Induction of Tier 1 HIV Neutralizing Antibodies by Envelope Trimers Incorporated into a Replication Competent Vesicular Stomatitis Virus Vector
Abstract
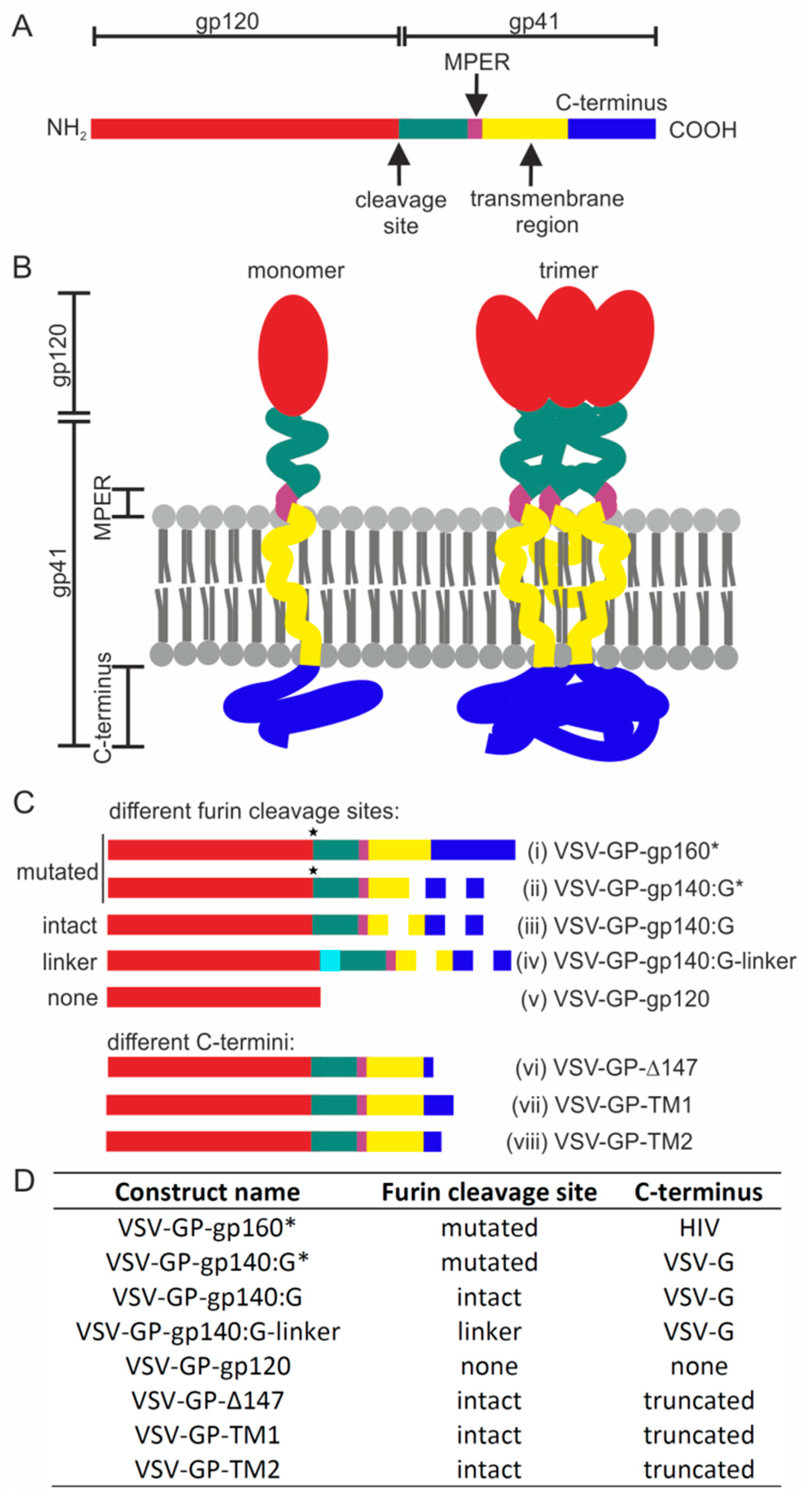
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Cell Lines
2.3. Viruses
2.4. TCID50 Assay
2.5. Replication Kinetics
2.6. Preparation of Lysates and Western Blot Analysis
2.7. Flow Cytometry Analysis
2.8. Electron Microscopic Pictures
2.9. Mouse Immunization Experiments
2.10. Rabbit Immunization Experiments
2.11. Anti-1086.C-gp140 Enzyme-Linked Immunosorbent Assay (ELISA)
2.12. HIV Neutralization Assays
2.13. Statistical Analysis
3. Results
3.1. Membrane-Anchored Env Induces Higher Antibody Titers Compared to Secreted Env
3.2. HIV Env is Expressed on the Surface of Infected Cells
3.3. HIV Env on the Surface of VSV-GP Particles Displays Crucial Epitopes for Binding of Neutralizing Antibodies
3.4. VSV-GP-Env Induces gp140-Specific Antibodies in Mice and Rabbits
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- UNAIDS. Global Aids Update; UNAIDS: Geneva, Switzerland, 2016. [Google Scholar]
- Hessell, A.J.; Poignard, P.; Hunter, M.; Hangartner, L.; Tehrani, D.M.; Bleeker, W.K.; Parren, P.W.; Marx, P.A.; Burton, D.R. Effective, low-titer antibody protection against low-dose repeated mucosal shiv challenge in macaques. Nat. Med. 2009, 15, 951–954. [Google Scholar] [CrossRef] [PubMed]
- Mascola, J.R.; Lewis, M.G.; Stiegler, G.; Harris, D.; VanCott, T.C.; Hayes, D.; Louder, M.K.; Brown, C.R.; Sapan, C.V.; Frankel, S.S.; et al. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6pd by passive transfer of neutralizing antibodies. J. Virol. 1999, 73, 4009–4018. [Google Scholar] [PubMed]
- Haynes, B.F.; Gilbert, P.B.; McElrath, M.J.; Zolla-Pazner, S.; Tomaras, G.D.; Alam, S.M.; Evans, D.T.; Montefiori, D.C.; Karnasuta, C.; Sutthent, R.; et al. Immune-correlates analysis of an hiv-1 vaccine efficacy trial. N. Engl. J. Med. 2012, 366, 1275–1286. [Google Scholar] [CrossRef] [PubMed]
- Rerks-Ngarm, S.; Pitisuttithum, P.; Nitayaphan, S.; Kaewkungwal, J.; Chiu, J.; Paris, R.; Premsri, N.; Namwat, C.; de Souza, M.; Adams, E.; et al. Vaccination with alvac and aidsvax to prevent hiv-1 infection in thailand. N. Engl. J. Med. 2009, 361, 2209–2220. [Google Scholar] [CrossRef] [PubMed]
- Robert-Guroff, M. Replicating and non-replicating viral vectors for vaccine development. Curr. Opin. Biotechnol. 2007, 18, 546–556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, J.E.; Schnell, M.J.; Buonocore, L.; Rose, J.K. Specific targeting to CD4+ cells of recombinant vesicular stomatitis viruses encoding human immunodeficiency virus envelope proteins. J. Virol. 1997, 71, 5060–5068. [Google Scholar] [PubMed]
