [130]. Conversely, in the case of NMSC, the most important environmental risk factor, namely UV exposure, was still unnoticed at that time. However, transgenic mice [131,132] or animals naturally infected with genuine papillomaviruses [133,134] recently filled this gap, focusing on the functional interaction between PV infection and UV exposure. While a synergistic effect of UV and wound healing could be shown in transgenic HPV8-CER (complete early region) or HPV8-E6 mice [135], the effect of UV exposure on a natural PV infection was investigated later after the development of sensitive tools and methods needed to measure the course of a natural infection with minor amounts of viral DNA together with solid prospective serological data.
5.2. Recent Contributions of Mastomys coucha to PV Research as a Preclinical Model
While previous reports mainly focused on the final manifestation of skin tumors and the viral presence in those lesions, the subsequent studies in this research field were also considering the immunological consequences and the molecular mechanisms of the interplay between virus and host. Experimental techniques, such as PCR and qPCR, DNA/RNA sequencing and ELISAs, to monitor the immune response against individual viral proteins were not available during that time. Remarkably, the systematic analysis of body compartments by PCR of these animals showed that besides the skin, also other organs, e.g., stomach, lungs and liver were positive for MnPV DNA, while fetuses or newborns were negative [105]. Although viral DNA could also be detected in the blood, inner organs were not necessarily positive in the same animal. MnPV presence in the blood must be merely transient (temporary viremia), but apparently is the only way of viral spread to inner organs, because no other routes are plausible [105]. It turned out that the incidence of tumors strongly depends on the number of MnPV genomes in healthy skin, which enrich as the animal becomes older [105].
Analysis of viral gene expression in skin samples with productive infection identified a complex MnPV transcription pattern, revealing novel splicing isoforms that have not yet been described for other papillomaviruses [136], but can be further studied in the meanwhile in recently established Mastomys-derived keratinocyte and fibroblast cell lines [137]. Moreover, comparable to HPVs, two promoters for early and late transcripts and two different polyadenylation sites were identified within the MnPV genome. Estimation of expression levels of each transcript showed that L1 and E1^E4 mRNAs were the most abundant [136]. Consistent with episomal replication under permissive conditions, RNAseq did not reveal any viral-cellular fusion transcripts. The comprehensive transcription map provides a basis for understanding MnPV pathogenesis and allows the identification of transcripts expressed within tissue samples on a spatial level.
Moreover, improved detection methods also identified a new papillomavirus (McPV2) (Figure 5A), which is phylogenetically distant from MnPV and mainly found in anogenital lesions (Figure 5B,D), but also in other organs and mucosal tissues, such as the oral cavity or the tongue (Figure 5C,E) [138]. In that way, Mastomys coucha is the only model where the biology of both a cutaneous (MnPV) and mucosal papillomavirus (McPV2) can be investigated in the same animal. Here, as measured with glutathione S-transferase (GST)-capture ELISAs, the seroreactivity against the L1 capsid proteins of both viruses was found to be significantly increased in animals bearing the respective skin or anogenital lesions [139].
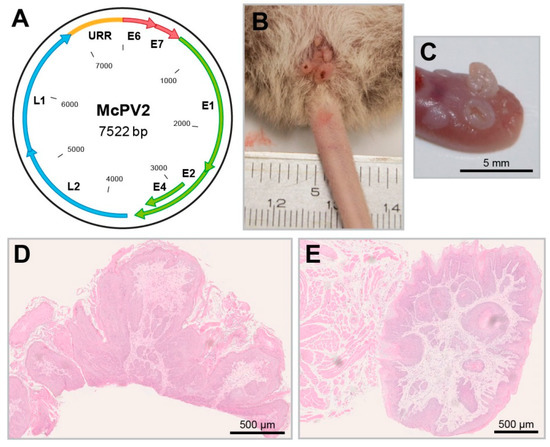
Figure 5.
The Mastomys coucha papillomavirus 2. (A) Schematic representation of the McPV2 genome. (B) Condylomas in the anogenital region induced by McPV2. (C) Tongue papillomas induced by McPV2 (these are frequently positive for MnPV as well). (D) HE
staining of a condyloma. (E) HE staining of a tongue papilloma.
Papillomavirus serology of individual viral proteins additionally allows prospective studies in terms of the time course of infection and skin lesion development. Particularly antibodies against the early E2 proteins of both viruses already appear in one month old animals [122], while the immune response against the late L1 viral proteins is delayed, appearing after approximately 2.5 months, but indicative for productive viral infections [122]. Notably, in humans, seropositivity against cutaneous HPVs can also be detected already in early childhood [121], which at least in the case of certain types is associated with an increased risk for development of SCCs [140]. Seroconversion against MnPV L1 and E2 correlates with the manifestation of lesions and can be used as a marker of a current or a preceding infection. Hence, both serology and the appearance of lesions in the immunocompetent host-virus system Mastomys coucha offer the opportunity to test prophylactic and therapeutic strategies against infection with cutaneous papillomaviruses.
Nevertheless, although Mastomys coucha serves as a suitable natural model for different diseases including cancer, as in every model, it also has its limitations. Here, it is mainly the lack of standardized molecular and immunological tools, e.g., primer sequences for the measurement of gene expression levels or different immune responses. Established antibodies, for instance, although specific for mouse or/and rat, do not necessarily cross-react with Mastomys proteins and need to be carefully tested. Therefore, before investigating certain scientific questions with this animal—e.g., how infections are controlled, including the immunological surveillance of a commensal virus like MnPV and how this virus finally contributes to NMSC—it may be a challenge to establish needed tools. The future lab work is simplified, since the genome of Mastomys coucha was recently sequenced at the Institute for Human Genetics at the UCSF, San Francisco [141].
6. Vaccination and Tumor Prevention in Mastomys coucha
The success seen after immunization with virus-like particles (VLPs) against mucosal high-risk HPVs causing anogenital tumors also prompted the development of prophylactic vaccines against cutaneous PVs [142]. Especially for immunosuppressed organ transplant recipients, vaccination would be of great benefit, since up to estimated 40% of these patients develop NMSC within 10 years after transplantation and up to 80% after 20 years [143,144,145]. Their risk of developing such lesions is correlated with the HPV load in plugged eyebrow hair follicles [146]. The efficacy of such a vaccine on either ongoing or newly established MnPV infections was examined in the Mastomys model. For this purpose, an MnPV-free Mastomys colony was generated via hysterectomy to allow infections under defined experimental conditions. Using MnPV-VLPs, a strong and long-lasting immune response was established in virus-infected and virus-free animals even under systemic long-term immunosuppression. It comprised of high titers of neutralizing antibodies, as measured by pseudovirion-based neutralization assays [147]. In all cases, the formation of both benign and malignant skin lesions was completely prevented and further led to a significant reduction of MnPV DNA loads in normal skin [148]. This study provided the first evidence that a VLP-based vaccine can trigger an effective immune response in the skin irrespective of the immune and infection status at the time of vaccination. Thinking in translatability, this was an important proof-of-principle for the clinical development and application of vaccines against skin tumors caused by cutaneous HPV infection, especially in patients awaiting an organ transplant.
