Molecular Survey and Phylogenetic Analysis of Atypical Porcine Pestivirus (APPV) Identified in Swine and Wild Boar from Northern Italy
Abstract
1. Introduction
2. Materials and Methods
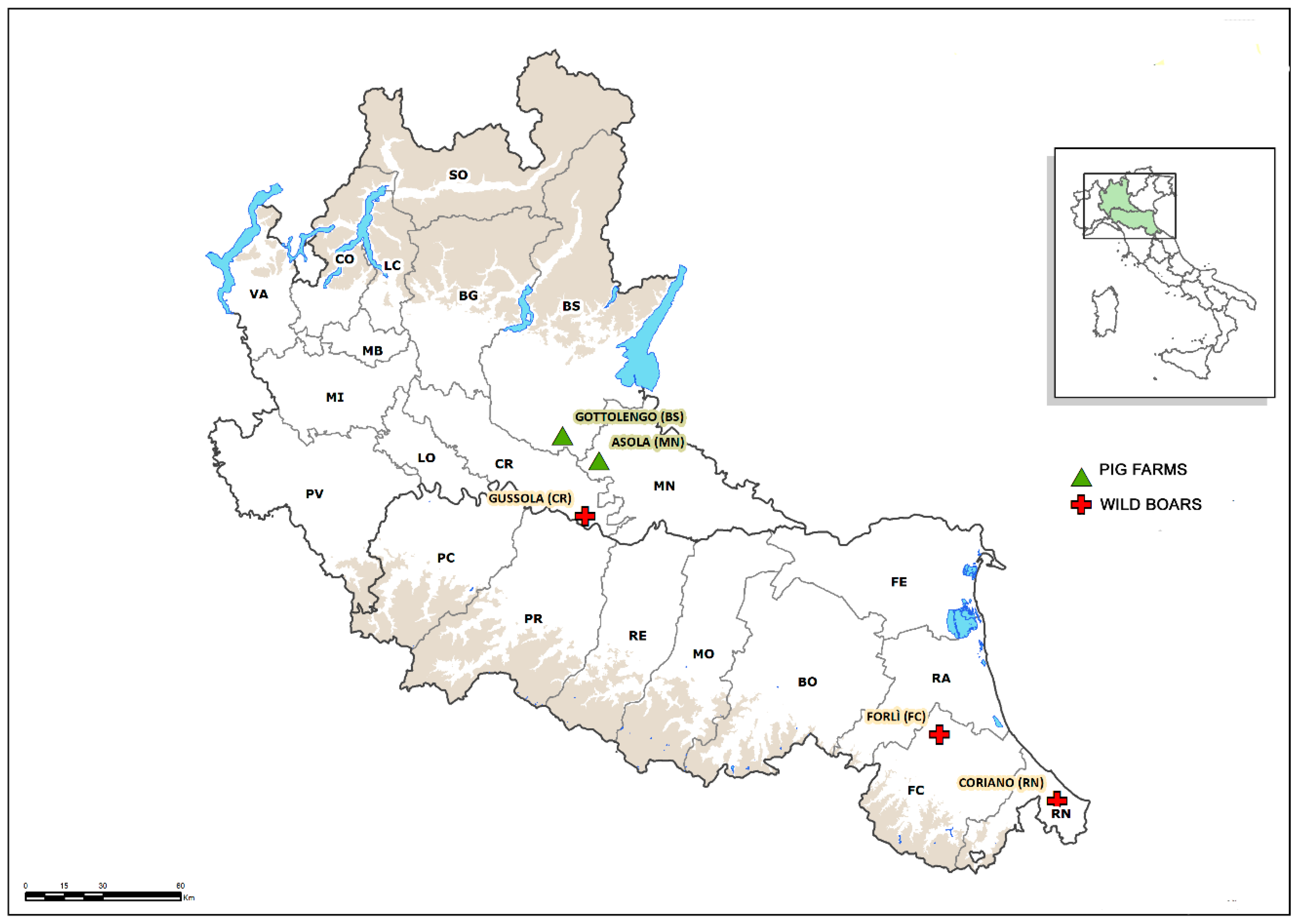
2.1. Pigs
2.2. Wild Boars
2.3. Identification and Genomic Characterization of Atypical Porcine Pestivirus (APPV)
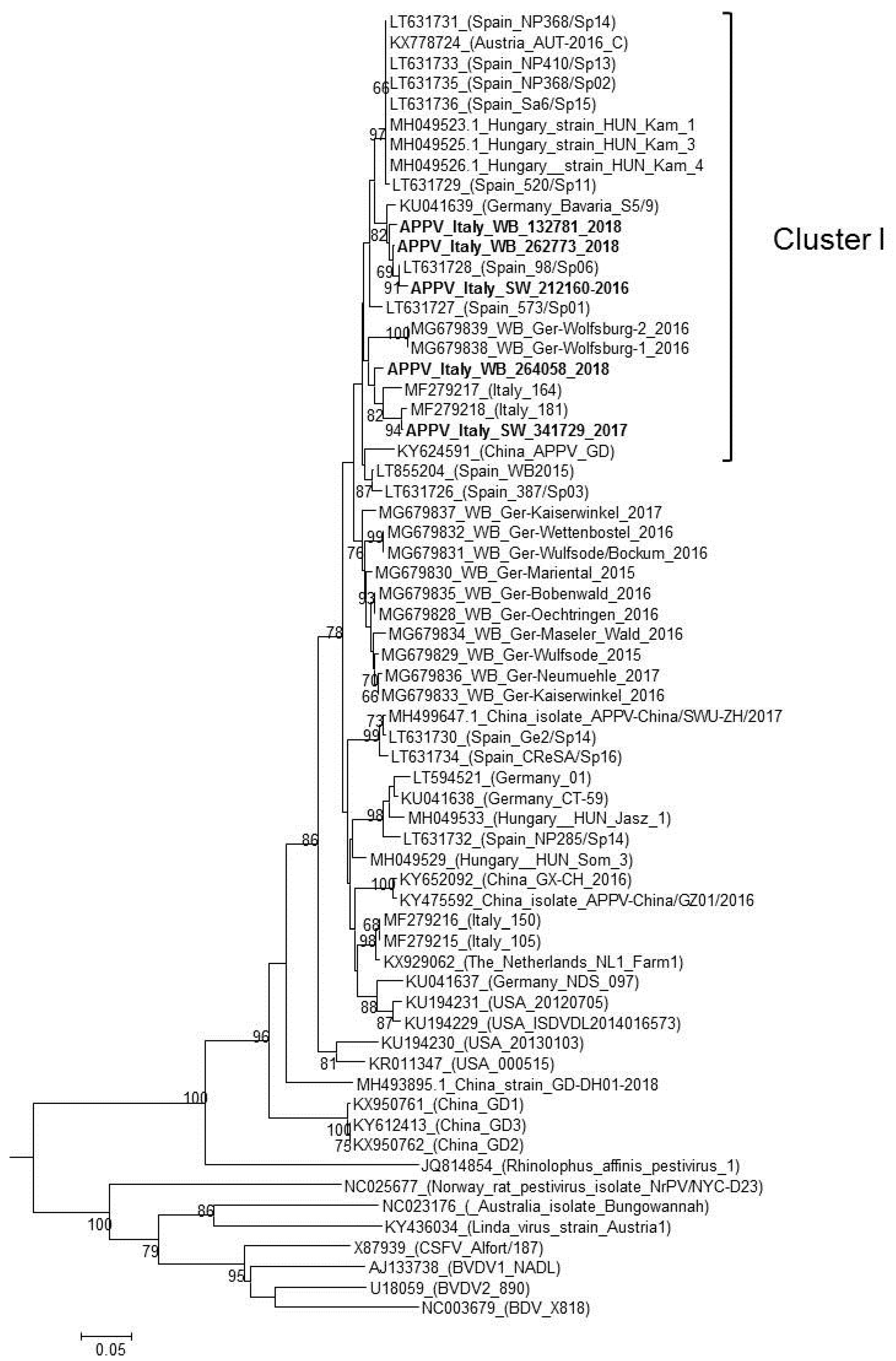
3. Results
3.1. Pigs
3.2. Wild Boars
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Smith, D.B.; Meyers, G.; Bukh, J.; Gould, E.A.; Monath, T.; Scott Muerhoff, A.; Pletnev, A.; Rico-Hesse, R.; Stapleton, J.T.; Simmonds, P.; et al. Proposed revision to the taxonomy of the genus Pestivirus, family Flaviviridae. J. Gen. Virol. 2017, 98, 2106–2112. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.Q.; Ren, X.W.; Yang, L.; Hu, Y.F.; Yang, J.; He, G.M.; Zhang, J.P.; Dong, J.; Sun, L.L.; Du, J.; et al. Virome analysis for identification of novel mammalian viruses in bat species from Chinese provinces. J. Virol. 2012, 86, 10999–11012. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.Q.; Liu, B.; Du, J.; Zhang, J.P.; Lu, L.; Zhu, G.J.; Han, Y.L.; Su, H.X.; Yang, L.; Zhang, S.Y.; et al. Discovery of diverse rodent and bat pestiviruses with distinct genomic and phylogenetic characteristics in several Chinese provinces. Front. Microbiol. 2018, 9, 2562. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.L.; Xia, H.; Wahlberg, N.; Belák, S.; Baule, C. Phylogeny, classification and evolutionary insights into pestiviruses. Virol. 2009, 385, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Lamp, B.; Schwarz, L.; Högler, S.; Riedel, C.; Sinn, L.; Rebel-Bauder, B.; Weissenböck, H.; Ladinig, A.; Rümenapf, T. Novel pestivirus species in pigs, Austria, 2015. Emerg. Infect. Dis. 2017, 23, 1176–1179. [Google Scholar] [CrossRef]