- Alam, S.M.; Morelli, M.; Dennison, S.M.; Liao, H.X.; Zhang, R.; Xia, S.M.; Rits-Volloch, S.; Sun, L.; Harrison, S.C.; Haynes, B.F.; et al. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc. Natl. Acad. Sci. USA 2009, 106, 20234–20239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asbach, B.; Wagner, R. Particle-based delivery of the hiv envelope protein. Curr. Opin. HIV AIDS 2017, 12, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Nabi, G.; Genannt Bonsmann, M.S.; Tenbusch, M.; Gardt, O.; Barouch, D.H.; Temchura, V.; Uberla, K. Gagpol-specific cd4(+) t-cells increase the antibody response to env by intrastructural help. Retrovirology 2013, 10, 117. [Google Scholar] [CrossRef]
- Rose, N.F.; Marx, P.A.; Luckay, A.; Nixon, D.F.; Moretto, W.J.; Donahoe, S.M.; Montefiori, D.; Roberts, A.; Buonocore, L.; Rose, J.K. An effective aids vaccine based on live attenuated vesicular stomatitis virus recombinants. Cell 2001, 106, 539–549. [Google Scholar] [CrossRef]
- Ramsburg, E.; Rose, N.F.; Marx, P.A.; Mefford, M.; Nixon, D.F.; Moretto, W.J.; Montefiori, D.; Earl, P.; Moss, B.; Rose, J.K. Highly effective control of an aids virus challenge in macaques by using vesicular stomatitis virus and modified vaccinia virus ankara vaccine vectors in a single-boost protocol. J. Virol. 2004, 78, 3930–3940. [Google Scholar] [CrossRef] [PubMed]
- Schell, J.; Rose, N.F.; Fazo, N.; Marx, P.A.; Hunter, M.; Ramsburg, E.; Montefiori, D.; Earl, P.; Moss, B.; Rose, J.K. Long-term vaccine protection from aids and clearance of viral DNA following shiv89.6p challenge. Vaccine 2009, 27, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Van Rompay, K.K.; Abel, K.; Earl, P.; Kozlowski, P.A.; Easlick, J.; Moore, J.; Buonocore-Buzzelli, L.; Schmidt, K.A.; Wilson, R.L.; Simon, I.; et al. Immunogenicity of viral vector, prime-boost siv vaccine regimens in infant rhesus macaques: Attenuated vesicular stomatitis virus (vsv) and modified vaccinia ankara (mva) recombinant siv vaccines compared to live-attenuated siv. Vaccine 2010, 28, 1481–1492. [Google Scholar] [CrossRef] [PubMed]
- Publicover, J.; Ramsburg, E.; Rose, J.K. Characterization of nonpathogenic, live, viral vaccine vectors inducing potent cellular immune responses. J. Virol. 2004, 78, 9317–9324. [Google Scholar] [CrossRef]
- Johnson, J.E.; Nasar, F.; Coleman, J.W.; Price, R.E.; Javadian, A.; Draper, K.; Lee, M.; Reilly, P.A.; Clarke, D.K.; Hendry, R.M.; et al. Neurovirulence properties of recombinant vesicular stomatitis virus vectors in non-human primates. Virology 2007, 360, 36–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooper, D.; Wright, K.J.; Calderon, P.C.; Guo, M.; Nasar, F.; Johnson, J.E.; Coleman, J.W.; Lee, M.; Kotash, C.; Yurgelonis, I.; et al. Attenuation of recombinant vesicular stomatitis virus-human immunodeficiency virus type 1 vaccine vectors by gene translocations and g gene truncation reduces neurovirulence and enhances immunogenicity in mice. J. Virol. 2008, 82, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Li, S.S.; Kochar, N.K.; Elizaga, M.; Hay, C.M.; Wilson, G.J.; Cohen, K.W.; De Rosa, S.C.; Xu, R.; Ota-Setlik, A.; Morris, D.; et al. DNA priming increases frequency of t-cell responses to a vesicular stomatitis virus hiv vaccine with specific enhancement of cd8(+) t-cell responses by interleukin-12 plasmid DNA. Clin. Vaccine Immunol. 2017, 24. [Google Scholar] [CrossRef]
