Abstract
Human noroviruses (HuNoVs) are a leading cause of acute gastroenteritis worldwide. HuNoV infections lead to substantial societal and economic burdens. There are currently no licensed vaccines or therapeutics for the prevention or treatment of HuNoVs. A lack of well-characterized in vitro and in vivo infection models has limited the development of HuNoV countermeasures. Experimental infection of human volunteers and the use of related viruses such as murine NoV have provided helpful insights into HuNoV biology and vaccine and therapeutic development. There remains a need for robust animal models and reverse genetic systems to further HuNoV research. This review summarizes available HuNoV animal models and reverse genetic systems, while providing insight into their usefulness for vaccine and therapeutic development.
1. Introduction
Human noroviruses (HuNoVs) are non-enveloped, single-stranded, positive-sense, RNA viruses belonging to the Caliciviridae family [1,2,3]. Their 7.5–7.7 kb genomes contain three open reading frames (ORFs) (Figure 1a) [4]. ORF1 codes for the six nonstructural proteins, in order from N-terminus to C-terminus: p48, NTPase, p22, VPg, 3C-like protease (3CLpro), and RNA dependent RNA polymerase (RdRp) [5]. Subgenomic RNA, containing ORFs 2 and 3, codes for the major and minor structural proteins, VP1 and VP2 (Figure 1a) [6]. The Norovirus (NoV) genus is divided into seven genogroups (GI–GVII) based on VP1 amino acid homology [7,8,9]. Each genogroup is made up of genotypes GI (n = 9), GII (n = 25), GIII (n = 2), GIV (n = 2), GV (n = 1), GVI (n = 2), and GVII (n = 1), which contain individual virus strains [8,10]. GI, GII, and to a lesser extent GIV NoVs cause disease in humans. GI NoVs only infect humans; however, GII and GIV contain NoVs that infect cats, dogs, pigs, and humans [8]. NoVs are understood to be species-specific [8,11]. Interestingly, GII HuNoVs bind to porcine gastric mucins [12,13,14] and can infect pigs, but robust zoonotic and reverse zoonotic transmissions have not been reported.
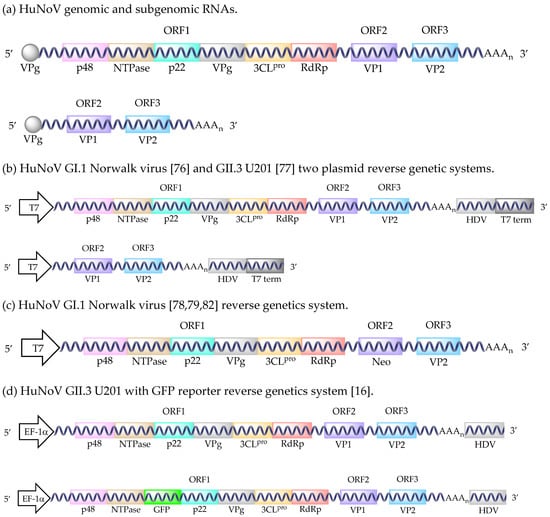
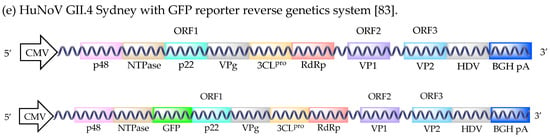
Figure 1.
HuNoV reverse genetic systems. HuNoV genomic and subgenomic RNAs (a). HuNoV GI.1 Norwalk virus and GII.3 U201 two plasmid reverse genetic systems (b). HuNoV GI.1 Norwalk virus reverse genetics system (c). HuNoV GII.3 U201 with GFP reporter reverse genetics system (d). HuNoV GII.4 Sydney with GFP reporter reverse genetics system (e). Abbreviations: T7 polymerase promoter sequence (T7), hepatitis delta virus ribozyme (HDV), T7 terminator sequence (T7 term), human elongation factor-1 alpha promoter sequence (EF-1α), cytomegalovirus promoter sequence (CMV), bovine growth hormone polyadenylation signal (BGH pA).
HuNoVs are the leading cause of acute gastroenteritis worldwide [15,16,17,18,19]. HuNoVs transmit through the fecal–oral route upon ingestion of the encapsidated virions. Following a 24–48 h incubation period, HuNoVs cause symptomatic diarrhea and vomiting for the next 12–60 h [20,21,22]. The infection is self-limiting within a few days, but the virus continues to be shed in the feces for the next few weeks in immunocompetent patients [23,24,25,26]. Annually, there are approximately 700 million infections that result in >200,000 deaths and have an economic burden of >$64 billion [27]. Humans of all age groups are susceptible to HuNoV infection, but children, the immunocompromised, and the elderly are more likely to develop severe disease and therefore are groups of interest for vaccination. Currently, there are no licensed vaccines or therapeutics for the prevention or treatment of HuNoV. Nearly all candidate HuNoV vaccines are subunit vaccines generated from virus-like particle (VLP) constructs. HuNoV VLPs assemble spontaneously after the expression of either VP1, or VP1 and VP2. The immunogenicity of HuNoV VLPs in BALB/c mice upon oral [28,29], intradermal [29,30], intramuscular [31], intranasal [31,32,33,34], and sublingual [32] administration have been studied. The immune responses following intranasal administration of HuNoV VLPs to guinea pigs have also been evaluated [34,35]. VLP vaccination using mucosal adjuvants in gnotobiotic (Gn), germ-free, piglets was evaluated after oral vaccination followed by two intranasal boost immunizations [36]. Following homologous GII.4 challenge, no VLP + adjuvant immunized piglets shed virus and only one had diarrhea [36]. The immunogenicity and protective efficacy of oral, intranasal, and intramuscular bovine NoV VLP immunizations were tested in Gn calves, providing partial protection from disease after homologous bovine NoV challenge [37]. Intramuscular vaccination of chimpanzees with GI.1 VLPs, but not GII.4 VLPs, protected them from homologous HuNoV challenge [38]. Despite the studies completed, no standard animal models have been established for testing HuNoV candidate vaccines. There are currently two VLP vaccines in clinical trials. One is an aluminum hydroxide adjuvanted bivalent GI.1 and GII.4 VLP vaccine [15,39,40]. It has been shown to be immunogenic in rabbits by intranasal and intramuscular administration routes [41]. The other is an adenovirus-vectored GI.1 VP1 VLP vaccine which completed phase I clinical trials [42], but was not tested in any animal models prior to experimental human immunizations. While HuNoV VLPs stimulate antibody responses in BALB/c mice and rabbits, it has not been reported whether or not these animals are susceptible to HuNoV challenge [43,44]. Thus, the implications of vaccination and protection in these studies are unclear. Animal models used in HuNoV studies are summarized in Table 1. Translatable animal models may be useful for evaluating candidate HuNoV vaccines and therapeutics.

