Abstract
Viroid discovery as well as the economic significance of viroids and biological properties are presented. Next-generation sequencing (NGS) technologies combined with informatics have been applied to viroid research and diagnostics for almost a decade. NGS provides highly efficient, rapid, low-cost high-throughput sequencing of viroid genomes and of the 21–24 nt vd-sRNAs generated by the RNA silencing defense of the host. NGS has been utilized in various viroid studies which are presented. The discovery during the last few years that prokaryotes have heritable adaptive immunity mediated through clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated Cas proteins, have led to transformative advances in molecular biology, notably genome engineering and most recently molecular diagnostics. The potential application of the CRISPR-Cas13a system for engineering viroid interference in plants is suggested by targeting specific motifs of three economically important viroids. The CRISPR-Cas13 system has been utilized recently for the accurate detection of human RNA viruses by visual read out in 90 min or less and by paper-based assay. Multitarget RNA tests by this technology have a good potential for application as a rapid and accurate diagnostic assay for known viroids. The CRISPR/Cas system will work only for known viroids in contrast to NGS, but it should be much faster.
1. Introduction
Theodor O. Diener discovered the first viroid in 1971, the causal agent of potato spindle tuber disease [1,2]. He showed that the agent is a free RNA of 25,000–110,000 Daltons, much smaller than a viral genome, and that no viral coat proteins were synthesized in infected plants. He concluded that the RNA is too small to contain the genetic information necessary for self-replication and it must rely on host enzymes for its replication.
Diener’s discovery took place 16 years after another virology landmark discovery in which Heinz Fraenkel-Conrat demonstrated that the genetic information controlling viral replication is carried in the nucleic acid core of each virus particle. He showed that he could actually reconstitute a complete infectious tobacco mosaic virus from the protein and RNA [3].
Viroids, the smallest known infectious agents (246–401 nt), comprise a novel class of infectious single-stranded RNA that replicates autonomously and exists as circular and linear forms with a high degree of base pairing. In contrast to viruses, viroids lack capsid proteins, do not code for proteins and completely dependent on host precellular RNA polymerases and processing enzymes for their replication.
In contrast to plant RNA and DNA viruses for which many species have been created, viroids currently comprise 32 recognized species and eight putative species [4]. Five and three of the putative species were discovered during the last and current decade, respectively [4]. Viroid species are divided into two families based on the presence or absence of the conserved central region (CCR) in their genome: Pospiviroidae (type species, Potato spindle tuber viroid) with CCR and members of its five genera replicate and accumulate in host nuclei; Avsunviroidae (type species, Avocado sunblotch viroid) lacks CCR and has hammerhead ribozymes in both genomic and antigenomic RNAs, and members of its three genera replicate and accumulate in host plastids, mainly chloroplasts [4].
Viroids are economically significant as when they infect susceptible plant hosts, they replicate and spread systemically resulting, in most cases, of specific diseases. The damage associated with plant virus infection [5] largely applies to viroids. However, viroid losses, as compared with viruses or other plant pathogens, are generally confined to a country or several countries and do not reach a global level [6]. With the exception of coconut cadang-cadang viroid (CCCVd) and coconut tinangaja viroid (CTiVd) that cause infected coconut palm trees to cease producing coconuts for many years before they die [6], the damage caused by several other viroids in their hosts can be: severe, such as avocado sunblotch viroid (ASBVd) and chrysanthemum stunt viroid (CSVd); moderate, such as potato spindle tuber viroid (PSTVd) and hop stunt viroid (HSVd); mild, such as pome fruit viroids; or variable, such as citrus viroids [6,7,8]. Quantitative data on losses due to viroid infections of susceptible hosts are scarce as only a few yield loss data are available [9]. The yield losses due to infection by PSTVd could range from 17 to 64% [10] and even higher by the third generation of growth [11]. The cone yields of hop plants infected with HSVd in Japan were reduced by 50% or more [12] and infected hop vines could be reduced in height by 35% at the end of the seventh growing season [13]. Infection of coconut palm trees by CCCVd is detrimental as it killed over 40 million coconut palm trees in the Philippines at a cost of four billion US dollars [6]. Citrus bark cracking viroid (CBCVd) infects citrus species with no measurable impact [14], however, when it infects hops it causes an aggressive disease that kills plants in 3–5 years [15,16].
The natural host range of viroids include vegetable and field crops, ornamentals as well as grapevine, fruit trees and palm species. All are members of angiosperms (flowering plants). No viroids have been reported to infect gymnosperms or animals, including primates. Some viroid species such as HSVd and PSTVd have a wide host range while others such as chrysanthemum chlorotic mottle viroid (CChMVd) and coleus blumei viroid (CbVd) 1 to 6 have a narrow one. When a viroid species infects susceptible host, it may cause an economically important disease as discussed above, or it may result in latent (symptomless) infection [7,8,17,18], but they may be pathogenic in other hosts and cause yield reduction and weakness of plants [7,8,17,18,19], depending on the viroid variant nucleotide sequence, host genotype and environmental conditions. Similar to RNA viruses, viroids exist as quasispecies composed of closely related sequence variants with differential properties resulting in heterogeneous progeny. A large number of viroid disease symptoms are similar to those of plant viruses and other pathogens, such as stunting and epinasty of infected plants and different leaf symptoms. Other symptoms may include bark cracking, fruit or tuber malformation, and reduction in number and size of produced flowers. Many factors may affect viroid symptom expression, most notably temperature and host genotype.
The pathogenicity of viroids at the subcellular level may include the formation of paramural bodies or plasmalemmasomes, cell walls and chloroplasts malformations, formation of electron dense deposits in the cytoplasm and chloroplasts. Viroid pathogenicity also involves changes in host metabolism as well as changes in biochemical, molecular and transcriptional mechanisms for viroid disease induction. RNA silencing has also been implicated in viroid pathogenicity. Consequently, single host genes that respond to signals originating from the viroid genome have been identified [7,8].
Viroids are transmitted by mechanical means, grafting, seed, pollen and/or insects [8]. All viroids are transmitted mechanically. With the exception of CCCVd and CTiVd that infect the monocot palm trees, all other viroids are transmitted by grafting. At least 14, seven and six viroids are transmitted by seed, pollen and insect, respectively. Seed-transmitted viroids are: ASBVd; apple scar skin viroid (ASSVd); CbVd; CCCVd; citrus exocortis viroid (CEVd); columnea latent viroid (CLVd); CSVd; eggplant latent viroid (ELVd); HSVd; grapevine yellow speckle viroid-1 (GYSVd-1); pepper chat fruit viroid (PCFVd); PSTVd; tomato apical stunt viroid (TASVd); tomato chlorotic dwarf viroid (TCDVd). Pollen-transmitted viroids include: ASBVd; CCCVd; HSVd; CSVd; PCFVd; peach latent mosaic viroid (PLMVd); PSTVd. Insect-transmitted viroids are: ASBVd; ASSVd; PSTVd; TASVd; TCDVd; tomato planta macho viroid (TPMVd).