- Fuchs, J.D.; Frank, I.; Elizaga, M.L.; Allen, M.; Frahm, N.; Kochar, N.; Li, S.; Edupuganti, S.; Kalams, S.A.; Tomaras, G.D.; et al. First-in-human evaluation of the safety and immunogenicity of a recombinant vesicular stomatitis virus human immunodeficiency virus-1 gag vaccine (hvtn 090). Open Forum Infect. Dis. 2015, 2, ofv082. [Google Scholar] [CrossRef]
- Zemp, F.; Rajwani, J.; Mahoney, D.J. Rhabdoviruses as vaccine platforms for infectious disease and cancer. Biotechnol. Genet. Eng. Rev. 2018, 34, 122–138. [Google Scholar] [CrossRef]
- Parks, C.L. G-106 mucosal vaccination with a replication-competent vsv-hiv chimera delivering env trimers protects rhesus macaques from rectal shiv infection. JAIDS J. Acquired Immune Defic. Syndr. 2017, 74. [Google Scholar] [CrossRef]
- Hoffmann, M.; Wu, Y.J.; Gerber, M.; Berger-Rentsch, M.; Heimrich, B.; Schwemmle, M.; Zimmer, G. Fusion-active glycoprotein g mediates the cytotoxicity of vesicular stomatitis virus m mutants lacking host shut-off activity. J. Gen. Virol. 2010, 91, 2782–2793. [Google Scholar] [CrossRef] [PubMed]
- Rose, N.F.; Roberts, A.; Buonocore, L.; Rose, J.K. Glycoprotein exchange vectors based on vesicular stomatitis virus allow effective boosting and generation of neutralizing antibodies to a primary isolate of human immunodeficiency virus type 1. J. Virol. 2000, 74, 10903–10910. [Google Scholar] [CrossRef] [PubMed]
- Tober, R.; Banki, Z.; Egerer, L.; Muik, A.; Behmuller, S.; Kreppel, F.; Greczmiel, U.; Oxenius, A.; von Laer, D.; Kimpel, J. Vsv-gp: A potent viral vaccine vector that boosts the immune response upon repeated applications. J. Virol. 2014, 88, 4897–4907. [Google Scholar] [CrossRef] [PubMed]
- Muik, A.; Stubbert, L.J.; Jahedi, R.Z.; Geibeta, Y.; Kimpel, J.; Dold, C.; Tober, R.; Volk, A.; Klein, S.; Dietrich, U.; et al. Re-engineering vesicular stomatitis virus to abrogate neurotoxicity, circumvent humoral immunity, and enhance oncolytic potency. Cancer Res. 2014, 74, 3567–3578. [Google Scholar] [CrossRef]
- Dold, C.; Rodriguez Urbiola, C.; Wollmann, G.; Egerer, L.; Muik, A.; Bellmann, L.; Fiegl, H.; Marth, C.; Kimpel, J.; von Laer, D. Application of interferon modulators to overcome partial resistance of human ovarian cancers to vsv-gp oncolytic viral therapy. Mol. Ther. Oncolytics 2016, 3, 16021. [Google Scholar] [CrossRef] [PubMed]
- Kimpel, J.; Urbiola, C.; Koske, I.; Tober, R.; Banki, Z.; Wollmann, G.; von Laer, D. The oncolytic virus vsv-gp is effective against malignant melanoma. Viruses 2018, 10, 108. [Google Scholar] [CrossRef] [PubMed]
- Witko, S.E.; Kotash, C.S.; Nowak, R.M.; Johnson, J.E.; Boutilier, L.A.; Melville, K.J.; Heron, S.G.; Clarke, D.K.; Abramovitz, A.S.; Hendry, R.M.; et al. An efficient helper-virus-free method for rescue of recombinant paramyxoviruses and rhadoviruses from a cell line suitable for vaccine development. J. Virol. Methods 2006, 135, 91–101. [Google Scholar] [CrossRef]