Table 1.
Animal models of human norovirus (HuNoV) infection and disease. Non-human primates (NHP), gnotobiotic (Gn) piglet, natural flora piglet, Gn calf, and mouse animal models are grouped by shading. Abbreviations: humanized (Hum.), intravenous (i.v.), intraperitoneal (i.p.), days post-infection (dpi), not applicable (N/A), not reported (NR), data not shown (DNS), respectively (resp.), abnormal histopathology (AH), antibodies (Abs).
HuNoV vaccine efforts are further impeded by an inability to grow HuNoV in a vaccine-approved cell line. This prevents the production of whole virus vaccines (inactivated or live-attenuated). Currently, B cell [45,46,47] and human intestinal enteroid [48,49,50,51] models are used for propagating HuNoV in vitro; however, these systems are not optimal for HuNoV vaccine production [52]. Work to develop a reverse genetics system in lieu of an in vitro cell culture system is in progress to bypass the requirement for natural infection. A reverse genetics system provides a platform for studying the role of individual HuNoV proteins in the context of a HuNoV infection, which may inform the generation of attenuated HuNoV vaccines.
2. Large Animal Models
2.1. Non-Human Primates (NHPs)
Initial efforts to develop large animal models began with unsuccessful attempts to infect non-human primates (NHPs), e.g., rhesus monkeys and baboons, with HuNoV [43]. It was then discovered that chimpanzees produce serum antibodies and shed virus upon oral HuNoV infection, but do not develop gastroenteritis [43]. A single passage of HuNoV in chimpanzees did not alter virus shedding, symptom presentation, or antibody production, indicating a lack of adaptation to the chimpanzee host [43]. Intravenous (i.v.) administration of HuNoV resulted in asymptomatic infection and antibody responses that were dominated by serum IgM and IgG [38]. HuNoV was shed in the feces at similar times following oral or i.v. infection, but the duration of shedding was increased in i.v. infected chimpanzees. Viremia was not detected at any timepoints post-infection (pi), although viral RNAs were detectable in liver tissue. Interestingly, HuNoV antigens were detected in the duodenum, jejunum, and lamina propria (dendritic cells) despite no histological changes to the intestines [38]. Lastly, an infectious dose 50% (ID50) of 4.0 × 107 genome equivalents (g.e.) was established for i.v. infection of chimpanzees [38]. A lack of disease presentation greatly limits the utility of the chimpanzee model for HuNoV studies.
Adult and neonatal pigtail macaques are also susceptible to HuNoV infection [53]. The onset of virus shedding and its duration, as well as the presence and duration of diarrhea are similar between adults and neonates [53]. Curiously, one adult pigtail macaque vomited during the study, a symptom not previously observed in any other HuNoV animal model [53]. Further evaluation of the pigtail macaque animal model may prove fruitful if these animals consistently display symptomatic diarrhea and vomiting as a result of HuNoV infection. Experimental oral infection of other NHPs (common marmosets, cotton-top tamarins, and cynomolgus macaques) did not induce the production of HuNoV-specific antibodies or robust virus shedding [54]. However, a single rhesus macaque from this study shed virus for over two weeks and maintained robust IgG titers until the end of the experimental timecourse [54].
2.2. Gnotobiotic (Gn) Piglets
Gn piglets develop diarrhea, shed virus, and have detectable levels of HuNoV in the intestines upon oral infection [55,56,57,58,59,60,61]. Although low, HuNoV-specific serum and mucosal antibodies have been reported in Gn piglets [61]. Additionally, HuNoV remains infectious after two passages in Gn piglets [55]. Like chimpanzees, the passage of HuNoV in Gn piglets did not improve any infection parameters. An ID50 for oral HuNoV infection of Gn piglets was established at <2.74 × 103 g.e. for 4–5 day old piglets and 6.43 × 104 g.e. for 33–34 day old piglets [57]. Experimentally, an ID50 of 18 virions was established for GI.1 Norwalk virus [62], while another group determined the Norwalk virus ID50 to range between 1.32 × 103–2.80 × 103 viral particles [24]. To date, no ID50 has been reported for any GII.4 HuNoV. The Gn piglet model has been used to test adjuvanted HuNoV VLPs [36] and the inactivation of HuNoV by high pressure processing [63]. Work with the Gn piglet model has also been expanded to include RAG2/IL2RG double knockout (KO) piglets that experience prolonged HuNoV antigen retention in the intestines and asymptomatic virus shedding [58]. In contrast, piglet models with natural flora, such as the miniature piglet model, lack substantial disease presentation [64]. Simvastatin, a cholesterol-reducing statin, has been shown to increase HuNoV infectivity and disease in the Gn piglet model [57,65]. These results strengthen a report that statin use is a risk factor for enhanced HuNoV disease in humans [66]. Simvastatin has immunosuppressive properties and has been linked to the downregulation of interferon-α (IFNα) and major histocompatibility complex II expression [57,65]. Oral administration of IFNα augmented the immune responses of HuNoV-infected Gn piglets, decreasing the onset of virus shedding [65]. The Gn piglet model has also been used to study how individual commensal bacteria affect HuNoV replication in vivo. The histo-blood group antigen (HBGA)-expressing Enterobacter cloacae inhibited HuNoV replication and virus shedding, possibly because HuNoV binds to HBGAs on the bacteria rather than their target eukaryotic cells [67]. Interestingly, this result contrasts with an in vitro study that demonstrated the enhancement of HuNoV infection by E. cloacae [46]. The effects of gastrointestinal bacteria (both commensal and pathogenic) on enteric viral infections represent important considerations relevant for HuNoV animal model development [68,69,70]. Additionally, the role of HBGAs as attachment factors influencing susceptibility for HuNoV infections is important and has been reviewed [71,72]. HuNoV infection of HBGA-typed Gn piglets has been studied [73], but the implications of HBGA expression for HuNoV infection have not been previously studied in the other animal models.
2.3. Gn Calves
Gn calves develop diarrhea and shed HuNoV for up to 6 days after oral infection [74]. Additionally, HuNoV antigen is readily detectable in enterocytes and the lamina propria of infected calves [74]. Notably, intestinal damage and intestinal/serum IgA and IgG production are observed in this animal model [74].