The volume and diversity of international exchanges of plant germplasm and newly desired plant cultivars have contributed significantly to international distribution of viroids. Some economically important viroids such as ASSVd, CCCVd, CSVd, HSVd, and PSTVd are transmitted by seed and pollen. The frequency of their occurrence in infected hosts and the harm they cause have created additional pathways for the introduction of these and other viroids in new areas as well as for the emergence of novel viroid variants. It has been shown that the utilization of next-generation sequencing (NGS) in viroid research and diagnosis is sensitive, accurate, and fast using different viroid host plant species, including woody perennial crops such as pome and stone fruits and citrus which have low titers of these pathogens. Moreover, NGS increased the number of novel viroids and variants being discovered and characterized in different plant hosts, including extending the host range of known viroids. Effective genome engineering editing methods, based on clustered regularly interspaced short palindromic repeats (CRISPR) and their associated Cas proteins (CRISPR-Cas), are expected to play a significant role in developing plant resistance to viroid infections as they did for developing transgene-free plants resistant to RNA and DNA plant viruses [20], and have the potential to develop sensitive diagnostic methods for viroids as shown for human viruses [21]. CRISPR/Cas 13 systems have the potential to engineer interference with viroid replication in infected plants as well as to detect viroids in a very short time—about 90 minutes. Thus, both NGS CRISPR/Cas 13 systems have the potential to be used in viroid research and diagnostics as both technologies will be a significant and powerful tool in controlling economically important viroid diseases. In this article we discuss NGS and CRISPR-Cas13 systems as related to viroid research and molecular diagnostics.
2. NGS and Viroids
2.1. Remarks
NGS platforms became available in the market for the first time in 2000, and their use since 2004 has changed the approach to both basic and applied research in many biological disciplines, including plant virology, which deals with viruses (and their satellites) and viroids. The major advance offered by NGS as compared to first-generation Sanger-based sequencing methods [22] (currently dideoxy chain termination sequencing combined with capillary electrophoresis), is the ability to generate an enormous volume of data, generally in excess of one billion short reads per instrument run, as well as its ability to deliver fast, cost-effective, and accurate genome information [20,23,24,25]. With new NGS platforms continually being developed, the nature of the generated sequence data and the associated costs will likely decrease. Bioinformatics’ software tools are required for NGS data analysis that may include, but are not limited to, alignment of sequence reads, base-calling and/or polymorphism detection, de novo, and genome browsing and annotation. A review of bioinformatics’ software tools available for NGS analysis is beyond the scope of this article but have been the subject of review articles, books as well as journal Bioinformatics. The bioinformatics are continuously being developed and improved to keep up with advances of NGS technologies.
2.2. Cost of NGS
The estimated costs of NGS of the human genome (3000 Mb) in 2017 are about $ 1000 (Figure 1) [26]. Thus, NGS of viroid genomes (246–401 nt) should be significantly cheaper.
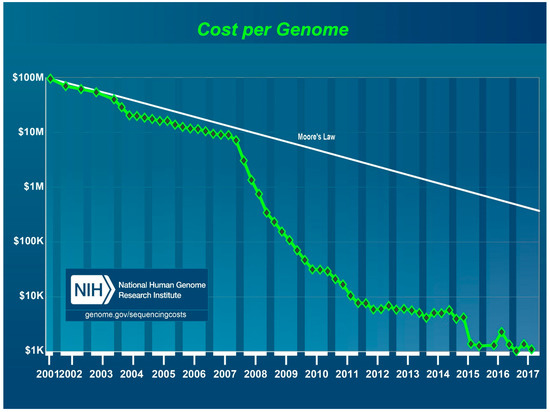
Figure 1.
Cost per human genome sequencing in US Dollars from 2001 to 2017 as estimated by the National Human Genome Research Institute, US National Institutes of Health, Bethesda, MD, USA. During this time period, first generation Sanger sequencing methods were used from 2001 through 2007 and NGS platforms from 2008 to 2017.
It is expected in the next few years that the third-generation sequencing (long-read sequencing) platforms will increase sequencing capacity and speed while reducing costs very significantly. Thus, viroid sequencing may be done with costs for less than $ 25–50.
2.3. Viroid Studies by NGS
Because of the small size of viroid genome, its complete genomic sequence can be determined in a single run by NGS as the sequence of hundreds of thousands to millions of viroid-derived small RNAs (vd-sRNA) of 21–24 nt, can be re-assembled to obtain the viroid genome(s) of interest [25]. The vd-sRNA sequences may also be compared to specific sequences of the host genome in order to identify genes that may be down-regulated upon viroid infection vía RNA silencing [8].
NGS of vd-sRNAs, viroid circular and oligomeric RNAs as well as infected host total RNA, DNase-treated total RNA, rRNA-depleted total RNA and rRNA-depleted dsRNA, combined with informatic and computational tools, have been used for almost a decade in viroid research and diagnostics. In most of these studies, NGS data were confirmed by different methods of RT-PCR to ensure that the viroid putative sequence exist in plants. NGS studies may include, but are not limited to, viroid de novo discovery, identification and detection as well as studies of viroid characterization, profiling, distribution, accumulation, biogenesis, strain differentiation, systemic movement, viroid-host interactions, viroid evolution, pathogenesis, mutation, mRNA targeting, extending host range and others as shown in Table 1, Table 2, Table 3, Table 4 and Table 5.

Table 1.
Discovery of novel viroids or variants by NGS.

Table 2.
Extending the host range of viroids by NGS.

Table 3.
Viroid mutants and quasi-species identified by NGS.

Table 4.
vd-sRNAs: characterization, biogenesis pathway and target host mRNAs identified by NGS.

Table 5.
Detection and identification of viroids in host plants by NGS.
2.4. NGS Identification of Transcriptional (Gene Expression) Changes Associated with Viroid Infection
Methods such as two-dimensional differential gel electrophoresis coupled with mass spectrometry, differential display and microarray have been used to study gene expression in viroid-infected plants [8]. NGS, as compared to these methods, is more sensitive in detecting and identifying transcriptional changes, has higher reproducibility and lower cost. The accurate detailed analyses of transcriptional changes in viroid-infected host are critical for understanding viroid pathogenesis and disease control.