- Kaerber, G. 50% end-point calculation. Arch. Exp. Pathol. Pharmakol. 1931, 162, 480–483. [Google Scholar]
- Liao, H.X.; Tsao, C.Y.; Alam, S.M.; Muldoon, M.; Vandergrift, N.; Ma, B.J.; Lu, X.; Sutherland, L.L.; Scearce, R.M.; Bowman, C.; et al. Antigenicity and immunogenicity of transmitted/founder, consensus, and chronic envelope glycoproteins of human immunodeficiency virus type 1. J. Virol. 2013, 87, 4185–4201. [Google Scholar] [CrossRef]
- Montefiori, D.C. Evaluating neutralizing antibodies against hiv, siv, and shiv in luciferase reporter gene assays. Curr. Protoc. Immunol. 2005. [Google Scholar] [CrossRef]
- Sharma, S.K.; de Val, N.; Bale, S.; Guenaga, J.; Tran, K.; Feng, Y.; Dubrovskaya, V.; Ward, A.B.; Wyatt, R.T. Cleavage-independent hiv-1 env trimers engineered as soluble native spike mimetics for vaccine design. Cell Rep. 2015, 11, 539–550. [Google Scholar] [CrossRef] [PubMed]
- Hogan, M.J.; Conde-Motter, A.; Jordan, A.P.O.; Yang, L.; Cleveland, B.; Guo, W.; Romano, J.; Ni, H.; Pardi, N.; LaBranche, C.C.; et al. Increased surface expression of hiv-1 envelope is associated with improved antibody response in vaccinia prime/protein boost immunization. Virology 2017, 514, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Hogan, M.C.A.; Jordan, A.P.O.; Elser, S.; Weissman, D.; Hoxie, J.A. Increase in hiv-1 envelope incorporation into virions mediated by genetic modification of the cytoplasmic tail of env. In Proceedings of the Keystone Symposia Conference on HIV Vaccines: Adaptive Immunity and Beyond, Banff, AB, Canada, 9–14 March 2014. [Google Scholar]
- Wyatt, R.; Sodroski, J. The hiv-1 envelope glycoproteins: Fusogens, antigens, and immunogens. Science 1998, 280, 1884–1888. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.E.; Rodgers, W.; Rose, J.K. A plasma membrane localization signal in the hiv-1 envelope cytoplasmic domain prevents localization at sites of vesicular stomatitis virus budding and incorporation into vsv virions. Virology 1998, 251, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.; Mendy, J.; Vang, L.; Avanzini, J.B.; Garduno, F.; Manayani, D.J.; Ishioka, G.; Farness, P.; Ping, L.H.; Swanstrom, R.; et al. Pre-clinical development of a recombinant, replication-competent adenovirus serotype 4 vector vaccine expressing hiv-1 envelope 1086 clade c. PLoS ONE 2013, 8, e82380. [Google Scholar] [CrossRef] [PubMed]
- Khattar, S.K.; Manoharan, V.; Bhattarai, B.; LaBranche, C.C.; Montefiori, D.C.; Samal, S.K. Mucosal immunization with newcastle disease virus vector coexpressing hiv-1 env and gag proteins elicits potent serum, mucosal, and cellular immune responses that protect against vaccinia virus env and gag challenges. mBio 2015, 6, e01005. [Google Scholar] [CrossRef] [PubMed]
- Gomez, C.E.; Esteban, M. Recombinant proteins produced by vaccinia virus vectors can be incorporated within the virion (imv form) into different compartments. Arch. Virol. 2001, 146, 875–892. [Google Scholar] [CrossRef]