7. The Role of UV Exposure and Papillomavirus Infection in NMSC Development
Numerous seroepidemiological studies already support an association between infection with certain cutaneous HPV types and NMSC [140,149]. However, the sporadic absence of viral DNA within malignant lesions still raises skepticism whether HPVs (i) act via a “hit-and-run” mechanism, (ii) are just opportunistic bystanders or (iii) there are two independent events in the formation of virus-positive and -negative SCCs in the context of cumulative UV exposure as environmental risk factor [128].
In recent experiments, Mastomys coucha served as a model to decipher this fundamental question by studying the effect of UV exposure on MnPV-positive and -negative skin [134]. For this purpose, virus-free and naturally infected animals were chronically exposed to UVB light and the incidence of skin tumors was monitored. Notably, UVB irradiation (corresponding to UV doses of different geographical regions) only led to significant tumor formation in MnPV-infected skin. Two distinct tumor types were induced within a time frame and histopathology similar to humans (Figure 6A,B): Well-differentiated keratinizing SCCs (KSCC) that still support productive infections with high viral DNA loads and transcription and poorly differentiated non-keratinizing SCCs (nKSCC) (Figure 6C,D). The latter type may cause a microenvironment that counteracts viral propagation, which explains the low viral loads or even the lack of MnPV DNA and transcription [134]. Nevertheless, all tumor-bearing animals developed MnPV-specific antibodies, directly corroborating a preceding infection, a scenario also reported in SCC patients [150].
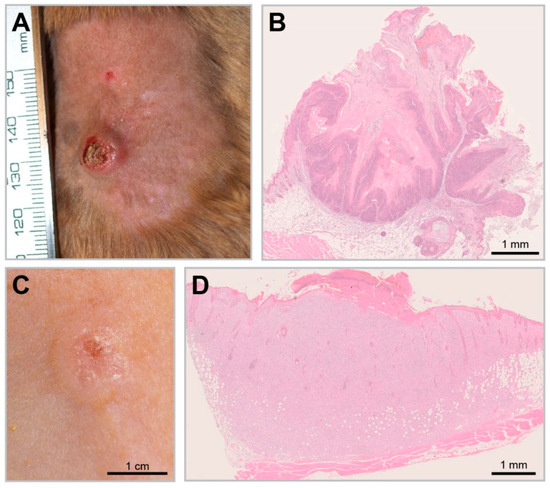
Figure 6.
UV-induced SCCs in the Mastomys model. (A) A UV-induced KSCC. (B) KSCCs are characterized by the growth of well-differentiated squamous cells and show strong keratinization comparable to spontaneous MnPV-induced tumors (HE staining). (C) A UV-induced nKSCC. (D) HE staining of a poorly differentiated nKSCC.
MnPV infection apparently leads to an attenuation of DNA repair and genomic stability and favors the accumulation of UV-induced mutations, including in Trp53. Especially two hot-spot positions—also well-known from human SCCs [151]—within the DNA-binding domain of p53 were found [134]. Since it is known that the loss of functional epidermal Trp53 in mice can favor the expansion of poorly differentiated SCCs, a decisive role of mutated p53 on the phenotype of squamous cells can be anticipated [152]. In other words, functional loss of epidermal p53 in Mastomys tumors may favor dedifferentiation [153], which counteracts the permissive cycle and explains the differences in viral load between KSCCs and nKSCCs. Such a “hit-and-run” mechanism is a reasonable explanation for dispensable continuous viral presence during skin carcinogenesis as observed in patients with NMSC [128,134]. It provides the basis to investigate the temporal and spatial order of events underlying papillomavirus-driven skin carcinogenesis and for the development of preventive or curative strategies against NMSC.
8. Summary
As outlined above, particular scientific questions often require suitable animal systems. Although paradigmatic models like mice may be attractive and “well-accepted” within the scientific community, one should be open for nontraditional models that already exist in nature with great similarities to human diseases. The African rodent Mastomys coucha has a long history in various research fields, including parasite, bacteria and virus research. In the latter context, due to the high degree of resemblance with humans in terms of viral skin infections, Mastomys coucha is an easy-to-handle laboratory animal that represents a unique and powerful model to investigate basic molecular interactions between papillomaviruses and their natural host. Moreover, due to the presence of two papillomaviruses in the same animal, broad-protective vaccines [154] can be tested under different conditions.
Author Contributions
This was an invited review to FR. All authors listed, have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
Infect-ERA III: HPV-MOTIVA “BETA-TYPE HPV: INFECTION AND VACCINES” 031L0095B to FR.
Acknowledgments
We thank K. Richter and M. Neßling (Central Unit Electron Microscopy, DKFZ) for the acquisition of EM images. Support and assistance by the animal technicians and veterinarians of the Center for Preclinical Research, DKFZ is also gratefully acknowledged.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Rheinberger, H.J. Preparations, models, and simulations. Hist. Philos. Life Sci. 2015, 36, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Wright, A. Milestone 3: (1925) the Spinning Electron. Available online: https://www.nature.com/milestones/milespin/pdf/milespin03.pdf (accessed on 22 January 2019).