3. Small Animal Models
Attempts to infect adult and suckling mice, kittens, guinea pigs, or rabbits with HuNoV have been unsuccessful [43]. To date, the only small animal model described is a recombination activation gene (Rag−/−) and common gamma chain (γc−/−) deficient BALB/c mouse [44]. The double KO supported a mixed culture of GII HuNoVs through intraperitoneal (i.p.), but not oral infection. Interestingly, dual administration (oral and i.p.) increased viral loads above those observed after i.p. injection alone [44]. The Rag−/−γc−/− BALB/c mouse model has been implemented to demonstrate the antiviral properties of the nucleoside analogue, 2′-C-methylcytidine (2CMC), in vivo [47]. Commercially available wildtype BALB/c and Rag−/−γc−/− C57BL/6J × C57BL/10SgSnAi mice do not support HuNoV, suggesting the requirement for a compromised immune system and possibly other unresolved host factors that may permit virus infection and replication [44].
4. Reverse Genetic Systems
It has been shown that HuNoV RNA isolated from stool can produce all of the structural and nonstructural proteins upon transfection into either Huh7 or Caco-2 cells [75]. However, only a single cycle of replication occurs due to blocks at the receptor binding or uncoating stages [75]. Although inefficient, 100–150 positive cells could be detected after transfecting 5.0 × 108 HuNoV RNA molecules into 105 cells [75]. This observation initiated the development of HuNoV reverse genetics. The first report of a HuNoV replicon system utilized a recombinant Vaccinia virus strain, modified Vaccinia Ankara (MVA), to drive a T7 promoter-controlled plasmid containing a GI.1 Norwalk virus full-length clone in HEK293T cells (Figure 1b) [76]. This system produced approximately 8.5 × 104 HuNoV particles from 12 T-75 flasks [76]. The first GII HuNoV infectious clone was created shortly after using a similar Vaccinia virus T7 promoter system (vTF7) with the GII.3 U201 strain in HEK293T cells (Figure 1b) [77]. However, the particles formed using this system were found to be less dense than naturally isolated HuNoV (1.32 g/cm3 versus 1.39–1.40 g/cm3). With both methods, Vaccinia caused HuNoV-independent cytopathic effects at 72 hpi, limiting the utility of these systems.
Subsequently, a helper virus-free GI.1 Norwalk virus system was developed (Figure 1c) [78]. In vitro transcription of a plasmid using Vaccinia virus-T7 produced a recombinant RNA that contained the full-length genome with a neomycin resistance gene (neo) inserted into the VP1 gene. Transfection of this recombinant RNA produced the nonstructural proteins and VP2. Disruption of the VP1 gene prevents the use of this system to isolate infectious virus, because the major capsid protein is compromised. This system was successfully used to transfect Huh7 and BHK21 cells, but not Vero, 293, 293T, or LLC-PK cells [78]. The transfected cells remained stably transfected after at least 100 passages, with levels of 2.6 × 1011 g.e./1 µg of RNA and 8.0 × 108 g.e./1 µg of RNA produced in Huh7 and BHK21 cells, respectively. The implementation of this system was used to determine that Norwalk virus was sensitive to 72 h treatment with IFNα (effective dose 50% (ED50) levels of 2 units/mL and 20 units/mL for Huh7 and BHK21 cells, respectively) [78]. Another study using this system demonstrated that Norwalk virus produced in Huh7 cells was also sensitive to IFNγ, ribavirin, and mycophenolic acid [79]. Nucleoside analogue inhibition of viral transcription by 2CMC [80] or 7-deaza-2′-C-methyladenosine [81] treatment decreased Norwalk virus production in this replicon system. Additionally, a GI.1 Norwalk virus replicon-expressing human gastric tumor-1 system was adapted from the Huh7 system to explore the development of resistance to rupintrivir, a viral protease inhibitor [82].
The first true helper virus-free system used the mammalian EF-1α promoter to drive HuNoV RNA production (Figure 1d) [16]. This system also supported infectious murine NoV (MuNoV) production [16]. Originally developed for the GII.3 U201 strain, the production of full-length genomes per 106 cells amounted to 8.0 × 104, 1.4 × 104, 2.4 × 102, and 1.3 × 101 copies in COS7, 293T, Huh7, and Caco-2 cells, respectively. As evidenced by MuNoV production using this system, HuNoV virion production may be up to 10-fold less than the corresponding RNA levels [16]. Differences in virus production levels can be explained by the transformed cell lines (COS7 and HEK293T) expressing the SV40 T antigen, which helps drive the SV40 promoter in the expression system [16]. The utility of this system was enhanced by adding a GFP gene between the NTPase and p22 viral genes, creating a GFP reporter infectious clone [16]. However, the insertion of the GFP gene resulted in up to 50-fold fewer HuNoV virions being produced [16]. Despite an inability to detect the capsid proteins by Western blot or immunofluorescence assays, an ORF2 GFP reporter plasmid indicated that the structural proteins were being produced [16]. Unfortunately, attempts to adapt the system to GI.1 NV68, GII.P4-GII.3 chimera TCH04-577, and GII.4 Saga1 viruses resulted in 10- to 1000-fold less HuNoV production [16]. Although the HuNoV RNA from these recombinant virions is infectious, upon transfection, the infectivity of the intact virions has not been tested [16].
The clinical relevancy of HuNoV reverse genetic systems was an issue, until the development of a GII.4 Sydney 2012 infectious clone (Figure 1e) [83]. Structural proteins were readily detected upon transfection of the plasmid into Caco-2 cells [83]. The insertion of a GFP sequence between the NTPase and p22 viral genes generated a fluorescent reporter system. However, this negatively affected viral structural protein synthesis through an unknown mechanism, as previously observed [16,83]. The utility and robustness of this system remain unclear as studies quantifying virus production and demonstrating the passage of the recombinant virions still need to be completed.