- Kirkland, P.D.; Frost, M.J.; Finlaison, D.S.; King, K.R.; Ridpath, J.F.; Gu, X. Identification of a novel virus in pigs—Bungowannah virus: A possible new species of pestivirus. Virus Res. 2007, 129, 26–34. [Google Scholar] [CrossRef]
- Arruda, B.L.; Arruda, P.H.; Magstadt, D.R.; Schwartz, K.J.; Dohlman, T.; Schleining, J.A.; Patterson, A.R.; Visek, C.A.; Victoria, J.G. Identification of a Divergent Lineage Porcine Pestivirus in Nursing Piglets with Congenital Tremors and Reproduction of Disease following Experimental Inoculation. PLoS ONE 2016, 11, e0150104. [Google Scholar] [CrossRef]
- Hause, B.M.; Collin, E.A.; Peddireddi, L.; Yuan, F.; Chen, Z.; Hesse, R.A.; Gauger, P.C.; Clement, T.; Fang, Y.; Anderson, G. Discovery of a novel putative atypical porcine pestivirus in pigs in the USA. J. Gen. Virol. 2015, 96, 2994–2998. [Google Scholar] [CrossRef]
- Gatto, I.R.H.; Harmon, K.; Bradner, L.; Silva, P.; Linhares, D.C.L.; Arruda, P.H.; de Oliveira, L.G.; Arruda, B.L. Detection of atypical porcine pestivirus in Brazil in the central nervous system of suckling piglets with congenital tremor. Transbound Emerg. Dis. 2018, 65, 375–380. [Google Scholar] [CrossRef]
- Mósena, A.C.S.; Weber, M.N.; da Cruz, R.A.S.; Cibulski, S.P.; da Silva, M.S.; Puhl, D.E.; Hammerschmitt, M.E.; Takeuti, K.L.; Driemeier, D.; de Barcellos, D.E.S.N.; et al. Presence of atypical porcine pestivirus (APPV) in Brazilian pigs. Transbound Emerg. Dis. 2018, 65, 22–26. [Google Scholar] [CrossRef]