- Van Regenmortel, M.H. Reductionism and complexity in molecular biology. Scientists now have the tools to unravel biological and overcome the limitations of reductionism. EMBO Rep. 2004, 5, 1016–1020. [Google Scholar] [PubMed]
- Sverdlov, E.D. Unsolvable problems of biology: It is impossible to create two identical organisms, to defeat cancer, or to map organisms onto their genomes. Biochemistry 2018, 83, 370–380. [Google Scholar] [CrossRef] [PubMed]
- Massoud, T.F.; Hademenos, G.J.; Young, W.L.; Gao, E.; Pile-Spellman, J.; Vinuela, F. Principles and philosophy of modeling in biomedical research. FASEB J. 1998, 12, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Shamir, E.R.; Ewald, A.J. Three-dimensional organotypic culture: Experimental models of mammalian biology and disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 647–664. [Google Scholar] [CrossRef] [PubMed]
- Horvath, P.; Aulner, N.; Bickle, M.; Davies, A.M.; Nery, E.D.; Ebner, D.; Montoya, M.C.; Ostling, P.; Pietiainen, V.; Price, L.S.; et al. Screening out irrelevant cell-based models of disease. Nat. Rev. Drug Discov. 2016, 15, 751–769. [Google Scholar] [CrossRef] [PubMed]
- Day, C.P.; Merlino, G.; van Dyke, T. Preclinical mouse cancer models: A maze of opportunities and challenges. Cell 2015, 163, 39–53. [Google Scholar] [CrossRef]
- Dolgos, H.; Trusheim, M.; Gross, D.; Halle, J.P.; Ogden, J.; Osterwalder, B.; Sedman, E.; Rossetti, L. Translational medicine guide transforms drug development processes: The recent merck experience. Drug Discov. Today 2016, 21, 517–526. [Google Scholar] [CrossRef]
- Delaney, A.; Angus, D.C.; Bellomo, R.; Cameron, P.; Cooper, D.J.; Finfer, S.; Harrison, D.A.; Huang, D.T.; Myburgh, J.A.; Peake, S.L.; et al. Bench-to-bedside review: The evaluation of complex interventions in critical care. Crit. Care 2008, 12, 210. [Google Scholar] [CrossRef]
- Hobin, J.A.; Galbraith, R.A. Engaging basic scientists in translational research. FASEB J. 2012, 26, 2227–2230. [Google Scholar] [CrossRef]
- Lazebnik, Y. Are scientists a workforce?—Or, how Dr. Frankenstein made biomedical research sick: A proposed plan to rescue us biomedical research from its current “malaise” will not be effective as it misdiagnoses the root cause of the disease. EMBO Rep. 2015, 16, 1592–1600. [Google Scholar] [CrossRef] [PubMed]
- Bentley, P.J.; Gulbrandsen, M.; Kyvik, S. The relationship between basic and applied research in universities. High Educ. 2015, 70, 689–709. [Google Scholar] [CrossRef]
- Reeves, M.Q.; Kandyba, E.; Harris, S.; Del Rosario, R.; Balmain, A. Multicolour lineage tracing reveals clonal dynamics of squamous carcinoma evolution from initiation to metastasis. Nat. Cell Biol. 2018, 20, 699–709. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, E.R.; Shmulevich, I. On the limitations of biological knowledge. Curr. Genom. 2012, 13, 574–587. [Google Scholar] [CrossRef]
- Anisimov, V.N.; Ukraintseva, S.V.; Yashin, A.I. Cancer in rodents: Does it tell us about cancer in humans? Nat. Rev. Cancer 2005, 5, 807–819. [Google Scholar] [CrossRef]
- Moher, D.; Glasziou, P.; Chalmers, I.; Nasser, M.; Bossuyt, P.M.; Korevaar, D.A.; Graham, I.D.; Ravaud, P.; Boutron, I. Increasing value and reducing waste in biomedical research: Who’s listening? Lancet 2016, 387, 1573–1586. [Google Scholar] [CrossRef]
- Chalmers, I.; Bracken, M.B.; Djulbegovic, B.; Garattini, S.; Grant, J.; Gulmezoglu, A.M.; Howells, D.W.; Ioannidis, J.P.; Oliver, S. How to increase value and reduce waste when research priorities are set. Lancet 2014, 383, 156–165. [Google Scholar] [CrossRef]
- Schneierson, S.S.; Perlman, E. Toxicity of penicillin for the syrian hamster. Proc. Soc. Exp. Biol. Med. 1956, 91, 229–230. [Google Scholar] [CrossRef] [PubMed]
- Green, R.H. The association of viral activation with penicillin toxicity in guinea pigs and hamsters. Yale J. Biol. Med. 1974, 47, 166–181. [Google Scholar]
- Rand, M.S. Selection of biomedical animal models. In Sourcebook of Models for Biomedical Research; Conn, P.M., Ed.; Humana Press Inc.: Totowa, NJ, USA, 2008; pp. 9–16. [Google Scholar]
- Sarasamma, S.; Lai, Y.H.; Liang, S.T.; Liu, K.; Hsiao, C.D. The power of fish models to elucidate skin cancer pathogenesis and impact the discovery of new therapeutic opportunities. Int. J. Mol. Sci. 2018, 19, 3929. [Google Scholar] [CrossRef]
- Zhao, S.; Huang, J.; Ye, J. A fresh look at zebrafish from the perspective of cancer research. J. Exp. Clin. Cancer Res. 2015, 34, 80. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.; Orme, I.M. Animal models of tuberculosis: An overview. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Christensen, N.D.; Budgeon, L.R.; Cladel, N.M.; Hu, J. Recent advances in preclinical model systems for papillomaviruses. Virus Res. 2017, 231, 108–118. [Google Scholar] [CrossRef] [PubMed]
- McGovern, J.A.; Griffin, M.; Hutmacher, D.W. Animal models for bone tissue engineering and modelling disease. Dis. Model. Mech. 2018, 11, dmm033084. [Google Scholar] [CrossRef]
- Workman, P.; Aboagye, E.O.; Balkwill, F.; Balmain, A.; Bruder, G.; Chaplin, D.J.; Double, J.A.; Everitt, J.; Farningham, D.A.; Glennie, M.J.; et al. Guidelines for the welfare and use of animals in cancer research. Br. J. Cancer 2010, 102, 1555–1577. [Google Scholar] [CrossRef] [PubMed]
- Denayer, T.; Stöhr, T.; van Roy, M. Animal models in translational medicine: Validation and prediction. New Horiz. Transl. Med. 2014, 2, 5–11. [Google Scholar] [CrossRef]
- Khaled, W.T.; Liu, P. Cancer mouse models: Past, present and future. Semin. Cell Dev. Biol. 2014, 27, 54–60. [Google Scholar] [CrossRef]