A combination of microarray and large-scale RNA sequence analysis have been used to study gene expression in two PSTVd-infected tomato cultivars: the sensitive cultivar “Rutgers” and the dwarf cultivar “MicroTom” [60]. “Rutgers” infection-related changes were extensive, more than 5, 000 genes with different cellular components were affected. Chloroplast genes were downregulated while many genes encoding proteins associated with nucleus, plasma membrane, ribosomes, cell wall and apoplast were upregulated. It was revealed that “MicroTom” cultivar has a defect in brassinosteroids synthesis and when the hormones were applied exogenously to infected plants, genes involved in stress and other stimuli were upregulated. This observation suggests that potato spindle tuber disease induction may involve brassinosteroid-mediated signaling [60].
NGS was used to identify 11, 600 expressed sequence tags (ESTs) from a cDNA library prepared from total RNA extracted from CSVd-infected chrysanthemum leaves which provided good information of the transcriptome in that system [61]. Approximately 70% of the chrysanthemum ESTs were orthologous to those of five representative plant species [61]. Orthologous genes are genes in different species that have similar functions and are similar in their nucleotide sequences which can be traced back to a common ancestral gene. The majority of the identified genes were responsible for diverse metabolic pathways, various stress responses, transcription, translation and transportation as well as genes coding for several cellular components [61].
The gene expression changes in CBCVd in infected hop were revealed by transcriptome analyses [62]. CBCVd infection resulted in extensive modulation of activity of over 200 genes [62]. Expression of genes associated with plant immune responses, hypersensitive responses, phytohormone signaling pathways, photosynthesis, protein metabolism and others were altered. Moreover, genes encoding RNA-dependent RNA polymerase, pathogenesis-related protein, chitinase, as well as those related to defense responses were upregulated.
2.5. Searching for and Identifying Ancient Viroids by NGS
For several years, due to advances in NGS technologies, the complete sequence of ancient genomes from both modern and archaic humans has revolutionized our understanding of human evolution and migration [63,64]. NGS analyses of ancient viroids in samples from herbarium, ancient plant samples, soil, air or water may allow us to gain knowledge on the evolutionary history of viroids over the past decades, centuries or even the past few, several or many millennia, as viroids may be “living fossils” of an ancient RNA world [65,66,67].
2.6. NGS in Viroid Quarantine and Certification Programs
All known viroids have been sequenced [4,8]. A prior knowledge of viroid sequence is required for RT-PCR detection and identification [68,69,70,71], which is in contrast to detection and identification of known and unknown viroids by NGS [23,24,25]. The cost of RT-PCR-based assay is less expensive than that of NGS analysis, however, the cost of NGS sequencing has become more competitive and affordable during the last couple of years [25]. NGS may be used as the primary diagnostic tool for plant viroids in quarantine and certification programs where various restrictions apply and viroid detection and identification are critical. Viroids of quarantine importance in North America include: ASSVd, PBCVd and PSTVd in Canada; ChCMVd, CSVd, CEVd, CCCVd, ELVd, HSVd (cachexia strain), PLMVd, PBCVd and PSTVd in Mexico; HSVd (cachexia strain), PBCVd and PSTVd in the United States [72]. Viroids of certification importance in propagative material in the European Union include ADFVd, ASSVd, CEVd, HSVd (cachexia strain), PLMVd, PBCVd [72] and CCCVd [73]. Other countries may have similar or different viroids of quarantine or certification importance. For example, all viroids are of quarantine importance in Australia while China considers HSVd (cachexia strain), CEVd and CCCVd of quarantine importance, and Chile considers ASBVd, CCCVd, PSTVd, TASVd and TCDVd of quarantine importance [72].
NGS revealed in 2015 that the causal agent of severe stunting and death of hop plants in Slovenia is CBCVd [16]. As a result, the European and Mediterranean Plant Protection Organization (EPPO) added the viroid to “The EPPO Alert List” [16], suggesting that NGS technology could/should be adopted as a certification or post-entry quarantine measure to detect and identify CBCVd in hops. NGS may become instrumental in releasing plants in quarantine and certification programs at a faster rate than current strategies while improving our ability to prevent the introduction of foreign viroids into new countries (this article, [48]).
3. Potential Utilization of CRISPR-Cas 13 Systems in Viroid Interference and Diagnostics
3.1. General Aspects
Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated Cas proteins comprise the CRISPR-Cas systems, which confer adaptive immunity against foreign genetic elements such as bacteriophages and plasmids in many bacteria and most archaea [74,75,76,77]. CRISPR-Cas systems act as RNA-guided programmable nucleases to degrade DNA and/or RNA derived from foreign nucleic acids by preserving molecular memory information of prior infections [78,79,80,81,82]. Three processes are involved in CRISPR/Cas-mediated immunity: adaptation, transcription and processing, and interference (for review, see Ref. [77,81]). Adaptation includes information of the most recent infection during which “spacer” sequences (short segments of foreign DNA) acquired from the invader genome, such as plasmids and bacteriophages [74,83,84], are integrated into the prokaryotic genome [78]. That followed by placing the spacer sequences into a CRISPR array in between pairs of short repeated sequences (for array review see Ref. [81]). The array is then transcribed to generate pre-crRNAs, which are further processed to generate the mature crRNAs. Thus, the spacer sequences provide the sequence specificity for interfering with invading DNA and/ or RNA. Interference, whereby CRISPR Cas enzymes are guided by the crRNAs to form “effector or interference complexes” that enables crRNAs to target, cleave and degrade complementary foreign nucleic acids in the respective invader genomes, thus preventing further infection [74,75,77,78,79,80,81,82,83,84,85,86,87,88,89].
The CRISPR-Cas systems are divided into two general classes on the basis of the composition of the interference complex [82]. Class 1 systems rely on multi-effector complexes mediate the interference whereas Class 2 systems utilize single multi-domain effectors to mediate the interference. These two classes are further divided into six types and 33 subtypes based on the genomic composition of the CRISPR array and the signature interference effector. Class 1 includes types I, III, and IV, and Class 2 includes types II, V and VI [82,86,87]. Types I, II and V target double-stranded DNA but types III and VI target single-stranded RNA [82,88,89,90,91].
Class 2 type VI systems, include a single “effector” protein designated Cas13a, formerly C2c2 [91], which when combined with crRNA forms a crRNA-guided RNA-targeting CRISPR effector complex [82,91,92]. Cas13a provides specificity through CRISPR RNA (crRNA)-target pairing and additional sensitivity due to signal amplification by Cas13a collateral cleavage activity [93]. Cas13a possesses two enzymatically distinct ribonucleases activities that are needed for optimum interference [92]. The first RNase is required for pre-crRNA processing to help formation of mature interference complexes while the second RNase, which is provided by the two Higher Eukaryotes and Prokaryotes Nucleotide-binding (HEPN) domains [94], is required for degradation of target RNA during viral interference. The RNase activity provided by HEPN in Cas13a is lacking in other known Cas proteins [95].