- Postel, A.; Hansmann, F.; Baechlein, C.; Fischer, N.; Alawi, M.; Grundhoff, A.; Derking, S.; Tenhündfeld, J.; Pfankuche, V.M.; Herder, V.; et al. Presence of atypical porcine pestivirus (APPV) genomes in newborn piglets correlates with congenital tremor. Sci. Rep. 2016, 6, 27735. [Google Scholar] [CrossRef] [PubMed]
- Beer, M.; Wernike, K.; Dräger, C.; Höper, D.; Pohlmann, A.; Bergermann, C.; Schröder, C.; Klinkhammer, S.; Blome, S.; Hoffmann, B. High Prevalence of Highly Variable Atypical Porcine Pestiviruses Found in Germany. Transbound Emerg. Dis. 2017, 64, e22–e26. [Google Scholar] [CrossRef] [PubMed]
- de Groof, A.; Deijs, M.; Guelen, L.; van Grinsven, L.; van Os-Galdos, L.; Vogels, W.; Derks, C.; Cruijsen, T.; Geurts, V.; Vrijenhoek, M.; et al. Atypical Porcine Pestivirus: A Possible Cause of Congenital Tremor Type A-II in Newborn Piglets. Viruses 2016, 8, 271. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Han, Z.; Li, J.; Huang, Y.; Yang, J.; Ding, H.; Zhang, J.; Zhu, M.; Zhang, Y.; Liao, J.; et al. Atypical Porcine Pestivirus as a Novel Type of Pestivirus in Pigs in China. Front. Microbiol. 2017, 8, 862. [Google Scholar] [CrossRef] [PubMed]
- Postel, A.; Meyer, D.; Cagatay, G.N.; Feliziani, F.; De Mia, G.M.; Fischer, N.; Grundhoff, A.; Milićević, V.; Deng, M.C.; Chang, C.Y.; et al. High Abundance and Genetic Variability of Atypical Porcine Pestivirus in Pigs from Europe and Asia. Emerg. Infect. Dis. 2017, 23, 2104–2107. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-González, S.; Canturri, A.; Pérez-Simó, M.; Bohórquez, J.A.; Rosell, R.; Cabezón, O.; Segalés, J.; Domingo, M.; Ganges, L. First report of the novel atypical porcine pestivirus in Spain and a retrospective study. Transbound Emerg. Dis. 2017, 64, 1645–1649. [Google Scholar] [CrossRef] [PubMed]
- Cagatay, G.N.; Antos, A.; Meyer, D.; Maistrelli, C.; Keuling, O.; Becher, P.; Postel, A. Frequent infection of wild boar with atypical porcine pestivirus (APPV). Transbound Emerg. Dis. 2018, 65, 1087–1093. [Google Scholar] [CrossRef]
- Colom-Cadena, A.; Ganges, L.A.; Muñoz-González, S.; Castillo-Contreras, R.; Alejandro Bohórquez, J.; Rosell, R.; Segalés, J.; Marco, I.; Cabezon, O. Atypical porcine pestivirus in wild boar (Sus scrofa), Spain. Vet. Rec. 2018, 183, 569. [Google Scholar]
- Schwarz, L.; Riedel, C.; Högler, S.; Sinn, L.J.; Voglmayr, T.; Wöchtl, B.; Dinhopl, N.; Rebel-Bauder, B.; Weissenböck, H.; Ladinig, A.; et al. Congenital infection with atypical porcine pestivirus (APPV) is associated with disease and viral persistence. Vet. Res. 2017, 48, 1. [Google Scholar] [CrossRef]
- OIE Manual for Terrestrial Animals 2016 cap 2.1.4 par B1, B1.1, B1.2, B1.3: Brucellosis [Brucella abortus, B. melitensis, B. suis (Infection with B. abortus, B. melitensis, B. suis)]—Diagnostic Techniques—Identification of the Agent—Staining Methods—Collection of Samples and Culture—Identification and Typing. Available online: https://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/3.01.04_BRUCELLOSIS.pdf (accessed on 15 October 2019).