- Justice, M.J.; Dhillon, P. Using the mouse to model human disease: Increasing validity and reproducibility. Dis. Model. Mech. 2016, 9, 101–103. [Google Scholar] [CrossRef]
- Beck, J.A.; Lloyd, S.; Hafezparast, M.; Lennon-Pierce, M.; Eppig, J.T.; Festing, M.F.; Fisher, E.M. Genealogies of mouse inbred strains. Nat. Genet. 2000, 24, 23–25. [Google Scholar] [CrossRef]
- Hau, J. Animal models for human diseases. In Sourcebook of Models for Biomedical Research; Conn, P.M., Ed.; Humana Press Inc.: Totowa, NJ, USA, 2008; pp. 3–8. [Google Scholar]
- Pelleitier, M.; Montplaisir, S. The nude mouse: A model of deficient t-cell function. Methods Achiev. Exp. Pathol. 1975, 7, 149–166. [Google Scholar]
- Geissler, E.K. Post-transplantation malignancies: Here today, gone tomorrow? Nat. Rev. Clin. Oncol. 2015, 12, 705–717. [Google Scholar] [CrossRef] [PubMed]
- Arroyo Muhr, L.S.; Bzhalava, Z.; Hortlund, M.; Lagheden, C.; Nordqvist Kleppe, S.; Bzhalava, D.; Hultin, E.; Dillner, J. Viruses in cancers among the immunosuppressed. Int. J. Cancer. 2017, 141, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Abolins, S.R.; Pocock, M.J.; Hafalla, J.C.; Riley, E.M.; Viney, M.E. Measures of immune function of wild mice, mus musculus. Mol. Ecol. 2011, 20, 881–892. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Wu, J.; Jin, D.; Wang, B.; Cao, H. Fecal microbiota transplantation in cancer management: Current status and perspectives. Int. J. Cancer. 2018. [Google Scholar] [CrossRef] [PubMed]
- Hamada, T.; Nowak, J.A.; Milner, D.A., Jr.; Song, M.; Ogino, S. Integration of microbiology, molecular pathology, and epidemiology: A new paradigm to explore the pathogenesis of microbiome-driven neoplasms. J. Pathol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.Z.; Ravindran, R.; Chassaing, B.; Carvalho, F.A.; Maddur, M.S.; Bower, M.; Hakimpour, P.; Gill, K.P.; Nakaya, H.I.; Yarovinsky, F.; et al. Tlr5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity 2014, 41, 478–492. [Google Scholar] [CrossRef]
- Kennedy, E.A.; King, K.Y.; Baldridge, M.T. Mouse microbiota models: Comparing germ-free mice and antibiotics treatment as tools for modifying gut bacteria. Front. Physiol. 2018, 9, 1534. [Google Scholar] [CrossRef]
- Conti, F.; Abnave, P.; Ghigo, E. Unconventional animal models: A booster for new advances in host-pathogen interactions. Front. Cell Infect. Microbiol. 2014, 4, 142. [Google Scholar] [CrossRef]
- Gladfelter, A.S. How nontraditional model systems can save us. Mol. Biol. Cell 2015, 26, 3687–3689. [Google Scholar] [CrossRef]
- Gutekunst, J.; Andriantsoa, R.; Falckenhayn, C.; Hanna, K.; Stein, W.; Rasamy, J.; Lyko, F. Clonal genome evolution and rapid invasive spread of the marbled crayfish. Nat. Ecol. Evol. 2018, 2, 567–573. [Google Scholar] [CrossRef]
- Petruseva, I.O.; Evdokimov, A.N.; Lavrik, O.I. Genome stability maintenance in naked mole-rat. Acta Nat. 2017, 9, 31–41. [Google Scholar]
- Zhao, Y.; Tyshkovskiy, A.; Munoz-Espin, D.; Tian, X.; Serrano, M.; de Magalhaes, J.P.; Nevo, E.; Gladyshev, V.N.; Seluanov, A.; Gorbunova, V. Naked mole rats can undergo developmental, oncogene-induced and DNA damage-induced cellular senescence. Proc. Natl. Acad. Sci. USA 2018, 115, 1801–1806. [Google Scholar] [CrossRef]
- Jansa, S.A.; Giarla, T.C.; Lim, B.K. The phylogenetic position of the rodent genus typhlomys and the geographic origin of muroidea. J. Mammal. 2009, 90, 1083–1094. [Google Scholar] [CrossRef]
- Lecompte, E.; Granjon, L.; Denys, C. The phylogeny of the praomys complex (rodentia: Muridae) and its phylogeographic implications. J. Zool. Syst. Evol. Res. 2002, 40, 8–25. [Google Scholar] [CrossRef]
- Chevret, P.; Granjon, L.; Duplantier, J.M.; Denys, C.; Catzeflis, F.M. Molecular phylogeny of the praomys complex (rodentia, murinae)—A study based on DNA/DNA hybridization experiments. Zool. J. Linn. Soc.-Lond. 1994, 112, 425–442. [Google Scholar] [CrossRef]
- Granjon, L.; Duplantier, J.M.; Catalan, J.; BrittonDavidian, J. Systematics of the genus mastomys (thomas, 1915) (rodentia: Muridae)—A review. Belg. J. Zool. 1997, 127, 7–18. [Google Scholar]
- Solleveld, H.A. The multimammate mouse. In The Ufaw Handbook on the Care and Management of Laboratory Animals, Subsequent edition (1 June 1987) ed.; Churchill Livingstone Inc.: New York, NY, USA, 1987; p. 948. [Google Scholar]
- Weiss, J.; Becker, K.; Bernsmann, E.; Chourbaji, S.; Dietrich, H. Versuchstierkunde: Tierpflege in Forschung und klinik, 4th ed.; Enke: Stuttgart, Germany, 2014. [Google Scholar]
- Smit, A.; van der Bank, H.; Falk, T.; de Castro, A. Biochemical genetic markers to identify two morphologically similar south african mastomys species (rodentia: Muridae). Biochem. Syst. Ecol. 2001, 29, 21–30. [Google Scholar] [CrossRef]
- Smit, A.A.; van der Bank, H.F. Isozyme and allozyme markers distinguishing two morphologically similar, medically important mastomys species (rodentia: Muridae). BMC Genet. 2001, 2, 15. [Google Scholar] [CrossRef]
- Kruppa, T.F.; Iglauer, F.; Ihnen, E.; Miller, K.; Kunstyr, I. Mastomys natalensis or mastomys coucha. Correct species designation in animal experiments. Trop. Med. Parasitol. 1990, 41, 219–220. [Google Scholar]
- Britton-Davidian, J.; Catalan, J.; Granjon, L.; Duplantier, J.M. Chromosomal phylogeny and evolution in the genus mastomys (mammalia, rodentia). J. Mammal. 1995, 76, 248–262. [Google Scholar] [CrossRef]
- Hallett, J.M. Cytological and Cytogenetical Studies on the Multimammate Mouse Praomys (Mastomys) Natalensis. Ph.D. Thesis, University of the Witwatersrand, Johannesburg, South Africa, 1977. [Google Scholar]
- Davis, D.H.S.; Heisch, R.B.; Mcneill, D.; Meyer, K.F. Serological survey of plague in rodents and other small mammals in kenya. Trans. R. Soc. Trop. Med. Hyg. 1968, 62, 838–861. [Google Scholar] [CrossRef]