Classical breeding and different molecular strategies to introduce resistance to viroids have been unsuccessful or partially successful in containing viroid infection [96]. A new molecular approach for controlling viroid infection by CRISPR-Cas13a system has the potential to be useful and successful. This system has been recently utilized successfully to engineer interference with an RNA plant virus, turnip mosaic virus, in Nicotiana benthamiana [95].
3.2. Application to Target Viroids for Inactivation
PSTVd, PLMVd and ASBVd are economically important and their genomes and replication have been studied extensively [8]. Selected motifs of each of the above viroids [97] could be specifically targeted by CRISPR Cas13a system. Briefly, the following viroid motifs could be targeted (as indicated in [97]):
“A-PSTVd terminal left domain: 1-Initiation site of minus PSTVd RNA synthesis at nt U359 or C1. 2-Three bulges associated with plus PSTVd RNA replication.
B-PSTVd center conserved region: 1- Hairpin I, delimited by nt 79 and 110 on the upper strand of the viroid RNA. 2- Hairpin II, delimited by nt 227 and 328 on the lower strand of the viroid RNA. 3-Loop E, located between nt 5’-G97 to C103-3’ and nt 5’-G255 to C262-3’.
C-PSTVd terminal right domain: RY motif, composed of two asymmetric internal loops, with sequence elements 5’-ACAGG-3’ (nt 173-177) in the upper strand and 3’-CUCUUCC-5’ (nt 190–184) in the lower strand.
D-PLMVd initiation sites: Such sites are located at nt C51 for the plus strand and at nt U286 for the minus strand, both mapping at similar double-stranded motifs of 6–7 bp that also contain the highly conserved GUC triplet preceding the self-cleavage site in both polarity strands. These motifs are located at the base of a similar long hairpin that presumably contains the promoters for a chloroplastic RNA polymerase. Since the in vivo initiation of PLMVd RNAs occurs near the self-cleavage (and self-ligation) sites, CRISPR-Cas13a editing in this region would attenuate both the transcription and processing of PLMVd RNA.
E-ASBVd initiation sites: These sites are located at nt U121 and at nt U119 for the plus and minus strands, respectively. The initiation sites are only two nucleotides apart and each site starts with the same sequence, UAAAA, which suggests that the viroid promoters are formed, at least in part, by the sequences flanking the two initiation sites.”
3.3. Application to Viroid Detection
The RNA guided nuclease activities of CRISPR-Cas13a, -Cas13b and -Cas12a, which are functionally distinct from -Cas9, have been recently utilized to develop sensitive detection methods for human viruses [98,99,100,101]. CRISPR-Cas13a and -Cas 13b were used for the detection of RNA viruses [98,99,100] and -Cas12a for DNA viruses [99,101].
It has been shown that the Cas13 SHERLOCK (specific high-sensitivity enzymatic reporter unlocking) platform can incorporate pre-amplified input material to develop a paper-based assay with improved sensitivity [98]. SHERLOCK conflates recombinase polymerase amplification (RPA) [102] with highly specific Cas13-based detection. Cas13a [91] and Cas13b [103,104] homologs have discrete crRNA and substrate preferences to simultaneously detect multiple transcripts [99,100,105]. When Cas13 was combined with Csm6, an auxiliary CRISPR-type III associated nuclease [106], an increase of signal sensitivity by approximately 3.5-fold was observed [99]. The revised platform, termed SHERLOCKv2, can simultaneously detect three ssRNA targets and one DNA target in a single reaction [99]. It is accurate and quantitative. By characterization of 17 CRISPR-Cas13a and -Cas13b enzymes and selecting three with distinct cleavage preferences and combining them with a Cas12a enzyme and RPA, synthetic ssRNA, dengue virus ssRNA, Zika virus ssRNA and synthetic dsDNA were accurately detected by a visual readout in less than 90 minutes [99] and in a readily deployable format via a paper-based assay [100].These results highlight the potential of such an approach as a multiplex, rapid and quantitative detection assay for viroid RNA. The CRISPR/Cas system will work only for known viroids, in contrast to NGS, but it should be much faster.
4. Final Remarks
CRISPR-Cas editing does not incorporate permanently foreign DNA into the host genome as in transgenic plants, and agricultural applications of this precise technology are already creating valuable products for various markets in the United States. For this reason, the U.S. Department of Agriculture does not regulate foods or plants developed by CRISPR-Cas editing [107,108,109]. Thus, this technology can be used in the US to develop plants resistant to viroids or other pathogens. In the European Union (EU), however, the highest court ruled on July 25, 2018 that CRISPR-Cas-edited plants are subject to the same regulations as those of conventional genetically modified organisms [110], which were imposed in EU about two decades ago. Consequently, CRISPR-Cas-edited plants need to go through a lengthy approval process.
Dedication
This article is dedicated to the memory of Dr. Heinz Fraenkel-Conrat with whom the author was NIH Postdoctoral Fellow from 1970 to 1973 at the Department of Molecular Biology, University of California, Berkeley.
Acknowledgments
The author would like to thank Ricardo Flores and John W. Randles for critical reading of the manuscript.
Conflicts of Interest
The author declares no conflicts of interest.