- Ehricht, R.; Slickers, P.; Goellner, S.; Hotzel, H.; Sachse, K. Optimized DNA microarray assay allows detection and genotyping of single PCR-amplifiable target copies. Mol. Cell Probes. 2006, 20, 60–66. [Google Scholar] [CrossRef]
- Vicari, N.; Santoni, R.; Vigo, P.G.; Magnino, S. A PCR-RFLP assay targeting the 16S ribosomal gene for the diagnosis of animal chlamydioses. In Proceedings of the 5th Meeting of the European Society for Chlamydia Research, Budapest, Hungary, 1–4 September 2004. [Google Scholar]
- Van Kuppeveld, F.J.; van der Logt, J.T.; Angulo, A.F.; van Zoest, M.J.; Quint, W.G.; Niesters, H.G.; Galama, J.M.; Melchers, W.J. Genus- and species-specific identification of mycoplasmas by 16S rRNA amplification. Appl. Environ. Microbiol. 1992, 58, 2606–2615, Erratum in: Appl. Environ. Microbiol. 1993, 59, 655. [Google Scholar]
- Olvera, A.; Sibila, M.; Calsamiglia, M.; Segalés, J.; Domingo, M. Comparison of porcine circovirus type 2 load in serum quantified by a real time PCR in postweaning multisystemic wasting syndrome and porcine dermatitis and nephropathy syndrome naturally affected pigs. J. Virol. Methods 2004, 117, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Choi, C.; Han, D.U.; Chae, C. Simultaneous detection of porcine circovirus type 2 and porcine parvovirus in pigs with PMWS by multiplex PCR. Vet. Rec. 2001, 149, 304–305. [Google Scholar] [CrossRef] [PubMed]
- Palinski, R.; Pineyro, P.; Shang, P.; Yuan, F.; Guo, R.; Fang, Y.; Byers, E.; Hause, B.M. A novel porcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure. J. Virol. 2016, 91, 16. [Google Scholar] [CrossRef] [PubMed]
- OIE Manual for Terrestrial Animals 2018 cap 3.4.7 par B1.2: Bovine Viral Diarrhea—Diagnostic Techniques—Detection of the Agent—Nucleic Acid Detection. Available online: https://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/3.04.07_BVD.pdf (accessed on 15 October 2019).
- Brocchi, E.; Cordioli, P.; Berlinzani, A.; Gamba, D.; De Simone, F. Development of a panel of anti-pestivirus monoclonal antibodies useful for virus identification and antibody assessment. In Proceedings of the second Symposium on Ruminant Pestiviruses, Annecy, France, 1–3 October 1992; Edwards, S., Ed.; Fondation Marcel Mérieux: Lyon, France, 1993; pp. 215–218. [Google Scholar]
- Grazioli, S.; Pezzoni, G.; Cordioli, P.; Brocchi, E. Validation of a competitive ELISA for serodiagnosis of PRRS based on recombinant n-protein and monoclonal antibody. In Proceedings of the 8th International Congress of Veterinary Virology, Budapest, Hungary, 24–27 August 2009; Benkõ, M., Harrach, B., Eds.; Hungarian Academy of Sciences: Budapest, Hungary, 2009; p. 199. [Google Scholar]
- Heckert, R.A.; Brocchi, E.; Berlinzani, A.; Mackay, D.K. An international comparative analysis of a competitive ELISA for the detection of antibodies to swine vesicular disease virus. J. Vet. Diagn. Invest. 1998, 10, 295–297. [Google Scholar] [CrossRef]
- OIE Manual for Terrestrial Animals 2018 cap 3.8.8 par B2.2: Swine Vesicular Disease—Diagnostic Techniques—Serological Tests—Enzyme-Linked Immunosorbent Assay. Available online: https://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/3.08.08_SVD.pdf (accessed on 15 October 2019).