- Pugh, A.O.; Parker, D.A. Plague: Rhodesia’s first recorded outbreak. Cent. Afr. J. Med. 1975, 21, 93–96. [Google Scholar]
- Hallett, A.F. Evaluation of live attenuated plague vaccines in praomys (mastomys) natalensis. Infect. Immun. 1977, 18, 8–13. [Google Scholar] [PubMed]
- Matthey, R. Cytogénétique et taxonomie des rats appartenant au sous-genre mastomys thomas (rodentia-muridae). Mammalia 1966, 30, 105–119. [Google Scholar] [CrossRef]
- Lyons, N.F.; Green, C.R.; Gordon, D.H.; Walters, C.R. G-banding chromosome analysis of Praomys natalensis-(smith) (Rodentia muridae) from rhodesia. 1. 36-chromosome population. Heredity 1977, 38, 197. [Google Scholar] [CrossRef]
- Isaacson, M.; Taylor, P.; Arntzen, L. Ecology of plague in africa: Response of indigenous wild rodents to experimental plague infection. Bull. World Health Organ. 1983, 61, 339–344. [Google Scholar]
- Arntzen, L.; Wadee, A.A.; Isaacson, M. Immune responses of two mastomys sibling species to yersinia pestis. Infect. Immun. 1991, 59, 1966–1971. [Google Scholar]
- Green, C.A.; Keogh, H.; Gordon, D.H.; Pinto, M.; Hartwig, E.K. The distribution, identification, and naming of the mastomys natalensis species complex in southern africa (rodentia: Muridae). J. Zool. 1980, 192, 17–23. [Google Scholar] [CrossRef]
- Frame, J.D.; Baldwin, J.M., Jr.; Gocke, D.J.; Troup, J.M. Lassa fever, a new virus disease of man from west africa. I. Clinical description and pathological findings. Am. J. Trop. Med. Hyg. 1970, 19, 670–676. [Google Scholar] [CrossRef]
- Monath, T.P.; Newhouse, V.F.; Kemp, G.E.; Setzer, H.W.; Cacciapuoti, A. Lassa virus isolation from mastomys natalensis rodents during an epidemic in sierra leone. Science 1974, 185, 263–265. [Google Scholar] [CrossRef]
- Lecompte, E.; Fichet-Calvet, E.; Daffis, S.; Koulemou, K.; Sylla, O.; Kourouma, F.; Dore, A.; Soropogui, B.; Aniskin, V.; Allali, B.; et al. Mastomys natalensis and lassa fever, west africa. Emerg. Infect. Dis. 2006, 12, 1971–1974. [Google Scholar] [CrossRef]
- Gryseels, S.; Baird, S.J.; Borremans, B.; Makundi, R.; Leirs, H.; Gouy de Bellocq, J. When viruses don’t go viral: The importance of host phylogeographic structure in the spatial spread of arenaviruses. PLoS Pathog. 2017, 13, e1006073. [Google Scholar] [CrossRef] [PubMed]
- Mari Saez, A.; Cherif Haidara, M.; Camara, A.; Kourouma, F.; Sage, M.; Magassouba, N.; Fichet-Calvet, E. Rodent control to fight lassa fever: Evaluation and lessons learned from a 4-year study in upper guinea. PLoS Negl. Trop. Dis. 2018, 12, e0006829. [Google Scholar] [CrossRef] [PubMed]
- Lämmler, G.; Zahner, H.; Texdorf, I. Infektionsversuche mit darmnematoden, cestoden und trematoden bei mastomys natalensis (smith, 1834). Zeitschrift für Parasitenkunde 1968, 31, 166–202. [Google Scholar]
- Lammler, G.; Petranyi, G. Chemotherapeutic studies on experimental schistosoma mansoni infection of mastomys natalensis. Bull. World Health Organ. 1971, 44, 739–750. [Google Scholar] [PubMed]
- Kushwaha, V.; Saxena, K.; Verma, R.; Verma, S.K.; Katoch, D.; Kumar, N.; Lal, B.; Murthy, P.K.; Singh, B. Antifilarial activity of diterpenoids from taxodium distichum. Parasites Vectors 2016, 9, 312. [Google Scholar] [CrossRef] [PubMed]
- McNulty, S.N.; Mitreva, M.; Weil, G.J.; Fischer, P.U. Inter and intra-specific diversity of parasites that cause lymphatic filariasis. Infect. Genet. Evol. 2013, 14, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Saunders, M.; Taubert, A.; Dafaalla, T.; Zahner, H. Effect of chemotherapeutic treatment on cytokine (IFN-γ, il-2, il-4, il-5, il-10) gene transcription in response to specific antigens in brugia malayi-infected mastomys coucha. Parasitol. Res. 2008, 103, 1163–1176. [Google Scholar] [CrossRef]
- Singh, P.K.; Kushwaha, S.; Rana, A.K.; Misra-Bhattacharya, S. Cofactor independent phosphoglycerate mutase of brugia malayi induces a mixed Th1/Th2 type immune response and inhibits larval development in the host. BioMed Res. Int. 2014, 2014, 590281. [Google Scholar]
- Verma, S.K.; Joseph, S.K.; Verma, R.; Kushwaha, V.; Parmar, N.; Yadav, P.K.; Thota, J.R.; Kar, S.; Murthy, P.K. Protection against filarial infection by 45–49 kda molecules of brugia malayi via IFN-γ-mediated inos induction. Vaccine 2015, 33, 527–534. [Google Scholar] [CrossRef]
- Verma, S.K.; Kushwaha, V.; Dubey, V.; Saxena, K.; Sharma, A.; Murthy, P.K. Inflammatory mediator release by brugia malayi from macrophages of susceptible host mastomys coucha and Thp-1 and raw 264.7 cell lines. Asian Pac. J. Trop. Med. 2011, 4, 92–96. [Google Scholar] [CrossRef]
- Madhumathi, J.; Prince, P.R.; Rao, D.N.; Karande, A.A.; Reddy, M.V.; Kaliraj, P. Epitope mapping of brugia malayi Alt-2 and the development of a multi-epitope vaccine for lymphatic filariasis. J. Helminthol. 2017, 91, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, S.; Singh, P.K.; Rana, A.K.; Misra-Bhattacharya, S. Immunization of mastomys coucha with brugia malayi recombinant trehalose-6-phosphate phosphatase results in significant protection against homologous challenge infection. PLoS ONE 2013, 8, e72585. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, V.; Kumar, V.; Verma, S.K.; Sharma, R.; Siddiqi, M.I.; Murthy, P.K. Disorganized muscle protein-1 (dim-1) of filarial parasite brugia malayi: Cdna cloning, expression, purification, structural modeling and its potential as vaccine candidate for human filarial infection. Vaccine 2014, 32, 1693–1699. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.K.; Arora, A.; Murthy, P.K. Recombinant calponin of human filariid brugia malayi: Secondary structure and immunoprophylactic potential. Vaccine 2017, 35, 5201–5208. [Google Scholar] [CrossRef] [PubMed]