- Bachmann, M.F.; Rohrer, U.H.; Kundig, T.M.; Burki, K.; Hengartner, H.; Zinkernagel, R.M. The influence of antigen organization on b cell responsiveness. Science 1993, 262, 1448–1451. [Google Scholar] [CrossRef]
- Russell, S.M.; Liew, F.Y. T cells primed by influenza virion internal components can cooperate in the antibody response to haemagglutinin. Nature 1979, 280, 147–148. [Google Scholar] [CrossRef]
- Sanders, R.W.; Derking, R.; Cupo, A.; Julien, J.P.; Yasmeen, A.; de Val, N.; Kim, H.J.; Blattner, C.; de la Pena, A.T.; Korzun, J.; et al. A next-generation cleaved, soluble hiv-1 env trimer, bg505 sosip.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013, 9, e1003618. [Google Scholar] [CrossRef]
- Sanders, R.W.; van Gils, M.J.; Derking, R.; Sok, D.; Ketas, T.J.; Burger, J.A.; Ozorowski, G.; Cupo, A.; Simonich, C.; Goo, L.; et al. Hiv-1 vaccines. Hiv-1 neutralizing antibodies induced by native-like envelope trimers. Science 2015, 349, aac4223. [Google Scholar] [CrossRef] [PubMed]
- Pauthner, M.; Havenar-Daughton, C.; Sok, D.; Nkolola, J.P.; Bastidas, R.; Boopathy, A.V.; Carnathan, D.G.; Chandrashekar, A.; Cirelli, K.M.; Cottrell, C.A.; et al. Elicitation of robust tier 2 neutralizing antibody responses in nonhuman primates by hiv envelope trimer immunization using optimized approaches. Immunity 2017, 46, 1073–1088.e6. [Google Scholar] [CrossRef]
- Negri, D.; Blasi, M.; LaBranche, C.; Parks, R.; Balachandran, H.; Lifton, M.; Shen, X.; Denny, T.; Ferrari, G.; Vescio, M.F.; et al. Immunization with an siv-based idlv expressing hiv-1 env 1086 clade c elicits durable humoral and cellular responses in rhesus macaques. Mol. Ther. 2016, 24, 2021–2032. [Google Scholar] [CrossRef] [PubMed]
- Kadelka, C.; Liechti, T.; Ebner, H.; Schanz, M.; Rusert, P.; Friedrich, N.; Stiegeler, E.; Braun, D.L.; Huber, M.; Scherrer, A.U.; et al. Distinct, igg1-driven antibody response landscapes demarcate individuals with broadly hiv-1 neutralizing activity. J. Exp. Med. 2018, 215, 1589–1608. [Google Scholar] [CrossRef] [PubMed]
- Escolano, A.; Dosenovic, P.; Nussenzweig, M.C. Progress toward active or passive hiv-1 vaccination. J. Exp. Med. 2017, 214, 3–16. [Google Scholar] [CrossRef]
- Mendoza, P.; Gruell, H.; Nogueira, L.; Pai, J.A.; Butler, A.L.; Millard, K.; Lehmann, C.; Suarez, I.; Oliveira, T.Y.; Lorenzi, J.C.C.; et al. Combination therapy with anti-hiv-1 antibodies maintains viral suppression. Nature 2018, 561, 479–484. [Google Scholar] [CrossRef]
- Bar-On, Y.; Gruell, H.; Schoofs, T.; Pai, J.A.; Nogueira, L.; Butler, A.L.; Millard, K.; Lehmann, C.; Suarez, I.; Oliveira, T.Y.; et al. Safety and antiviral activity of combination hiv-1 broadly neutralizing antibodies in viremic individuals. Nat. Med. 2018, 24, 1701–1707. [Google Scholar] [CrossRef] [PubMed]
- Burton, D.R.; Mascola, J.R. Antibody responses to envelope glycoproteins in hiv-1 infection. Nat. Immunol. 2015, 16, 571–576. [Google Scholar] [CrossRef]