References
- Diener, T.O. Potato spindle tuber virus with properties of a free nucleic acid. III. Subcellular location of PSTV-RNA and the question of whether virions exist in extracts or in situ. Virology 1971, 43, 75–89. [Google Scholar] [CrossRef]
- Diener, T.O. Potato spindle tuber “virus”. IV. A replicating, low molecular weight RNA. Virology 1971, 45, 411–428. [Google Scholar] [CrossRef]
- Fraenkel-Conrat, H.; Williams, R.C. Reconstitution of active tobacco mosaic virus from its inactive protein and nucleic acid components. Proc. Natl. Acad. Sci. USA 1955, 41, 690–698. [Google Scholar] [CrossRef] [PubMed]
- Di Serio, F.; Li, S.-F.; Pallas, V.; Owens, R.A.; Randles, J.W.; Sano, T.; Verhoeven, T.J.; Vidalakis, G.; Flores, R. Viroid taxonomy. In Viroids and Satellites; Hadidi, A., Flores, R., Randles, J.W., Palukaitis, P., Eds.; Academic Press: Oxford, UK, 2017; pp. 135–146. ISBN 978-0-12-801498-1. [Google Scholar]
- Waterworth, H.; Hadidi, A. Economic losses due to plant viruses. In Plant Virus Disease Control; Hadidi, A., Khetarpal, R.K., Koganezawa, H., Eds.; APS Press: St. Paul, MN, USA, 1998; pp. 1–13. ISBN 0-89054-191-4. [Google Scholar]
- Randles, J.W. Economic impact of viroid diseases. In Viroids; Hadidi, A., Flores, R., Randles, J.W., Semancik, J.S., Eds.; CSIRO Publishing: Collingwood, Australia, 2003; pp. 3–11. ISBN 1-57808-272-2. [Google Scholar]
- Hadidi, A.; Flores, R.; Randles, J.W.; Semancik, J.S. Viroids; CSIRO Publishing: Collingwood, Australia, 2003; ISBN 1-57808-272-2. [Google Scholar]
- Hadidi, A.; Flores, R.; Randles, J.W.; Palukaitis, P. Viroids and Satellites; Academic Press: Cambridge, MA, USA; Elsevier: Oxford, UK, 2017; ISBN 978-0-12-801498-1. [Google Scholar]
- Singh, R.P.; Randles, J.W.; Hadidi, A. Strategies for the control of viroid diseases. In Viroids; Hadidi, A., Flores, R., Randles, J.W., Semancik, J.S., Eds.; CSIRO Publishing: Collingwood, Australia, 2003; pp. 295–302. ISBN 1-57808-272-2. [Google Scholar]
- Singh, R.P.; Finnie, R.E.; Bagnal, R.H. Losses due to the potato spindle virus. Am. Potato J. 1971, 48, 266–267. [Google Scholar] [CrossRef]
- Pfannensteil, M.A.; Slack, S.A. Response of potato cultivars to infection by the potato spindle tuber viroid. Phytopathology 1980, 70, 922–926. [Google Scholar] [CrossRef]
- Sasaki, M.; Shikata, E. Studies on the host range of hop stunt disease in Japan. Proc. Jpn. Acad. 1977, 53, 103–108. [Google Scholar] [CrossRef]
- Sano, T. Hop stunt viroid. In Viroids; Hadidi, A., Flores, R., Randles, J.W., Semancik, J.S., Eds.; CSIRO Publishing: Collingwood, Australia, 2003; pp. 207–212. ISBN 1-57808-272-2. [Google Scholar]
- Hadidi, A.; Vidalakis, G.; Sano, T. Economic significance of fruit tree and grapevine viroids. In Viroids and Satellites; Hadidi, A., Flores, R., Randles, J.W., Palukaitis, P., Eds.; Academic Press: Cambridge, MA, USA; Elsevier: Oxford, UK, 2017; pp. 15–25. ISBN 978-0-12-801498-1. [Google Scholar]
- Lavagi, I.; Matousek, J.; Vidalakis, G. Other cocadviroids. In Viroids and Satellites; Hadidi, A., Flores, R., Randles, J.W., Palukaitis, P., Eds.; Academic Press: Cambridge, MA, USA; Elsevier: Oxford, UK, 2017; pp. 275–287. ISBN 978-0-12-801498-1. [Google Scholar]
- Jakse, J.; Radisek, S.; Pokorn, T.; Matousek, J.; Javornik, B. Deep sequencing revealed Citrus bark cracking viroid (CBCVd) as a highly aggressive pathogen on hop. Plant Pathol. 2015, 64, 831–842. [Google Scholar] [CrossRef]
- Hadidi, A.; Khetarpal, R.K.; Koganezawa, H. Plant Virus Disease Control; APS Press: St. Paul, MN, USA, 1998; ISBN 0-89054-191-4. [Google Scholar]
- Hadidi, A.; Barba, M.; Candresse, T.; Jelkmann, W. Virus and Virus-Like Diseases of Pome and Stone Fruits; APS Press: St. Paul, MN, USA, 1998; ISBN 0-89054-191-4. [Google Scholar]
- Barba, M.; Ilardi, V.; Pasquini, G. Control of pome and stone fruit virus diseases. Adv. Virus Res. 2015, 91, 47–61. [Google Scholar] [CrossRef]
- Hadidi, A.; Flores, R.; Candresse, T.; Barba, M. Next-generation sequencing and genome editing in plant virology. Front. Microbiol. 2016, 7, 1325. [Google Scholar] [CrossRef]
- Chertow, D.S. Next-generation diagnostics with CRISPR. Science 2018, 360, 381–382. [Google Scholar] [CrossRef]
- Sanger, F.; Nicklen, S.; Coulsen, A.R. DNA sequencing with chain-terminator inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef]
- Barba, M.; Czosnek, H.; Hadidi, A. Historical perspective, development and applications of next-generation sequencing in plant virology. Viruses 2014, 6, 106–136. [Google Scholar] [CrossRef] [PubMed]
- Barba, M.; Hadidi, A. An overview of plant pathology and application of next-generation sequencing technologies. CAB Rev. 2015, 10, 1–10. [Google Scholar] [CrossRef]
- Barba, A.; Hadidi, A. Application of next-generation sequencing technologies to viroids. In Viroids and Satellites; Hadidi, A., Flores, R., Randles, J.W., Palukaitis, P., Eds.; Academic Press: Cambridge, MA, USA; Elsevier: Oxford, UK, 2017; pp. 401–412. ISBN 978-0-12-801498-1. [Google Scholar]
- Wetterstrand, K.A. DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP). Available online: www.genome.gov/sequencingcostsdata (accessed on 21 January 2019).