- Gupta, J.; Misra, S.; Misra-Bhattacharya, S. Immunization with brugia malayi myosin as heterologous DNA prime protein boost induces protective immunity against b. Malayi infection in mastomys coucha. PLoS ONE 2016, 11, e0164991. [Google Scholar] [CrossRef] [PubMed]
- Modlin, I.M.; Esterline, W.; Kim, H.; Goldenring, J.R. Enterochromaffin-like cells and gastric argyrophil carcinoidosis. Acta Oncol. 1991, 30, 493–498. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, O.; Wangberg, B.; Johansson, L.; Modlin, I.M.; Ahlman, H. Praomys (mastomys) natalensis: A model for gastric carcinoid formation. Yale J. Biol. Med. 1992, 65, 741–751, discussion 827–749. [Google Scholar]
- Oettle, A.G. Spontaneous carcinoma of the glandular stomach in rattus (mastomys) natalensis, an african rodent. Br. J. Cancer 1957, 11, 415–433. [Google Scholar] [CrossRef]
- Randeria, J.D. Animal model: Carcinoids and adenocarcinoma of the glandular stomach of praomys (mastomys) natalensis. Am. J. Pathol. 1979, 96, 359–362. [Google Scholar]
- Wardlaw, R.; Smith, J.W. Gastric carcinoid tumors. Ochsner J. 2008, 8, 191–196. [Google Scholar] [PubMed]
- Simmers, M.H.; Ibsen, K.H.; Berk, J.E. Concerning the incidence of “spontaneous” stomach cancer in praomys (mastomys) natalensis. Cancer Res. 1968, 28, 1573–1576. [Google Scholar] [PubMed]
- Hooi, J.K.Y.; Lai, W.Y.; Ng, W.K.; Suen, M.M.Y.; Underwood, F.E.; Tanyingoh, D.; Malfertheiner, P.; Graham, D.Y.; Wong, V.W.S.; Wu, J.C.Y.; et al. Global prevalence of helicobacter pylori infection: Systematic review and meta-analysis. Gastroenterology 2017, 153, 420–429. [Google Scholar] [CrossRef] [PubMed]
- De Martel, C.; Ferlay, J.; Franceschi, S.; Vignat, J.; Bray, F.; Forman, D.; Plummer, M. Global burden of cancers attributable to infections in 2008: A review and synthetic analysis. Lancet Oncol. 2012, 13, 607–615. [Google Scholar] [CrossRef]
- Moss, S.F. The clinical evidence linking helicobacter pylori to gastric cancer. Cell. Mol. Gastroenterol. Hepatol. 2017, 3, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Koga, T.; Takahashi, K.; Sato, K.; Kikuchi, I.; Okazaki, Y.; Miura, T.; Katsuta, M.; Narita, T. The effect of colonisation by helicobacter pylori in praomys (mastomys) natalensis on the incidence of carcinoids. J. Med. Microbiol. 2002, 51, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Kidd, M.; Modlin, I.M.; Eick, G.N.; Champaneria, M.C. Isolation, functional characterization, and transcriptome of mastomys ileal enterochromaffin cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2006, 291, G778–G791. [Google Scholar] [CrossRef]
- Kidd, M.; Modlin, I.M.; Eick, G.N.; Camp, R.L.; Mane, S.M. Role of CCN2/CTGF in the proliferation of mastomys enterochromaffin-like cells and gastric carcinoid development. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 292, G191–G200. [Google Scholar] [CrossRef]
- Kidd, M.; Tang, L.H.; Schmid, S.; Lauffer, J.; Louw, J.A.; Modlin, I.M. Helicobacter pylori lipopolysaccharide alters ecl cell DNA synthesis via a CD14 receptor and polyamine pathway in mastomys. Digestion 2000, 62, 217–224. [Google Scholar] [CrossRef]
- Shen, Z.; Xu, S.; Dewhirst, F.E.; Paster, B.J.; Pena, J.A.; Modlin, I.M.; Kidd, M.; Fox, J.G. A novel enterohepatic helicobacter species “helicobacter mastomyrinus” isolated from the liver and intestine of rodents. Helicobacter 2005, 10, 59–70. [Google Scholar] [CrossRef]
- Eaton, K.A.; Opp, J.S.; Gray, B.M.; Bergin, I.L.; Young, V.B. Ulcerative typhlocolitis associated with helicobacter mastomyrinus in telomerase-deficient mice. Vet. Pathol. 2011, 48, 713–725. [Google Scholar] [CrossRef] [PubMed]
- Harvey, J.J. Susceptibility of praomys (mastomys) natalensis to the murine sarcoma virus-harvey (MSV-h). Int. J. Cancer. 1968, 3, 634–643. [Google Scholar] [CrossRef]
- Haag, A.; Wayss, K.; Rommelaere, J.; Cornelis, J.J. Experimentally induced infection with autonomous parvoviruses, minute virus of mice and h-1, in the african multimammate mouse (mastomys coucha). Comp. Med. 2000, 50, 613–621. [Google Scholar] [PubMed]
- Orba, Y.; Kobayashi, S.; Nakamura, I.; Ishii, A.; Hang’ombe, B.M.; Mweene, A.S.; Thomas, Y.; Kimura, T.; Sawa, H. Detection and characterization of a novel polyomavirus in wild rodents. J. Gen. Virol. 2011, 92, 789–795. [Google Scholar] [CrossRef] [PubMed]
- Gardner, S.D.; Field, A.M.; Coleman, D.V.; Hulme, B. New human papovavirus (b.K.) isolated from urine after renal transplantation. Lancet 1971, 1, 1253–1257. [Google Scholar] [CrossRef]
- Padgett, B.L.; Walker, D.L.; ZuRhein, G.M.; Eckroade, R.J.; Dessel, B.H. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet 1971, 1, 1257–1260. [Google Scholar] [CrossRef]
- Feng, H.; Shuda, M.; Chang, Y.; Moore, P.S. Clonal integration of a polyomavirus in human merkel cell carcinoma. Science 2008, 319, 1096–1100. [Google Scholar] [CrossRef]
- Polyomaviridae Study Group of the International Committee on Taxonomy of Viruses; Calvignac-Spencer, S.; Feltkamp, M.C.; Daugherty, M.D.; Moens, U.; Ramqvist, T.; Johne, R.; Ehlers, B. A taxonomy update for the family polyomaviridae. Arch. Virol. 2016, 161, 1739–1750. [Google Scholar] [CrossRef]
- Nafz, J.; Kohler, A.; Ohnesorge, M.; Nindl, I.; Stockfleth, E.; Rösl, F. Persistence of mastomys natalensis papillomavirus in multiple organs identifies novel targets for infection. J. Gen. Virol. 2007, 88, 2670–2678. [Google Scholar] [CrossRef]
- Burtscher, H.; Grunberg, W.; Meingassner, G. Infectious keratoacanthomas of the epidermis in praomys (mastomys) natalensis. Die Nat. 1973, 60, 209–210. [Google Scholar] [CrossRef]