5. Conclusions
The development of animal models and reverse genetics for HuNoV are significant milestones that are needed to drive HuNoV pathogenesis studies and vaccine development. Currently, the pigtail macaque is the most promising NHP infection model for HuNoV. Expanded studies to test levels of virus replication, the detection of HuNoV in tissues, the presence of antibodies at mucosal surfaces, and infection with clinically relevant GII.4 HuNoVs are needed. Vomiting as a result of HuNoV infection is a unique observation for this animal model. Epidemiologically, one criterion for the determination of HuNoV as the etiological agent causing an outbreak is that ≥50% of affected persons present with vomiting [21,22]. The precise cause of HuNoV-induced vomiting is unclear and has not been as readily investigated as HuNoV-associated diarrhea. Of the models described, only NHPs and piglets have an emetic response, while calves and mice are understood to have no or a greatly limited emetic response. This precludes HuNoV-associated vomiting from being studied in calves or mice. Even so, both the Gn piglet and the under-studied Gn calf model are encouraging large animal models for HuNoV infection. Prolonged virus shedding and diarrhea in response to infection and the production of serum and mucosal antibodies underscore the usefulness of these models. Further development of the pigtail macaque, Gn piglet, and Gn calf models may yield robust models of infection that can bolster experimental human infection studies. A drawback to the current large animal models for HuNoV infection is their accessibility. NHPs are costly and studies involving such models raise ethical concerns, potentially limiting their utility. Likewise, Gn animals require specific handling and facilities in order to maintain bacteria-free animals [84,85]. Lack of commensal bacteria in these animals impacts HuNoV infection [64,67] and likely impairs mucosal immune responses especially in the gastrointestinal tract. However, Gn models provide the opportunity to study HuNoV infections in the context of controlled bacterial populations. Gn animals can be colonized with single bacterial species or complex bacterial populations by fecal transplants, from similar or even dissimilar mammalian hosts. Therefore, the reconstitution of Gn piglets or calves with a human microbiota favorable for HuNoV infection may enhance these models. The benefits of large animal models for HuNoV infection include more extensive tissue sampling and ease of test subject procurement, as compared to human experimental infection studies. Further advantages to accessibility and cost are realized when working with a small animal model such as the Rag−/−γc−/− BALB/c mouse. Unfortunately, transmission studies cannot be completed in these mice because they cannot be infected orally. Also, Rag−/−γc−/− mice lack the ability to produce numerous cytokines and mature B and T cells. Immune responses to infection are therefore not representative of typical HuNoV infection. Expansion of studies with the large animal models (pigtail macaque, Gn piglet, and Gn calf) and the development of small animal models beyond the Rag−/−γc−/− BALB/c mouse will provide opportunities to standardize the preclinical HuNoV therapeutic and vaccine pipelines.
HuNoV reverse genetic systems provide a platform for in vitro infection in lieu of a classical infection model. However, the adoption of all current HuNoV reverse genetic systems is impeded by their inefficiency compared to the recovery of virus from stool or MuNoV reverse genetic systems [86,87,88]. Additionally, lack of a recognized receptor for HuNoV infection and other gaps in the understanding of HuNoV biology have limited the development of both reverse genetic systems and cell culture models. The discovery of CD300lf as a proteinaceous receptor for MuNoV [89,90] has greatly advanced the study of MuNoV. This type of breakthrough for HuNoV is poised to revolutionize the development of HuNoV in vitro propagation systems. Further studies are needed to optimize the HuNoV reverse genetic systems, which may provide a platform for developing live-attenuated HuNoV vaccines, an untapped and under-investigated area. MuNoV work in this sector has previously identified sites for genetic engineering that may prove useful for HuNoV vaccine studies [91,92].
Author Contributions
Writing—original draft, K.V.T.; review and editing, R.A.T.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Green, K.Y. Caliciviridae: The Noroviruses, 6th ed.; Wolters Kluwer Health/Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; Volume 1, pp. 582–608. [Google Scholar]
- Dolin, R.; Blacklow, N.R.; DuPont, H.; Buscho, R.F.; Wyatt, R.G.; Kasel, J.A.; Hornick, R.; Chanock, R.M. Biological properties of Norwalk agent of acute infectious nonbacterial gastroenteritis. Proc. Soc. Exp. Biol. Med. 1972, 140, 578–583. [Google Scholar] [CrossRef]
- Thorne, L.G.; Goodfellow, I.G. Norovirus gene expression and replication. J. Gen. Virol. 2014, 95, 278–291. [Google Scholar] [CrossRef] [PubMed]
- Lambden, P.R.; Caul, E.O.; Ashley, C.R.; Clarke, I.N. Sequence and genome organization of a human small round-structured (Norwalk-like) virus. Science 1993, 259, 516–519. [Google Scholar] [CrossRef]
- Hardy, M.E. Norovirus protein structure and function. FEMS Microbiol. Lett. 2005, 253, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Wang, M.; Graham, D.Y.; Estes, M.K. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J. Virol. 1992, 66, 6527–6532. [Google Scholar] [PubMed]