- Ito, T.; Suzaki, K.; Nakano, M.; Sato, A. Characterization of a new apscaviroid from American persimmon. Arch. Virol. 2013, 158, 2629–2631. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Qi, S.; Tang, N.; Zhang, X.; Chen, S.; Zhu, P.; Ma, L.; Ching, J.; Xu, Y.; Lu, M.; et al. Discovery of replicating circular RNAs by RNA-seq and computational algorithms. PLoS Pathog. 2014, 10, e1004553. [Google Scholar] [CrossRef] [PubMed]
- Chiumenti, M.; Torchetti, E.M.; Di Serio, F.; Minafra, A. Identification and characterization of a viroid resembling apple dimple fruit viroid in fig (Ficus carica L.) by next generation sequencing of small RNAs. Virus Res. 2014, 188, 54–59. [Google Scholar] [CrossRef]
- Wang, Y.; Atta, S.; Wang, X.; Yang, F.; Zhou, C.; Cao, M. Transcriptome sequencing reveals novel Citrus bark cracking viroid (CBCVd) variants from citrus and their molecular characterization. PLoS ONE 2018, 13, e0198022. [Google Scholar] [CrossRef]
- Suzuki, T.; Fujibayashi, M.; Hataya, T.; Taneda, A.; He, Y.-H.; Tsushima, T.; Duraisamy, G.S.; Siglova, K.; Matoušek, J.; Sano, T. Characterization of host-dependent mutations of apple fruit crinkle viroid replicating in newly identified experimental hosts suggests maintenance of stem-loop structures in the left-hand half of the molecule is important for replication. J. Gen. Virol. 2017, 98, 506–516. [Google Scholar] [CrossRef]
- Pirovano, W.; Miozzi, L.; Boetzer, M.; Pantaleo, V. Bioinformatics approaches for viral metagenomics in plants using short RNAs: Model case of study and application to a Cicer arietinum population. Front. Microbiol. 2015, 5, 790. [Google Scholar] [CrossRef]
- Glouzon, J.P.-S.; Bolduc, F.; Wang, S.; Najmanovich, R.J.; Perreault, J.-P. Deep-sequencing of the peach latent mosaic viroid reveals new aspects of population heterogeneity. PLoS ONE 2014, 9, e87297. [Google Scholar] [CrossRef]
- Brass, J.R.J.; Owens, R.A.; Matoušek, J.; Steger, G. Viroid quasispecies revealed by deep sequencing. RNA Biol. 2017, 14, 317–325. [Google Scholar] [CrossRef]
- Adkar-Purushothama, C.R.; Perreault, J.-P. Alterations of the viroid regions that interact with the host defense genes attenuate viroid infection in host plant. RNA Biol. 2018. [Google Scholar] [CrossRef] [PubMed]
- López-Carrasco, A.; Ballesteros, C.; Sentandreu, V.; Delgado, S.; Gago-Zachert, S.; Flores, R.; Sanjuán, R. Different rates of spontaneous mutation of chloroplastic and nuclear viroids as determined by high-fidelity ultra-deep sequencing. PLoS Pathog. 2017, 13, e1006547. [Google Scholar] [CrossRef] [PubMed]
- Navarro, B.; Pantaleo, V.; Gisel, A.; Moxon, S.; Dalmay, T.; Bistray, G.; di Serio, F.; Burgyan, J. Deep sequencing of viroid-derived small RNAs from grapevine provides new insight on the role of RNA silencing in plant-viroid interaction. PLoS ONE 2009, 4, e7686. [Google Scholar] [CrossRef] [PubMed]
- Alabi, O.J.; Zheng, Y.; Jagadeeswaran, G.; Sunkar, R.; Naidu, R. High-throughput sequence analysis of small RNAs in grapevine (Vitis. vinifera L.) affected by grapevine leafroll disease. In Proceedings of the 17th Congress of ICVG, Davis, CA, USA, 7–14 October 2012. [Google Scholar]
- Martínez, G.; Donaire, L.; Llave, C.; Pallás, V.; Gómez, G. High-throughput sequencing of Hop stunt viroid-derived small RNAs from cucumber leaves and phloem. Mol. Plant Pathol. 2010, 11, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Di Serio, F.; Gisel, A.; Navarro, B.; Delgado, S.; Martínez de Alba, A.-E.; Donvito, G.; Flores, R. Deep sequencing of the small RNAs derived from two symptomatic variants of a chloroplastic viroid: Implications, for their genesis and for pathogenesis. PLoS ONE 2009, 4, e7539. [Google Scholar] [CrossRef] [PubMed]
- Navarro, B.; Gisel, A.; Rodio, M.E.; Delgado, S.; Flores, R.; Di Serio, F. Small RNAs containing the pathogenic determinant of a chloroplast-replicating viroid guide the degradation of a host mRNA as predicted by RNA silencing. Plant J. 2012, 70, 991–1003. [Google Scholar] [CrossRef] [PubMed]
- Di Serio, F.; Martínez de Alba, A.E.; Navarro, B.A.; Gisel, A.; Flores, R. RNA-dependent RNA polymerase 6 delays accumulation and precludes meristem invasion of a viroid that replicates in the nucleus. J. Virol. 2010, 84, 2477–2489. [Google Scholar] [CrossRef]
- Adkar-Purushothama, C.R.; Brosseau, C.; Giguère, T.; Sano, T.; Mofett, P.; Perreault, J.P. Small RNA derived from the virulence modulating region of the potato spindle tuber viroid silences callose synthase genes of tomato plants. Plant Cell 2015, 27, 2178–2194. [Google Scholar] [CrossRef]
- Wang, Y.; Shibuya, M.; Taneda, A.; Kurauchi, T.; Senda, M.; Owens, R.A.; Sano, T. Accumulation of potato spindle tuber viroid-specific small RNAs is accompanied by specific changes in gene expression in two tomato cultivars. Virology 2011, 413, 72–83. [Google Scholar] [CrossRef]
- Piernikarczyk, R.; Matousek, J.; Riesner, D.; Steger, G. Potential mRNA targets of viroid-specific small RNA. In Proceedings of the International Workshop on Viroids and Satellite RNAs, Beijing, China, 23–25 August 2013. [Google Scholar]
- Adkar-Purushothama, C.R.; Iyer, P.S.; Perreault, J.-P. Potato spindle tuber viroid infection triggers degradation of chloride channel protein CLC-b-like and ribosomal protein S3a-like mRNAs in tomato plants. Sci. Rep. 2017, 7, 8341. [Google Scholar] [CrossRef]
- Wu, Q.; Wang, Y.; Cao, M.; Pantaleo, V.; Burgyan, J.; Li, W.X.; Ding, S.W. Homology-independent discovery of replicating pathogenic circular RNAs by deep sequencing and a new computational algorithm. Proc. Natl. Acad. Sci. USA 2012, 109, 3938–3943. [Google Scholar] [CrossRef]
- Rott, M.; Xiang, Y.; Boyes, I.; Belton, M.; Saeed, H.; Kesanakurti, P.; Hayes, S.; Lawrence, T.; Birch, C.; Bhagwat, B.; et al. Application of next generation sequencing for diagnostic testing of tree fruit viruses and viroids. Plant Dis. 2017, 101, 1489–1499. [Google Scholar] [CrossRef]
- Al Rwahnih, M.; Daubert, S.; Golino, D.; Rowhani, A. Deep sequencing analysis of RNAs from a grapevine showing Syrah decline symptoms reveals a multiple virus infection that includes a novel virus. Virology 2009, 387, 395–401. [Google Scholar] [CrossRef]