- Brocchi, E.; Carra, E.; Koenen, F.; De Simone, F. Sviluppo di metodi ELISA basati sull’impiego di anticorpi monoclonali per la dimostrazione del virus della encefalomiocardite (EMCV) e dei relativi anticorpi; La Selezione Veterinaria—Supplemento; IZSLER: Brescia, Italy, 2000; pp. 207–215. [Google Scholar]
- OIE Manual for Terrestrial Animals 2018 cap 3.1.2 par B2.2: Aujeszky’s Disease (Infection with Aujeszky’s Disease Virus)—Diagnostic Techniques—Serological Tests—Enzyme-Linked Immunosorbent Assay (a Prescribed Test for International Trade). Available online: https://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/3.01.02_AUJESZKYS.pdf (accessed on 15 October 2019).
- OIE Manual for Terrestrial Animals 2019 cap 3.8.3 par B2.3: Classical Swine Fever (Infection with Classical Swine Fever Virus)—Diagnostic Techniques—Serological Tests—Enzyme-Linked Immunosorbent Assay (a Prescribed Test for International Trade). Available online: https://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/3.08.03_CSF.pdf (accessed on 15 October 2019).
- Kendall, A.P.; Pereira, M.S.; Skehel, J.J. Concepts and Procedures for Laboratory-Based Influ-Enza Surveillance; World Health Organization: Geneva, Switzerland, 1982; (Copies available from the WHO Collaborating Centre for Surveillance, Epidemiology and Control of Influenza, CDC, Atlanta, GA). [Google Scholar]
- OIE Manual for Terrestrial Animals 2018 cap 3.8.7 par B2.1: Influenza A Virus of Swine–Diagnostic Techniques—Serological Tests—Haemagglutination Inhibition Test. Available online: https://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/3.08.07_INF_A_SWINE.pdf (accessed on 15 October 2019).
- GenBank® (NCBI). Available online: www.ncbi.nlm.nih.gov (accessed on 22 November 2019).
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sozzi, E.; Salogni, C.; Lelli, D.; Barbieri, I.; Moreno, A.; Alborali, G.L.; Lavazza, A. Molecular Survey and Phylogenetic Analysis of Atypical Porcine Pestivirus (APPV) Identified in Swine and Wild Boar from Northern Italy. Viruses 2019, 11, 1142. https://doi.org/10.3390/v11121142
Sozzi E, Salogni C, Lelli D, Barbieri I, Moreno A, Alborali GL, Lavazza A. Molecular Survey and Phylogenetic Analysis of Atypical Porcine Pestivirus (APPV) Identified in Swine and Wild Boar from Northern Italy. Viruses. 2019; 11(12):1142. https://doi.org/10.3390/v11121142
Chicago/Turabian StyleSozzi, Enrica, Cristian Salogni, Davide Lelli, Ilaria Barbieri, Ana Moreno, Giovanni Loris Alborali, and Antonio Lavazza. 2019. "Molecular Survey and Phylogenetic Analysis of Atypical Porcine Pestivirus (APPV) Identified in Swine and Wild Boar from Northern Italy" Viruses 11, no. 12: 1142. https://doi.org/10.3390/v11121142
APA StyleSozzi, E., Salogni, C., Lelli, D., Barbieri, I., Moreno, A., Alborali, G. L., & Lavazza, A. (2019). Molecular Survey and Phylogenetic Analysis of Atypical Porcine Pestivirus (APPV) Identified in Swine and Wild Boar from Northern Italy. Viruses, 11(12), 1142. https://doi.org/10.3390/v11121142