- Proceedings of the Minutes of the 5th Meeting of the ICTV, Strasbourg, France, 4 August 1981; pp. 1–8.
- Vinje, J. Advances in laboratory methods for detection and typing of norovirus. J. Clin. Microbiol. 2015, 53, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Kroneman, A.; Vega, E.; Vennema, H.; Vinje, J.; White, P.A.; Hansman, G.; Green, K.; Martella, V.; Katayama, K.; Koopmans, M. Proposal for a unified norovirus nomenclature and genotyping. Arch. Virol. 2013, 158, 2059–2068. [Google Scholar] [CrossRef] [PubMed]
- Chhabra, P.; Aswath, K.; Collins, N.; Ahmed, T.; Olortegui, M.P.; Kosek, M.; Cebelinski, E.; Cooper, P.J.; Bucardo, F.; Lopez, M.R.; et al. Near-Complete Genome Sequences of Several New Norovirus Genogroup II Genotypes. Genome Announce. 2018, 6. [Google Scholar] [CrossRef]
- Taube, S.; Wobus, C.E. A novel reverse genetics system for human norovirus. Trends Microbiol. 2014, 22, 604–606. [Google Scholar] [CrossRef]
- Tian, P.; Brandl, M.; Mandrell, R. Porcine gastric mucin binds to recombinant norovirus particles and competitively inhibits their binding to histo-blood group antigens and Caco-2 cells. Lett. Appl. Microbiol. 2005, 41, 315–320. [Google Scholar] [CrossRef]
- Tian, P.; Jiang, X.; Zhong, W.; Jensen, H.M.; Brandl, M.; Bates, A.H.; Engelbrektson, A.L.; Mandrell, R. Binding of recombinant norovirus like particle to histo-blood group antigen on cells in the lumen of pig duodenum. Res. Vet. Sci. 2007, 83, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Tian, P.; Yang, D.; Jiang, X.; Zhong, W.; Cannon, J.L.; Burkhardt, W., 3rd; Woods, J.W.; Hartman, G.; Lindesmith, L.; Baric, R.S.; et al. Specificity and kinetics of norovirus binding to magnetic bead-conjugated histo-blood group antigens. J. Appl. Microbiol. 2010, 109, 1753–1762. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, D.I.; Atmar, R.L.; Lyon, G.M.; Treanor, J.J.; Chen, W.H.; Jiang, X.; Vinje, J.; Gregoricus, N.; Frenck, R.W., Jr.; Moe, C.L.; et al. Norovirus vaccine against experimental human GII.4 virus illness: A challenge study in healthy adults. J. Infect. Dis. 2015, 211, 870–878. [Google Scholar] [CrossRef] [PubMed]
- Katayama, K.; Murakami, K.; Sharp, T.M.; Guix, S.; Oka, T.; Takai-Todaka, R.; Nakanishi, A.; Crawford, S.E.; Atmar, R.L.; Estes, M.K. Plasmid-based human norovirus reverse genetics system produces reporter-tagged progeny virus containing infectious genomic RNA. Proc. Natl. Acad. Sci. USA 2014, 111, E4043–E4052. [Google Scholar] [CrossRef] [PubMed]
- Batz, M.B.; Hoffmann, S.; Morris, J.G., Jr. Ranking the disease burden of 14 pathogens in food sources in the United States using attribution data from outbreak investigations and expert elicitation. J. Food Prot. 2012, 75, 1278–1291. [Google Scholar] [CrossRef] [PubMed]
- Bartsch, S.M.; Lopman, B.A.; Hall, A.J.; Parashar, U.D.; Lee, B.Y. The potential economic value of a human norovirus vaccine for the United States. Vaccine 2012, 30, 7097–7104. [Google Scholar] [CrossRef] [PubMed]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States—Major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- Devasia, T.; Lopman, B.; Leon, J.; Handel, A. Association of host, agent and environment characteristics and the duration of incubation and symptomatic periods of norovirus gastroenteritis. Epidemiol. Infect. 2015, 143, 2308–2314. [Google Scholar] [CrossRef]
- Kaplan, J.E.; Feldman, R.; Campbell, D.S.; Lookabaugh, C.; Gary, G.W. The frequency of a Norwalk-like pattern of illness in outbreaks of acute gastroenteritis. Am. J. Public. Health 1982, 72, 1329–1332. [Google Scholar] [CrossRef]
- Turcios, R.M.; Widdowson, M.A.; Sulka, A.C.; Mead, P.S.; Glass, R.I. Reevaluation of epidemiological criteria for identifying outbreaks of acute gastroenteritis due to norovirus: United States, 1998–2000. Clin. Infect. Dis. 2006, 42, 964–969. [Google Scholar] [CrossRef]
- Atmar, R.L.; Opekun, A.R.; Gilger, M.A.; Estes, M.K.; Crawford, S.E.; Neill, F.H.; Graham, D.Y. Norwalk virus shedding after experimental human infection. Emerg. Infect. Dis. 2008, 14, 1553–1557. [Google Scholar] [CrossRef] [PubMed]
- Atmar, R.L.; Opekun, A.R.; Gilger, M.A.; Estes, M.K.; Crawford, S.E.; Neill, F.H.; Ramani, S.; Hill, H.; Ferreira, J.; Graham, D.Y. Determination of the 50% human infectious dose for Norwalk virus. J. Infect. Dis. 2014, 209, 1016–1022. [Google Scholar] [CrossRef] [PubMed]
- Roth, A.N.; Karst, S.M. Norovirus mechanisms of immune antagonism. Curr. Opin. Virol. 2016, 16, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Gustavsson, L.; Skovbjerg, S.; Lindh, M.; Westin, J.; Andersson, L.M. Low serum levels of CCL5 are associated with longer duration of viral shedding in norovirus infection. J. Clin. Virol. 2015, 69, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Bartsch, S.M.; Lopman, B.A.; Ozawa, S.; Hall, A.J.; Lee, B.Y. Global Economic Burden of Norovirus Gastroenteritis. PLoS ONE 2016, 11, e0151219. [Google Scholar] [CrossRef] [PubMed]
- Ball, J.M.; Hardy, M.E.; Atmar, R.L.; Conner, M.E.; Estes, M.K. Oral immunization with recombinant Norwalk virus-like particles induces a systemic and mucosal immune response in mice. J. Virol. 1998, 72, 1345–1353. [Google Scholar] [PubMed]
- Harrington, P.R.; Lindesmith, L.; Yount, B.; Moe, C.L.; Baric, R.S. Binding of Norwalk virus-like particles to ABH histo-blood group antigens is blocked by antisera from infected human volunteers or experimentally vaccinated mice. J. Virol. 2002, 76, 12335–12343. [Google Scholar] [CrossRef]