- Poojari, S.; Alabi, O.J.; Fofanov, V.Y.; Naidu, R.A. A leafhopper-transmissible DNA virus with novel evolutionary lineage in the family Geminiviridae implicated in grapevine redleaf disease by next-generation sequencing. PLoS ONE 2013, 8, e64194. [Google Scholar] [CrossRef]
- Pecman, A.; Kutnjak, D.; Gutiérrez-Aguirre, I.; Adams, I.; Fox, A.; Boonham, N.; Ravnikar, M. Next generation sequencing for detection and discovery of plant viruses and viroids: Comparison of two approaches. Front. Microbiol. 2017, 8, 1998. [Google Scholar] [CrossRef] [PubMed]
- Giampetruzzi, A.; Roumi, V.; Roberto, R.; Malossini, U.; Yoshikawa, N.; la Notte, P.; Terlizzi, F.; Credi, R.; Saldarelli, P. A new grapevine virus discovered by deep sequencing of virus- and viroid-derived small small RNAs in Cv Pinot gris. Virus Res. 2012, 163, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Seguin, J.; Rajeswaran, R.; Malpica-López, N.; Martin, R.R.; Kasschau, K.; Dolja, V.V.; Otten, P.; Farinelli, L.; Pooggin, M.M. De novo reconstruction of consensus master genomes of plant RNA and DNA viruses from siRNAs. PLoS ONE 2014, 9, e88513. [Google Scholar] [CrossRef]
- Chiumenti, M.; Giampetruzzi, A.; Pirolo, C.; Morelli, M.; Saldarelli, P.; Minafra, A.; Bottalico, G.; la Notte, P.; Campanale, A.; Savino, V.; et al. Approaches of next generation sequencing to investigate grapevine diseases of unknown etiology. In Proceedings of the 17th Congress of ICVG, Davis, CA, USA, 7–14 October 2012. [Google Scholar]
- Jo, Y.; Choi, H.; Yoon, J.-Y.; Choi, S.-K.; Cho, W.K. De novo genome assembly of grapevine yellow speckle viroid 1 from a grapevine transcriptome. Genome Announc. 2015, 3, e496. [Google Scholar] [CrossRef] [PubMed]
- Hily, J.-M.; Candresse, T.; Garcia, S.; Vigne, E.; Tannière, M.; Komar, V.; Barnabé, G.; Alliaume, A.; Gilg, S.; Hommay, G.; et al. High-throughput sequencing and the viromic study of grapevine leaves: From the detection of grapevine-infecting viruses to the description of a new environmental Tymovirales member. Front. Microbiol. 2018, 9, 1782. [Google Scholar] [CrossRef] [PubMed]
- Bolduc, F.; Hoareau, C.; St-Pierre, P.; Perreault, J.P. In-depth sequencing of the siRNAs associated with peach latent mosaic viroid infection. BMC Mol. Biol. 2010, 11, 16. [Google Scholar] [CrossRef] [PubMed]
- Diermann, N.; Matousek, J.; Junge, M.; Riesner, D.; Steger, G. Characterization of plant miRNAs and small RNAs derived from potato spindle tuber viroid (PSTVd) in infected tomato. Biol. Chem. 2010, 391, 1379–1390. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Gao, S.; Hernandez, A.G.; Wechter, W.P.; Fei, Z.; Ling, K.-S. Deep sequencing of small RNAs in tomato for virus and viroid identification and strain differentiation. PLoS ONE 2012, 7, e37127. [Google Scholar] [CrossRef] [PubMed]
- Owens, R.A.; Teck, K.B.; Shao, J.Y.; Sano, T.; Baker, C.J. Global analysis of tomato gene expression during potato spindle tuber viroid infection reveals a complex array of changes affecting hormone signaling. Mol. Plant Microbe Interact. 2012, 25, 582–598. [Google Scholar] [CrossRef]
- Jo, Y.-H.; Jo, K.-M.; Park, S.-H.; Kim, K.-H.; Cho, W.-K. Transcriptomic landscape of chrysanthemum infected by chrysanthemum stunt viroid. Plant Omics J. 2014, 7, 1–11. [Google Scholar]
- Mishra, A.K.; Kumar, A.; Mishra, D.; Nath, V.S.; Jakše, J.; Kocábek, T.; Killi, U.K.; Morina, F.; Matoušek, J. Genome-wide transcriptomic analysis reveals insights into the response to citrus bark cracking viroid (CBCVd) in hop (Humulus lupulus L.). Viruses 2018, 10, 570. [Google Scholar] [CrossRef]
- Pääbo, S. The human condition-a molecular approach. Cell 2014, 157, 216–226. [Google Scholar] [CrossRef]
- Wolf, A.B.; Akey, J.M. Outstanding questions in the study of archaic hominin admixture. PLoS Genet 2018, 14, e1007349. [Google Scholar] [CrossRef]
- Diener, T.O. Circular RNAs: Relics of precellular evolution? Proc. Natl. Acad. Sci. USA 1989, 86, 9370–9374. [Google Scholar] [CrossRef]
- Diener, T.O. Origin and evolution of viroids and viroid-like satellite RNAs. Virus Genes 1995, 11, 119–131. [Google Scholar] [CrossRef]
- Flores, R.; Gago-Zachert, S.; Serra, P.; Sanjuan, R.; Elena, S.F. Viroids-survivors from the RNA world? Annu. Rev. Microbiol. 2014, 68, 395–414. [Google Scholar] [CrossRef] [PubMed]
- Hadidi, A.; Yang, X. Detection of pome fruit viroids by enzymatic cDNA amplification. J. Virol. Methods 1990, 30, 261–270. [Google Scholar] [CrossRef]
- Hadidi, A.; Olmos, A.; Pasquini, G.; Barba, M.; Martin, R.R.; Shamloul, A.M. Polymerase chain reaction for detection of systemic plant pathogens. In Virus and Virus-Like Diseases of Pome and Stone Fruits; Hadidi, A., Barba, M., Candresse, T., Jelkmann, W., Eds.; APS Press: St. Paul, MN, USA, 2011; pp. 341–359. ISBN 978-0-89054-396-2. [Google Scholar]
- Faggioli, F.; Luigi, M.; Boubourakis, I.N. Viroid amplification methods: RT-PCR, Real-Time RT-PCR, and RT-LAMP. In Viroids and Satellites; Hadidi, A., Flores, R., Randles, J.W., Palukaitis, P., Eds.; Academic Press: Cambridge, MA, USA; Elsevier: Oxford, UK, 2017; pp. 381–391. ISBN 978-0-12-801498-1. [Google Scholar]
- Pallás, V.; Sánchez-Navarro, J.A.; James, D. Recent advances on the multiplex molecular detection of plant viruses and viroids. Front. Microbiol. 2018, 9, 2087. [Google Scholar] [CrossRef] [PubMed]
- Barba, M.; James, D. Quarantine and certification for viroids and viroid diseases. In Viroids and Satellites; Hadidi, A., Flores, R., Randles, J.W., Palukaitis, P., Eds.; Academic Press: Cambridge, MA, USA; Elsevier: Oxford, UK, 2017; pp. 415–424. ISBN 978-0-12-801498-1. [Google Scholar]
- Jeger, M.; Bragard, C.; Caffier, D.; Candresse, T.; Dehnen-Schmutz, K.; Gilioli, G.; Gregoire, J.-C.; Miret, J.A.J.; MacLeod, A.; Navarro, M.N.; et al. Pest categorization of cadang-cadang viroid. EFSA J. 2017. [Google Scholar] [CrossRef]
- Mójica, F.J.M.; Díez-Villasenor, C.; García-Martínez, J.; Soria, E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005, 60, 174–182. [Google Scholar] [CrossRef]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef]