- Zhu, P.; Liu, J.; Bess, J., Jr.; Chertova, E.; Lifson, J.D.; Grise, H.; Ofek, G.A.; Taylor, K.A.; Roux, K.H. Distribution and three-dimensional structure of aids virus envelope spikes. Nature 2006, 441, 847–852. [Google Scholar] [CrossRef]
- Haynes, B.F.; Burton, D.R. Developing an hiv vaccine. Science 2017, 355, 1129–1130. [Google Scholar] [CrossRef]
- Klein, F.; Diskin, R.; Scheid, J.F.; Gaebler, C.; Mouquet, H.; Georgiev, I.S.; Pancera, M.; Zhou, T.; Incesu, R.B.; Fu, B.Z.; et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent hiv-1 neutralization. Cell 2013, 153, 126–138. [Google Scholar] [CrossRef] [PubMed]
- Mouquet, H.; Scheid, J.F.; Zoller, M.J.; Krogsgaard, M.; Ott, R.G.; Shukair, S.; Artyomov, M.N.; Pietzsch, J.; Connors, M.; Pereyra, F.; et al. Polyreactivity increases the apparent affinity of anti-hiv antibodies by heteroligation. Nature 2010, 467, 591–595. [Google Scholar] [CrossRef] [PubMed]
- Sadanand, S.; Suscovich, T.J.; Alter, G. Broadly neutralizing antibodies against hiv: New insights to inform vaccine design. Annu. Rev. Med. 2016, 67, 185–200. [Google Scholar] [CrossRef] [PubMed]
- Kelsoe, G.; Haynes, B.F. Host controls of hiv broadly neutralizing antibody development. Immunol. Rev. 2017, 275, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Alsahafi, N.; Anand, S.P.; Castillo-Menendez, L.; Verly, M.M.; Medjahed, H.; Prevost, J.; Herschhorn, A.; Richard, J.; Schon, A.; Melillo, B.; et al. Sosip changes affect human immunodeficiency virus (hiv-1) envelope glycoprotein conformation and cd4 engagement. J. Virol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Herschhorn, A.; Ma, X.; Gu, C.; Ventura, J.D.; Castillo-Menendez, L.; Melillo, B.; Terry, D.S.; Smith, A.B., 3rd; Blanchard, S.C.; Munro, J.B.; et al. Release of gp120 restraints leads to an entry-competent intermediate state of the hiv-1 envelope glycoproteins. mBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Lu, M.; Gorman, J.; Terry, D.S.; Hong, X.; Zhou, Z.; Zhao, H.; Altman, R.B.; Arthos, J.; Blanchard, S.C.; et al. Hiv-1 env trimer opens through an asymmetric intermediate in which individual protomers adopt distinct conformations. eLife 2018, 7. [Google Scholar] [CrossRef]
- Wang, H.; Barnes, C.O.; Yang, Z.; Nussenzweig, M.C.; Bjorkman, P.J. Partially open hiv-1 envelope structures exhibit conformational changes relevant for coreceptor binding and fusion. Cell Host Microbe 2018, 24, 579–592.e4. [Google Scholar] [CrossRef]
- Zhang, P.; Gorman, J.; Geng, H.; Liu, Q.; Lin, Y.; Tsybovsky, Y.; Go, E.P.; Dey, B.; Andine, T.; Kwon, A.; et al. Interdomain stabilization impairs cd4 binding and improves immunogenicity of the hiv-1 envelope trimer. Cell Host Microbe 2018, 23, 832–844.e6. [Google Scholar] [CrossRef]