- LoBue, A.D.; Lindesmith, L.; Yount, B.; Harrington, P.R.; Thompson, J.M.; Johnston, R.E.; Moe, C.L.; Baric, R.S. Multivalent norovirus vaccines induce strong mucosal and systemic blocking antibodies against multiple strains. Vaccine 2006, 24, 5220–5234. [Google Scholar] [CrossRef]
- Heinimaki, S.; Malm, M.; Vesikari, T.; Blazevic, V. Parenterally Administered Norovirus GII.4 Virus-Like Particle Vaccine Formulated with Aluminum Hydroxide or Monophosphoryl Lipid A Adjuvants Induces Systemic but Not Mucosal Immune Responses in Mice. J. Immunol. Res. 2018, 2018, 3487095. [Google Scholar] [CrossRef]
- Hwang, H.S.; Puth, S.; Tan, W.; Verma, V.; Jeong, K.; Lee, S.E.; Rhee, J.H. More robust gut immune responses induced by combining intranasal and sublingual routes for prime-boost immunization. Hum. Vaccin. Immunother. 2018, 1–9. [Google Scholar] [CrossRef]
- Guerrero, R.A.; Ball, J.M.; Krater, S.S.; Pacheco, S.E.; Clements, J.D.; Estes, M.K. Recombinant Norwalk virus-like particles administered intranasally to mice induce systemic and mucosal (fecal and vaginal) immune responses. J. Virol. 2001, 75, 9713–9722. [Google Scholar] [CrossRef]
- Velasquez, L.S.; Shira, S.; Berta, A.N.; Kilbourne, J.; Medi, B.M.; Tizard, I.; Ni, Y.; Arntzen, C.J.; Herbst-Kralovetz, M.M. Intranasal delivery of Norwalk virus-like particles formulated in an in situ gelling, dry powder vaccine. Vaccine 2011, 29, 5221–5231. [Google Scholar] [CrossRef] [PubMed]
- Springer, M.J.; Ni, Y.; Finger-Baker, I.; Ball, J.P.; Hahn, J.; DiMarco, A.V.; Kobs, D.; Horne, B.; Talton, J.D.; Cobb, R.R. Preclinical dose-ranging studies of a novel dry powder norovirus vaccine formulation. Vaccine 2016, 34, 1452–1458. [Google Scholar] [CrossRef] [PubMed]
- Souza, M.; Costantini, V.; Azevedo, M.S.; Saif, L.J. A human norovirus-like particle vaccine adjuvanted with ISCOM or mLT induces cytokine and antibody responses and protection to the homologous GII.4 human norovirus in a gnotobiotic pig disease model. Vaccine 2007, 25, 8448–8459. [Google Scholar] [CrossRef] [PubMed]
- Han, M.G.; Cheetham, S.; Azevedo, M.; Thomas, C.; Saif, L.J. Immune responses to bovine norovirus-like particles with various adjuvants and analysis of protection in gnotobiotic calves. Vaccine 2006, 24, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Bok, K.; Parra, G.I.; Mitra, T.; Abente, E.; Shaver, C.K.; Boon, D.; Engle, R.; Yu, C.; Kapikian, A.Z.; Sosnovtsev, S.V.; et al. Chimpanzees as an animal model for human norovirus infection and vaccine development. Proc. Natl. Acad. Sci. USA 2011, 108, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Treanor, J.J.; Atmar, R.L.; Frey, S.E.; Gormley, R.; Chen, W.H.; Ferreira, J.; Goodwin, R.; Borkowski, A.; Clemens, R.; Mendelman, P.M. A novel intramuscular bivalent norovirus virus-like particle vaccine candidate—Reactogenicity, safety, and immunogenicity in a phase 1 trial in healthy adults. J. Infect. Dis. 2014, 210, 1763–1771. [Google Scholar] [CrossRef] [PubMed]
- Leroux-Roels, G.; Cramer, J.P.; Mendelman, P.M.; Sherwood, J.; Clemens, R.; Aerssens, A.; De Coster, I.; Borkowski, A.; Baehner, F.; Van Damme, P. Safety and Immunogenicity of Different Formulations of Norovirus Vaccine Candidate in Healthy Adults: A Randomized, Controlled, Double-Blind Clinical Trial. J. Infect. Dis. 2018, 217, 597–607. [Google Scholar] [CrossRef]
- Parra, G.I.; Bok, K.; Taylor, R.; Haynes, J.R.; Sosnovtsev, S.V.; Richardson, C.; Green, K.Y. Immunogenicity and specificity of norovirus Consensus GII.4 virus-like particles in monovalent and bivalent vaccine formulations. Vaccine 2012, 30, 3580–3586. [Google Scholar] [CrossRef]
- Kim, L.; Liebowitz, D.; Lin, K.; Kasparek, K.; Pasetti, M.F.; Garg, S.J.; Gottlieb, K.; Trager, G.; Tucker, S.N. Safety and immunogenicity of an oral tablet norovirus vaccine, a phase I randomized, placebo-controlled trial. JCI Insight 2018, 3. [Google Scholar] [CrossRef]
- Wyatt, R.G.; Greenberg, H.B.; Dalgard, D.W.; Allen, W.P.; Sly, D.L.; Thornhill, T.S.; Chanock, R.M.; Kapikian, A.Z. Experimental infection of chimpanzees with the Norwalk agent of epidemic viral gastroenteritis. J. Med. Virol. 1978, 2, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Taube, S.; Kolawole, A.O.; Hohne, M.; Wilkinson, J.E.; Handley, S.A.; Perry, J.W.; Thackray, L.B.; Akkina, R.; Wobus, C.E. A mouse model for human norovirus. mBio 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.K.; Grau, K.R.; Costantini, V.; Kolawole, A.O.; de Graaf, M.; Freiden, P.; Graves, C.L.; Koopmans, M.; Wallet, S.M.; Tibbetts, S.A.; et al. Human norovirus culture in B cells. Nat. Prot. 2015, 10, 1939–1947. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.K.; Watanabe, M.; Zhu, S.; Graves, C.L.; Keyes, L.R.; Grau, K.R.; Gonzalez-Hernandez, M.B.; Iovine, N.M.; Wobus, C.E.; Vinje, J.; et al. Enteric bacteria promote human and mouse norovirus infection of B cells. Science 2014, 346, 755–759. [Google Scholar] [CrossRef] [PubMed]
- Kolawole, A.O.; Rocha-Pereira, J.; Elftman, M.D.; Neyts, J.; Wobus, C.E. Inhibition of human norovirus by a viral polymerase inhibitor in the B cell culture system and in the mouse model. Antivir. Res. 2016, 132, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Ettayebi, K.; Crawford, S.E.; Murakami, K.; Broughman, J.R.; Karandikar, U.; Tenge, V.R.; Neill, F.H.; Blutt, S.E.; Zeng, X.L.; Qu, L.; et al. Replication of human noroviruses in stem cell-derived human enteroids. Science 2016, 353, 1387–1393. [Google Scholar] [CrossRef] [PubMed]