- van der Oost, J.; Westra, E.R.; Jackson, R.N.; Wiedenheft, B. Unraveling the structural and mechanistic basis of CRISPR-Cas systems. Nat. Rev. Microbiol. 2014, 12, 479–492. [Google Scholar] [CrossRef] [PubMed]
- Wright, A.V.; Nunez, J.K.; Doudna, J.A. Biology and application of CRISPR systems: Harnessing nature’s toolbox for genome engineering. Cell 2016, 164, 29–44. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Fermaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef] [PubMed]
- Mójica, F.J.M.; Díez-Villasenor, C.; García-Martínez, J.; Almendros, C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiol. SGM 2009, 155, 733–740. [Google Scholar] [CrossRef]
- Hale, C.R.; Zhao, P.; Olson, S.; Duff, M.O.; Graveley, L.; Wells, L.; Terns, R.M.; Terns, M.P. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 2009, 139, 945–956. [Google Scholar] [CrossRef] [PubMed]
- Sternberg, S.H.; Richter, H.; Charpentier, E.; Qimron, U. Adaptation in CRISPR-Cas systems. Mol. Cell 2016, 61, 797–808. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, M.R. Molecular mechanisms of RNA targeting by Cas13-containg type VI CRISPR-Cas systems. J. Mol. Biol. 2018. [Google Scholar] [CrossRef]
- Bolotin, A.; Quinquis, B.; Sorokin, A.; Ehrlich, S.D. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 2005, 151, 2551–2561. [Google Scholar] [CrossRef]
- Pourcel, C.; Salvignol, G.; Vergnaud, G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA and provide additional tools for evolutionary studies. Microbiology 2005, 151, 653–663. [Google Scholar] [CrossRef]
- Brouns, S.J.; Jore, M.M.; Lundgren, M.; Westra, E.R.; Slijkhuis, R.J.; Snijders, A.P.; Dickman, M.J.; Makarova, K.S.; Koonin, E.V.; van der Oost, J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 2008, 321, 960–964. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Haft, D.H. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015, 13, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Shmakov, S.; Abudayyeh, O.O.; Makarova, K.S.; Wolf, Y.I.; Gootenberg, J.S.; Semenova, E.; Minakhin, L.; Joung, J.; Konermann, S.; Severinov, K. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol. Cell 2015, 60, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Samai, P.; Marraffini, L.A. Degradation of phage transcripts by CRISPR-associated RNases enables type III CRISPR-Cas immunity. Cell 2016, 164, 710–721. [Google Scholar] [CrossRef]
- Plagens, A.; Richter, H.; Charpentier, E.; Randau, L. DNA and RNA interference mechanisms by CRISPR-Cas surveillance complexes. FEMS Microbiol. Rev. 2015, 39, 442–463. [Google Scholar] [CrossRef]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef] [PubMed]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konemann, S.; Joung, J.; Staymaker, I.M.; Cox, D.B.; Shmakov, S.; Makarova, K.S.; Ekaterina Semenova, E.; Minakhin, L.; et al. C2c2 is a single component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef]
- East-Seletsky, A.; O’Connell, M.R.; Knight, S.C.; Burstein, D.; Cate, J.H.; Tijan, R.; Doudna, J.A. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 2016, 538, 270–273. [Google Scholar] [CrossRef]
- Knott, C.J.; Doudna, J. CRISPR-Cas guides the future of genetic engineering. Science 2018, 361, 866–869. [Google Scholar] [CrossRef] [PubMed]
- Anantharman, V.; Makarova, K.S.; Burroughs, A.M.; Koonin, E.V.; Aravind, L. Comprehensive analysis of the HEPN superfamily: Identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing. Biol. Direct 2013, 8, 15. [Google Scholar] [CrossRef] [PubMed]
- Aman, R.; Ali, Z.; Butt, H.; Mahas, A.; Aljedaani, F.; Khan, M.Z.; Ding, S.; Mahfouz, M. RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol. 2018, 19, 1. [Google Scholar] [CrossRef] [PubMed]
- Hammond, R.W.; Kovalskaya, N. Strategies to introduce resistance to viroids. In Viroids and Satellites; Hadidi, A., Flores, R., Randles, J.W., Palukaitis, P., Eds.; Academic Press: Cambridge, MA, USA; Elsevier: Oxford, UK, 2017; pp. 447–455. ISBN 978-0-12-801498-1. [Google Scholar]
- Hadidi, A.; Flores, R. Genome editing by CRISPR-based technology: Potential application for viroids. In Viroids and Satellites; Hadidi, A., Flores, R., Randles, J.W., Palukaitis, P., Eds.; Academic Press: Cambridge, MA, USA; Elsevier: Oxford, UK, 2017; pp. 531–540. ISBN 978-0-12-801498-1. [Google Scholar]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Myhrvold, M.; Freijie, C.A.; Gootenberg, J.S.; Abudayyeh, O.O.; Metsky, H.C.; Durbin, A.F.; Kellner, M.J.; Tan, A.L.; Paul, L.M.; Parham, L.A.; et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science 2018, 360, 444–448. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.S.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Piepenburg, O.; Williams, C.H.; Stemple, D.L.; Armes, N.A. DNA detection using recombination proteins. PLOS Biol. 2006, 4, e204. [Google Scholar] [CrossRef] [PubMed]
- Smargon, A.A.; Cox, D.B.T.; Pyzocha, N.K.; Zheng, K.; Slaymaker, I.M.; Gootenberg, J.S.; Abudayyeh, O.A.; Essletzbichler, P.; Shmakov, S.; Makarova, K.S.; et al. Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol. Cell 2017, 65, 618–630. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Gersbach, C.A. Expanding the CRISPR toolbox: Targeting RNA with Cas13b. Mol. Cell 2017, 65, 582–584. [Google Scholar] [CrossRef] [PubMed]
- East-Seletsky, A.; O’Connell, M.R.; Burstein, D.; Knott, G.J.; Doudna, J.A. RNA targeting by functionally orthogonal type VI-A CRISPR-Cas enzymes. Mol. Cell 2017, 66, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Niewoehner, O.; Jinek, M. Structural basis for the endoribonuclease activity of the type III-A CRISPR-associated protein Csm6. RNA 2016, 22, 318–329. [Google Scholar] [CrossRef] [PubMed]
- Anonymous. CRISPR foods dodge USDA. Science 2016, 352, 388. [Google Scholar]
- Waltz, E. CRISPR-edited crops free to enter market, skip regulation. Nat. Biotechnol. 2016, 34, 582. [Google Scholar] [CrossRef]
- Waltz, E. With a free pass, CRISPR-edited plants reach market in record time. Nat. Biotechnol. 2018, 36, 6–7. [Google Scholar] [CrossRef]
- Callaway, E. CRISPR plants now subject to tough GM laws in European Union. Nature 2018, 560, 16. [Google Scholar] [CrossRef]
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).