- Amtmann, E.; Wayss, K. Papillomaviruses and carcinogenic progression ii. In The Papovaviridae: The Papillomaviruses; Salzman, N.P., Howley, P.M., Eds.; Springer: Boston, MA, USA, 1987; pp. 187–198. [Google Scholar]
- Inglis, T.J. Principia aetiologica: Taking causality beyond koch’s postulates. J. Med. Microbiol. 2007, 56, 1419–1422. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, R.; Hundeiker, M. Keratoacanthomas in mastomys natalensis. Arch. Dermatol. Res. 1975, 254, 239–243. [Google Scholar] [CrossRef]
- Rudolph, R.; Müller, H. Induktion von epidermalem tumorwachstum in der haut von mastomys natalensis durch übertragung virushaltigen tumorgewebes eines plattenepithelkarzinoms. Zentralblatt für Veterinärmedizin Reihe B 1976, 23, 143–150. [Google Scholar] [CrossRef]
- Rudolph, R.; Thiel, W. Pathological anatomy and histology of spontaneous, epithelial skin tumors in mastomys natalensis. Zentralblatt für Veterinärmedizin Reihe A 1976, 23, 429–441. [Google Scholar] [CrossRef] [PubMed]
- Müller, H.; Gissmann, L. Mastomys natalensis papilloma virus (mnpv), the causative agent of epithelial proliferations: Characterization of the virus particle. J. Gen. Virol. 1978, 41, 315–323. [Google Scholar] [CrossRef]
- Reinacher, M.; Müller, H.; Thiel, W.; Rudolph, R.L. Localization of papillomavirus and virus-specific antigens in the skin of tumor-bearing mastomys natalensis (gra giessen). Med. Microbiol. Immunol. 1978, 165, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.H.; Tachezy, R.; Van Ranst, M.; Chan, S.Y.; Bernard, H.U.; Burk, R.D. The mastomys natalensis papillomavirus: Nucleotide sequence, genome organization, and phylogenetic relationship of a rodent papillomavirus involved in tumorigenesis of cutaneous epithelia. Virology 1994, 198, 534–541. [Google Scholar] [CrossRef] [PubMed]
- De Villiers, E.M.; Fauquet, C.; Broker, T.R.; Bernard, H.U.; zur Hausen, H. Classification of papillomaviruses. Virology 2004, 324, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, J.C.; Calonje, E. Cutaneous manifestations of human papillomaviruses: A review. Acta Dermatovenerol. Alp. Pannon. Adriat. 2011, 20, 145–154. [Google Scholar]
- Wang, J.; Aldabagh, B.; Yu, J.; Arron, S.T. Role of human papillomavirus in cutaneous squamous cell carcinoma: A meta-analysis. J. Am. Acad. Dermatol. 2014, 70, 621–629. [Google Scholar] [CrossRef]
- Pfister, H.; Fuchs, P.G.; Majewski, S.; Jablonska, S.; Pniewska, I.; Malejczyk, M. High prevalence of epidermodysplasia verruciformis-associated human papillomavirus DNA in actinic keratoses of the immunocompetent population. Arch. Dermatol. Res. 2003, 295, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Takai, T. Advances in histopathological diagnosis of keratoacanthoma. J. Dermatol. 2017, 44, 304–314. [Google Scholar] [CrossRef] [PubMed]
- Hannigan, G.D.; Meisel, J.S.; Tyldsley, A.S.; Zheng, Q.; Hodkinson, B.P.; SanMiguel, A.J.; Minot, S.; Bushman, F.D.; Grice, E.A. The human skin double-stranded DNA virome: Topographical and temporal diversity, genetic enrichment, and dynamic associations with the host microbiome. MBio 2015, 6, e01578-15. [Google Scholar] [CrossRef] [PubMed]
- Antonsson, A.; Karanfilovska, S.; Lindqvist, P.G.; Hansson, B.G. General acquisition of human papillomavirus infections of skin occurs in early infancy. J. Clin. Microbiol. 2003, 41, 2509–2514. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, K.; Neumann, J.; Waterboer, T.; Rösl, F. Serological markers for papillomavirus infection and skin tumour development in the rodent model mastomys coucha. J. Gen. Virol. 2011, 92, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Wayss, K.; Reyes-Mayes, D.; Volm, M. Chemical carcinogenesis by the two-stage protocol in the skin ofmastomys natalensis (muridae) using topical initiation with 7, 12-dimethylbenz (a) anthracene and topical promotion with 12-0-tetradecanoylphorbol-13-acetate. Virchows Arch. B Cell Pathol. 1981, 38, 13–21. [Google Scholar] [CrossRef]
- Amtmann, E.; Volm, M.; Wayss, K. Tumour induction in the rodent mastomys natalensis by activation of endogenous papilloma virus genomes. Nature 1984, 308, 291–292. [Google Scholar] [CrossRef] [PubMed]
- Helfrich, I.; Chen, M.; Schmidt, R.; Furstenberger, G.; Kopp-Schneider, A.; Trick, D.; Grone, H.J.; Zur Hausen, H.; Rösl, F. Increased incidence of squamous cell carcinomas in mastomys natalensis papillomavirus e6 transgenic mice during two-stage skin carcinogenesis. J. Virol. 2004, 78, 4797–4805. [Google Scholar] [CrossRef]
- Abel, E.L.; Angel, J.M.; Kiguchi, K.; DiGiovanni, J. Multi-stage chemical carcinogenesis in mouse skin: Fundamentals and applications. Nat. Protoc. 2009, 4, 1350–1362. [Google Scholar] [CrossRef]
- Holderfield, M.; Lorenzana, E.; Weisburd, B.; Lomovasky, L.; Boussemart, L.; Lacroix, L.; Tomasic, G.; Favre, M.; Vagner, S.; Robert, C.; et al. Vemurafenib cooperates with hpv to promote initiation of cutaneous tumors. Cancer Res. 2014, 74, 2238–2245. [Google Scholar] [CrossRef]
- Hasche, D.; Vinzon, S.E.; Rosl, F. Cutaneous papillomaviruses and non-melanoma skin cancer: Causal agents or innocent bystanders? Front. Microbiol. 2018, 9, 874. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, M.; Munzel, P.A.; Braeuning, A. Non-melanoma skin cancer in mouse and man. Arch. Toxicol. 2013, 87, 783–798. [Google Scholar] [CrossRef]
- Siegsmund, M.; Wayss, K.; Amtmann, E. Activation of latent papillomavirus genomes by chronic mechanical irritation. J. Gen. Virol. 1991, 72, 2787–2789. [Google Scholar] [CrossRef] [PubMed]
- Hufbauer, M.; Cooke, J.; van der Horst, G.T.; Pfister, H.; Storey, A.; Akgül, B. Human papillomavirus mediated inhibition of DNA damage sensing and repair drives skin carcinogenesis. Mol. Cancer 2015, 14, 183. [Google Scholar] [CrossRef]