- Costantini, V.; Morantz, E.K.; Browne, H.; Ettayebi, K.; Zeng, X.L.; Atmar, R.L.; Estes, M.K.; Vinje, J. Human Norovirus Replication in Human Intestinal Enteroids as Model to Evaluate Virus Inactivation. Emerg. Infect. Dis. 2018, 24, 1453–1464. [Google Scholar] [CrossRef] [PubMed]
- Alvarado, G.; Ettayebi, K.; Atmar, R.L.; Bombardi, R.G.; Kose, N.; Estes, M.K.; Crowe, J.E., Jr. Human Monoclonal Antibodies That Neutralize Pandemic GII.4 Noroviruses. Gastroenterology 2018. [Google Scholar] [CrossRef]
- Santiana, M.; Ghosh, S.; Ho, B.A.; Rajasekaran, V.; Du, W.L.; Mutsafi, Y.; De Jesus-Diaz, D.A.; Sosnovtsev, S.V.; Levenson, E.A.; Parra, G.I.; et al. Vesicle-Cloaked Virus Clusters Are Optimal Units for Inter-organismal Viral Transmission. Cell Host Microbe 2018, 24, 208–220.e208. [Google Scholar] [CrossRef]
- WHO. Recommendations for the Evaluation of Animal Cell Cultures as Substrates for the Manufacture of Biological Medicinal Products and for the Characterization of Cells Banks; WHO Press: Geneva, Switzerland, 2013; pp. 79–187. [Google Scholar]
- Subekti, D.S.; Tjaniadi, P.; Lesmana, M.; McArdle, J.; Iskandriati, D.; Budiarsa, I.N.; Walujo, P.; Suparto, I.H.; Winoto, I.; Campbell, J.R.; et al. Experimental infection of Macaca nemestrina with a Toronto Norwalk-like virus of epidemic viral gastroenteritis. J. Med. Virol. 2002, 66, 400–406. [Google Scholar] [CrossRef]
- Rockx, B.H.; Bogers, W.M.; Heeney, J.L.; van Amerongen, G.; Koopmans, M.P. Experimental norovirus infections in non-human primates. J. Med. Virol. 2005, 75, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Cheetham, S.; Souza, M.; Meulia, T.; Grimes, S.; Han, M.G.; Saif, L.J. Pathogenesis of a genogroup II human norovirus in gnotobiotic pigs. J. Virol. 2006, 80, 10372–10381. [Google Scholar] [CrossRef] [PubMed]
- Takanashi, S.; Wang, Q.; Chen, N.; Shen, Q.; Jung, K.; Zhang, Z.; Yokoyama, M.; Lindesmith, L.C.; Baric, R.S.; Saif, L.J. Characterization of emerging GII.g/GII.12 noroviruses from a gastroenteritis outbreak in the United States in 2010. J. Clin. Microbiol. 2011, 49, 3234–3244. [Google Scholar] [CrossRef] [PubMed]
- Bui, T.; Kocher, J.; Li, Y.; Wen, K.; Li, G.; Liu, F.; Yang, X.; LeRoith, T.; Tan, M.; Xia, M.; et al. Median infectious dose of human norovirus GII.4 in gnotobiotic pigs is decreased by simvastatin treatment and increased by age. J. Gen. Virol. 2013, 94, 2005–2016. [Google Scholar] [CrossRef] [PubMed]
- Lei, S.; Ryu, J.; Wen, K.; Twitchell, E.; Bui, T.; Ramesh, A.; Weiss, M.; Li, G.; Samuel, H.; Clark-Deener, S.; et al. Increased and prolonged human norovirus infection in RAG2/IL2RG deficient gnotobiotic pigs with severe combined immunodeficiency. Sci. Rep. 2016, 6, 25222. [Google Scholar] [CrossRef] [PubMed]
- Park, B.J.; Jung, S.T.; Choi, C.S.; Myoung, J.J.; Ahn, H.S.; Han, S.H.; Kim, Y.H.; Go, H.J.; Lee, J.B.; Park, S.Y.; et al. Pathogenesis of human norovirus genogroup II genotype 4 in post-weaning gnotobiotic pigs. J. Microl. Biotechnol. 2018, 28, 2133–2140. [Google Scholar] [CrossRef]
- Kocher, J.; Bui, T.; Giri-Rachman, E.; Wen, K.; Li, G.; Yang, X.; Liu, F.; Tan, M.; Xia, M.; Zhong, W.; et al. Intranasal P particle vaccine provided partial cross-variant protection against human GII.4 norovirus diarrhea in gnotobiotic pigs. J. Virol. 2014, 88, 9728–9743. [Google Scholar] [CrossRef]
- Souza, M.; Cheetham, S.M.; Azevedo, M.S.; Costantini, V.; Saif, L.J. Cytokine and antibody responses in gnotobiotic pigs after infection with human norovirus genogroup II.4 (HS66 strain). J. Virol. 2007, 81, 9183–9192. [Google Scholar] [CrossRef]
- Teunis, P.F.; Moe, C.L.; Liu, P.; Miller, S.E.; Lindesmith, L.; Baric, R.S.; Le Pendu, J.; Calderon, R.L. Norwalk virus: How infectious is it? J. Med. Virol. 2008, 80, 1468–1476. [Google Scholar] [CrossRef]
- Lou, F.; Ye, M.; Ma, Y.; Li, X.; DiCaprio, E.; Chen, H.; Krakowka, S.; Hughes, J.; Kingsley, D.; Li, J. A Gnotobiotic Pig Model for Determining Human Norovirus Inactivation by High-Pressure Processing. Appl. Environ. Microbiol. 2015, 81, 6679–6687. [Google Scholar] [CrossRef]
- Seo, D.J.; Jung, D.; Jung, S.; Ha, S.K.; Ha, S.D.; Choi, I.S.; Myoung, J.; Choi, C. Experimental miniature piglet model for the infection of human norovirus GII. J. Med. Virol. 2017. [Google Scholar] [CrossRef]
- Jung, K.; Wang, Q.; Kim, Y.; Scheuer, K.; Zhang, Z.; Shen, Q.; Chang, K.O.; Saif, L.J. The effects of simvastatin or interferon-alpha on infectivity of human norovirus using a gnotobiotic pig model for the study of antivirals. PLoS ONE 2012, 7, e41619. [Google Scholar] [CrossRef]
- Rondy, M.; Koopmans, M.; Rotsaert, C.; Van Loon, T.; Beljaars, B.; Van Dijk, G.; Siebenga, J.; Svraka, S.; Rossen, J.W.; Teunis, P.; et al. Norovirus disease associated with excess mortality and use of statins: A retrospective cohort study of an outbreak following a pilgrimage to Lourdes. Epidemiol. Infect. 2011, 139, 453–463. [Google Scholar] [CrossRef]
- Lei, S.; Samuel, H.; Twitchell, E.; Bui, T.; Ramesh, A.; Wen, K.; Weiss, M.; Li, G.; Yang, X.; Jiang, X.; et al. Enterobacter cloacae inhibits human norovirus infectivity in gnotobiotic pigs. Sci. Rep. 2016, 6, 25017. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, J.K.; Virgin, H.W. Viral immunity. Transkingdom control of viral infection and immunity in the mammalian intestine. Science 2016, 351. [Google Scholar] [CrossRef] [PubMed]
- Erickson, A.K.; Jesudhasan, P.R.; Mayer, M.J.; Narbad, A.; Winter, S.E.; Pfeiffer, J.K. Bacteria Facilitate Enteric Virus Co-infection of Mammalian Cells and Promote Genetic Recombination. Cell Host Microbe 2018, 23, 77–88.e75. [Google Scholar] [CrossRef]