- Pauthner, M.G.; Nkolola, J.P.; Havenar-Daughton, C.; Murrell, B.; Reiss, S.M.; Bastidas, R.; Prevost, J.; Nedellec, R.; von Bredow, B.; Abbink, P.; et al. Vaccine-induced protection from homologous tier 2 shiv challenge in nonhuman primates depends on serum-neutralizing antibody titers. Immunity 2019, 50, 241–252.e6. [Google Scholar] [CrossRef]
- Aldon, Y.; McKay, P.F.; Allen, J.; Ozorowski, G.; Felfodine Levai, R.; Tolazzi, M.; Rogers, P.; He, L.; de Val, N.; Fabian, K.; et al. Rational design of DNA-expressed stabilized native-like hiv-1 envelope trimers. Cell Rep. 2018, 24, 3324–3338.e5. [Google Scholar] [CrossRef] [PubMed]
- Saunders, K.O.; Verkoczy, L.K.; Jiang, C.; Zhang, J.; Parks, R.; Chen, H.; Housman, M.; Bouton-Verville, H.; Shen, X.; Trama, A.M.; et al. Vaccine induction of heterologous tier 2 hiv-1 neutralizing antibodies in animal models. Cell Rep. 2017, 21, 3681–3690. [Google Scholar] [CrossRef] [PubMed]
- Moody, M.A.; Pedroza-Pacheco, I.; Vandergrift, N.A.; Chui, C.; Lloyd, K.E.; Parks, R.; Soderberg, K.A.; Ogbe, A.T.; Cohen, M.S.; Liao, H.X.; et al. Immune perturbations in hiv-1-infected individuals who make broadly neutralizing antibodies. Sci. Immunol. 2016, 1, aag0851. [Google Scholar] [CrossRef] [PubMed]
- Bradley, T.; Peppa, D.; Pedroza-Pacheco, I.; Li, D.; Cain, D.W.; Henao, R.; Venkat, V.; Hora, B.; Chen, Y.; Vandergrift, N.A.; et al. Rab11fip5 expression and altered natural killer cell function are associated with induction of hiv broadly neutralizing antibody responses. Cell 2018, 175, 387–399. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bresk, C.A.; Hofer, T.; Wilmschen, S.; Krismer, M.; Beierfuß, A.; Effantin, G.; Weissenhorn, W.; Hogan, M.J.; Jordan, A.P.O.; Gelman, R.S.; et al. Induction of Tier 1 HIV Neutralizing Antibodies by Envelope Trimers Incorporated into a Replication Competent Vesicular Stomatitis Virus Vector. Viruses 2019, 11, 159. https://doi.org/10.3390/v11020159
Bresk CA, Hofer T, Wilmschen S, Krismer M, Beierfuß A, Effantin G, Weissenhorn W, Hogan MJ, Jordan APO, Gelman RS, et al. Induction of Tier 1 HIV Neutralizing Antibodies by Envelope Trimers Incorporated into a Replication Competent Vesicular Stomatitis Virus Vector. Viruses. 2019; 11(2):159. https://doi.org/10.3390/v11020159
Chicago/Turabian StyleBresk, C. Anika, Tamara Hofer, Sarah Wilmschen, Marina Krismer, Anja Beierfuß, Grégory Effantin, Winfried Weissenhorn, Michael J. Hogan, Andrea P. O. Jordan, Rebecca S. Gelman, and et al. 2019. "Induction of Tier 1 HIV Neutralizing Antibodies by Envelope Trimers Incorporated into a Replication Competent Vesicular Stomatitis Virus Vector" Viruses 11, no. 2: 159. https://doi.org/10.3390/v11020159
APA StyleBresk, C. A., Hofer, T., Wilmschen, S., Krismer, M., Beierfuß, A., Effantin, G., Weissenhorn, W., Hogan, M. J., Jordan, A. P. O., Gelman, R. S., Montefiori, D. C., Liao, H.-X., Schmitz, J. E., Haynes, B. F., von Laer, D., & Kimpel, J. (2019). Induction of Tier 1 HIV Neutralizing Antibodies by Envelope Trimers Incorporated into a Replication Competent Vesicular Stomatitis Virus Vector. Viruses, 11(2), 159. https://doi.org/10.3390/v11020159