- Viarisio, D.; Müller-Decker, K.; Accardi, R.; Robitaille, A.; Durst, M.; Beer, K.; Jansen, L.; Flechtenmacher, C.; Bozza, M.; Harbottle, R.; et al. Beta hpv38 oncoproteins act with a hit-and-run mechanism in ultraviolet radiation-induced skin carcinogenesis in mice. PLoS Pathog. 2018, 14, e1006783. [Google Scholar] [CrossRef] [PubMed]
- Uberoi, A.; Yoshida, S.; Frazer, I.H.; Pitot, H.C.; Lambert, P.F. Role of ultraviolet radiation in papillomavirus-induced disease. PLoS Pathog. 2016, 12, e1005664. [Google Scholar] [CrossRef] [PubMed]
- Hasche, D.; Stephan, S.; Braspenning-Wesch, I.; Mikulec, J.; Niebler, M.; Gröne, H.J.; Flechtenmacher, C.; Akgül, B.; Rösl, F.; Vinzón, S.E. The interplay of UV and cutaneous papillomavirus infection in skin cancer development. PLoS Pathog. 2017, 13, e1006723. [Google Scholar] [CrossRef]
- Marcuzzi, G.P.; Hufbauer, M.; Kasper, H.U.; Weissenborn, S.J.; Smola, S.; Pfister, H. Spontaneous tumour development in human papillomavirus type 8 e6 transgenic mice and rapid induction by UV-light exposure and wounding. J. Gen. Virol. 2009, 90, 2855–2864. [Google Scholar] [CrossRef]
- Salvermoser, M.; Chotewutmontri, S.; Braspenning-Wesch, I.; Hasche, D.; Rösl, F.; Vinzon, S.E. Transcriptome analysis of mastomys natalensis papillomavirus in productive lesions after natural infection. J. Gen. Virol. 2016, 97, 1658–1669. [Google Scholar] [CrossRef]
- Hasche, D.; Stephan, S.; Savelyeva, L.; Westermann, F.; Rösl, F.; Vinzón, S.E. Establishment of an immortalized skin keratinocyte cell line derived from the animal model mastomys coucha. PLoS ONE 2016, 11, e0161283. [Google Scholar] [CrossRef]
- Nafz, J.; Schäfer, K.; Chen, S.F.; Bravo, I.G.; Ibberson, M.; Nindl, I.; Stockfleth, E.; Rösl, F. A novel rodent papillomavirus isolated from anogenital lesions in its natural host. Virology 2008, 374, 186–197. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, K.; Waterboer, T.; Rösl, F. A capture elisa for monitoring papillomavirus-induced antibodies in mastomys coucha. J. Virol. Methods 2010, 163, 216–221. [Google Scholar] [CrossRef] [PubMed]
- Andersson, K.; Michael, K.M.; Luostarinen, T.; Waterboer, T.; Gislefoss, R.; Hakulinen, T.; Forslund, O.; Pawlita, M.; Dillner, J. Prospective study of human papillomavirus seropositivity and risk of nonmelanoma skin cancer. Am. J. Epidemiol. 2012, 175, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Ahituv, N. Welcome to the Mastomys Genome Project. Available online: http://mastomys.ucsf.edu (accessed on 12 February 2019).
- Vinzón, S.E.; Rösl, F. Hpv vaccination for prevention of skin cancer. Hum. Vaccines Immunother. 2015, 11, 353–357. [Google Scholar] [CrossRef] [PubMed]
- Garrett, G.L.; Blanc, P.D.; Boscardin, J.; Lloyd, A.A.; Ahmed, R.L.; Anthony, T.; Bibee, K.; Breithaupt, A.; Cannon, J.; Chen, A.; et al. Incidence of and risk factors for skin cancer in organ transplant recipients in the united states. JAMA Dermatol. 2017, 153, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Garrett, G.L.; Yuan, J.T.; Shin, T.M.; Arron, S.T.; Transplant Skin Cancer, N. Validity of skin cancer malignancy reporting to the organ procurement transplant network: A cohort study. J. Am. Acad. Dermatol. 2018, 78, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Rangwala, S.; Tsai, K.Y. Roles of the immune system in skin cancer. Br. J. Dermatol. 2011, 165, 953–965. [Google Scholar] [CrossRef]
- Neale, R.E.; Weissenborn, S.; Abeni, D.; Bavinck, J.N.; Euvrard, S.; Feltkamp, M.C.; Green, A.C.; Harwood, C.; de Koning, M.; Naldi, L.; et al. Human papillomavirus load in eyebrow hair follicles and risk of cutaneous squamous cell carcinoma. Cancer Epidemiol. Prev. Biomark. 2013, 22, 719–727. [Google Scholar] [CrossRef]
- Buck, C.B.; Pastrana, D.V.; Lowy, D.R.; Schiller, J.T. Generation of hpv pseudovirions using transfection and their use in neutralization assays. Methods Mol. Med. 2005, 119, 445–462. [Google Scholar]
- Vinzón, S.E.; Braspenning-Wesch, I.; Muller, M.; Geissler, E.K.; Nindl, I.; Grone, H.J.; Schafer, K.; Rösl, F. Protective vaccination against papillomavirus-induced skin tumors under immunocompetent and immunosuppressive conditions: A preclinical study using a natural outbred animal model. PLoS. Pathog. 2014, 10, e1003924. [Google Scholar] [CrossRef]
- Nagarajan, P.; Asgari, M.M.; Green, A.C.; Guhan, S.M.; Arron, S.T.; Proby, C.M.; Rollison, D.E.; Harwood, C.A.; Toland, A.E. Keratinocyte carcinomas: Current concepts and future research priorities. Clin. Cancer Res. 2018. [Google Scholar] [CrossRef] [PubMed]
- Andersson, K.; Waterboer, T.; Kirnbauer, R.; Slupetzky, K.; Iftner, T.; de Villiers, E.M.; Forslund, O.; Pawlita, M.; Dillner, J. Seroreactivity to cutaneous human papillomaviruses among patients with nonmelanoma skin cancer or benign skin lesions. Cancer Epidemiol. Prev. Biomark. 2008, 17, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Missero, C.; Antonini, D. Crosstalk among p53 family members in cutaneous carcinoma. Exp. Dermatol. 2014, 23, 143–146. [Google Scholar] [CrossRef] [PubMed]
- Flores, A.; Grant, W.; White, A.C.; Scumpia, P.; Takahashi, R.; Lowry, W.E. Tumor suppressor identity can contribute to heterogeneity of phenotype in hair follicle stem cell induced squamous cell carcinoma. Exp. Dermatol. 2016, 25, 733. [Google Scholar] [CrossRef] [PubMed]
- Muller, P.A.; Vousden, K.H.; Norman, J.C. P53 and its mutants in tumor cell migration and invasion. J. Cell Biol. 2011, 192, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Schellenbacher, C.; Roden, R.B.S.; Kirnbauer, R. Developments in l2-based human papillomavirus (HPV) vaccines. Virus Res. 2017, 231, 166–175. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).