- Almand, E.A.; Moore, M.D.; Outlaw, J.; Jaykus, L.A. Human norovirus binding to select bacteria representative of the human gut microbiota. PLoS ONE 2017, 12, e0173124. [Google Scholar] [CrossRef]
- Tan, M.; Jiang, X. Histo-blood group antigens: A common niche for norovirus and rotavirus. Expert Rev. Mol. Med. 2014, 16, e5. [Google Scholar] [CrossRef]
- Schroten, H.; Hanisch, F.G.; Hansman, G.S. Human Norovirus Interactions with Histo-Blood Group Antigens and Human Milk Oligosaccharides. J. Virol. 2016, 90, 5855–5859. [Google Scholar] [CrossRef]
- Cheetham, S.; Souza, M.; McGregor, R.; Meulia, T.; Wang, Q.; Saif, L.J. Binding patterns of human norovirus-like particles to buccal and intestinal tissues of gnotobiotic pigs in relation to A/H histo-blood group antigen expression. J. Virol. 2007, 81, 3535–3544. [Google Scholar] [CrossRef] [PubMed]
- Souza, M.; Azevedo, M.S.; Jung, K.; Cheetham, S.; Saif, L.J. Pathogenesis and immune responses in gnotobiotic calves after infection with the genogroup II.4-HS66 strain of human norovirus. J. Virol. 2008, 82, 1777–1786. [Google Scholar] [CrossRef] [PubMed]
- Guix, S.; Asanaka, M.; Katayama, K.; Crawford, S.E.; Neill, F.H.; Atmar, R.L.; Estes, M.K. Norwalk virus RNA is infectious in mammalian cells. J. Virol. 2007, 81, 12238–12248. [Google Scholar] [CrossRef] [PubMed]
- Asanaka, M.; Atmar, R.L.; Ruvolo, V.; Crawford, S.E.; Neill, F.H.; Estes, M.K. Replication and packaging of Norwalk virus RNA in cultured mammalian cells. Proc. Natl. Acad. Sci. USA 2005, 102, 10327–10332. [Google Scholar] [CrossRef] [PubMed]
- Katayama, K.; Hansman, G.S.; Oka, T.; Ogawa, S.; Takeda, N. Investigation of norovirus replication in a human cell line. Arch. Virol. 2006, 151, 1291–1308. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.O.; Sosnovtsev, S.V.; Belliot, G.; King, A.D.; Green, K.Y. Stable expression of a Norwalk virus RNA replicon in a human hepatoma cell line. Virology 2006, 353, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.O.; George, D.W. Interferons and ribavirin effectively inhibit Norwalk virus replication in replicon-bearing cells. J. Virol. 2007, 81, 12111–12118. [Google Scholar] [CrossRef]
- Rocha-Pereira, J.; Jochmans, D.; Debing, Y.; Verbeken, E.; Nascimento, M.S.; Neyts, J. The viral polymerase inhibitor 2′-C-methylcytidine inhibits Norwalk virus replication and protects against norovirus-induced diarrhea and mortality in a mouse model. J. Virol. 2013, 87, 11798–11805. [Google Scholar] [CrossRef]
- Van Dycke, J.; Arnoldi, F.; Papa, G.; Vandepoele, J.; Burrone, O.R.; Mastrangelo, E.; Tarantino, D.; Heylen, E.; Neyts, J.; Rocha-Pereira, J. A Single Nucleoside Viral Polymerase Inhibitor Against Norovirus, Rotavirus, and Sapovirus-Induced Diarrhea. J. Infect. Dis. 2018, 218, 1753–1758. [Google Scholar] [CrossRef]
- Kitano, M.; Hosmillo, M.; Emmott, E.; Lu, J.; Goodfellow, I. Selection and Characterization of Rupintrivir-Resistant Norwalk Virus Replicon Cells in vitro. Antimicrob. Agents Chemother. 2018. [Google Scholar] [CrossRef]
- Oliveira, L.M.; Blawid, R.; Orilio, A.F.; Andrade, B.Y.G.; Souza, A.C.A.; Nagata, T. Development of an infectious clone and replicon system of norovirus GII.4. J. Virol. Methods 2018, 258, 49–53. [Google Scholar] [CrossRef]
- Meyer, R.C.; Bohl, E.H.; Kohler, E.M. Procurement and Maintenance Of Germ-Free Seine For Microbiological Investigations. Appl. Microbiol. 1964, 12, 295–300. [Google Scholar] [PubMed]
- Yuan, L.; Jobst, P.M.; Weiss, M. Chapter 5—Gnotobiotic Pigs: From Establishing Facility to Modeling Human Infectious Diseases. In Gnotobiotics; Schoeb, T.R., Eaton, K.A., Eds.; Academic Press: Cambridge, MA, USA, 2017; pp. 349–368. [Google Scholar]
- Chaudhry, Y.; Skinner, M.A.; Goodfellow, I.G. Recovery of genetically defined murine norovirus in tissue culture by using a fowlpox virus expressing T7 RNA polymerase. J. Gen. Virol. 2007, 88, 2091–2100. [Google Scholar] [CrossRef] [PubMed]
- Ward, V.K.; McCormick, C.J.; Clarke, I.N.; Salim, O.; Wobus, C.E.; Thackray, L.B.; Virgin, H.W.t.; Lambden, P.R. Recovery of infectious murine norovirus using pol II-driven expression of full-length cDNA. Proc. Natl. Acad. Sci. USA 2007, 104, 11050–11055. [Google Scholar] [CrossRef] [PubMed]
- Yunus, M.A.; Chung, L.M.; Chaudhry, Y.; Bailey, D.; Goodfellow, I. Development of an optimized RNA-based murine norovirus reverse genetics system. J. Virol. Methods 2010, 169, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Orchard, R.C.; Wilen, C.B.; Doench, J.G.; Baldridge, M.T.; McCune, B.T.; Lee, Y.C.; Lee, S.; Pruett-Miller, S.M.; Nelson, C.A.; Fremont, D.H.; et al. Discovery of a proteinaceous cellular receptor for a norovirus. Science 2016, 353, 933–936. [Google Scholar] [CrossRef] [PubMed]
- Haga, K.; Fujimoto, A.; Takai-Todaka, R.; Miki, M.; Doan, Y.H.; Murakami, K.; Yokoyama, M.; Murata, K.; Nakanishi, A.; Katayama, K. Functional receptor molecules CD300lf and CD300ld within the CD300 family enable murine noroviruses to infect cells. Proc. Natl. Acad. Sci. USA 2016, 113, E6248–E6255. [Google Scholar] [CrossRef] [PubMed]
- Thorne, L.; Bailey, D.; Goodfellow, I. High-resolution functional profiling of the norovirus genome. J. Virol. 2012, 86, 11441–11456. [Google Scholar] [CrossRef]
- Thorne, L.; Lu, J.; Chaudhry, Y.; Bailey, D.; Goodfellow, I. Targeting macrophage- and intestinal epithelial cell-specific microRNAs against norovirus restricts replication in vivo. J. Gen. Virol. 2018. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).