Detection of Multiple Variants of Grapevine Fanleaf Virus in Single Xiphinema index Nematodes
Abstract
1. Introduction
2. Materials and Methods
2.1. Grapevines and Nematode Rearings
2.2. Total RNA Extraction from Grapevine Leaves, Roots Samples and Nematodes
2.3. High Throughput Sequencing (HTS), Bioinformatics and Phylogenetic Analyses
2.4. RT-PCR for Virus Detection
2.5. RT-qPCR
2.6. Restriction Fragment Length Polymorphism (RFLP)
2.7. Cloning and Sanger Sequencing
3. Results
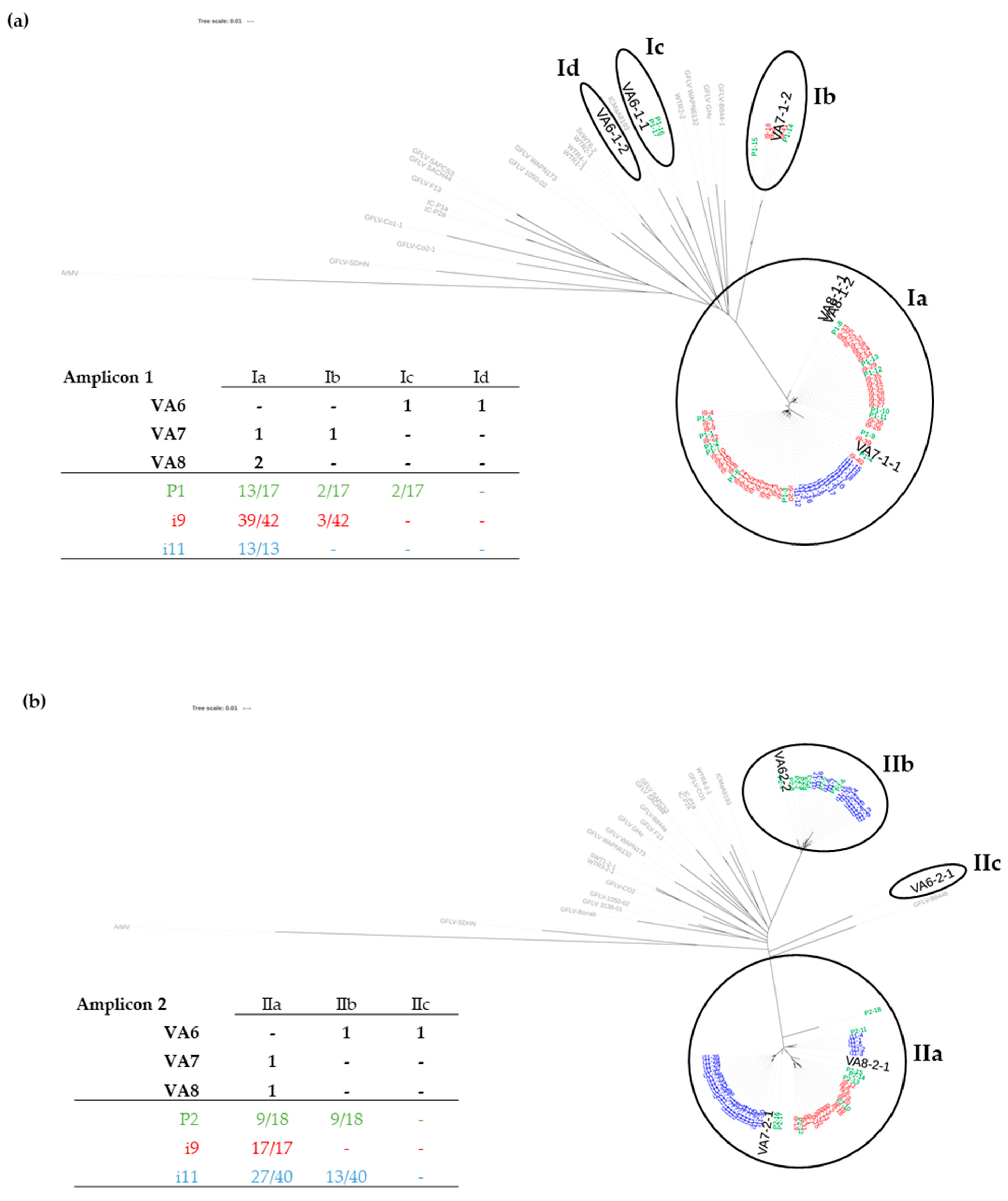
3.1. Sanitary Status of Grapevine Material and Identification of Multiple Variants of GFLV by HTS
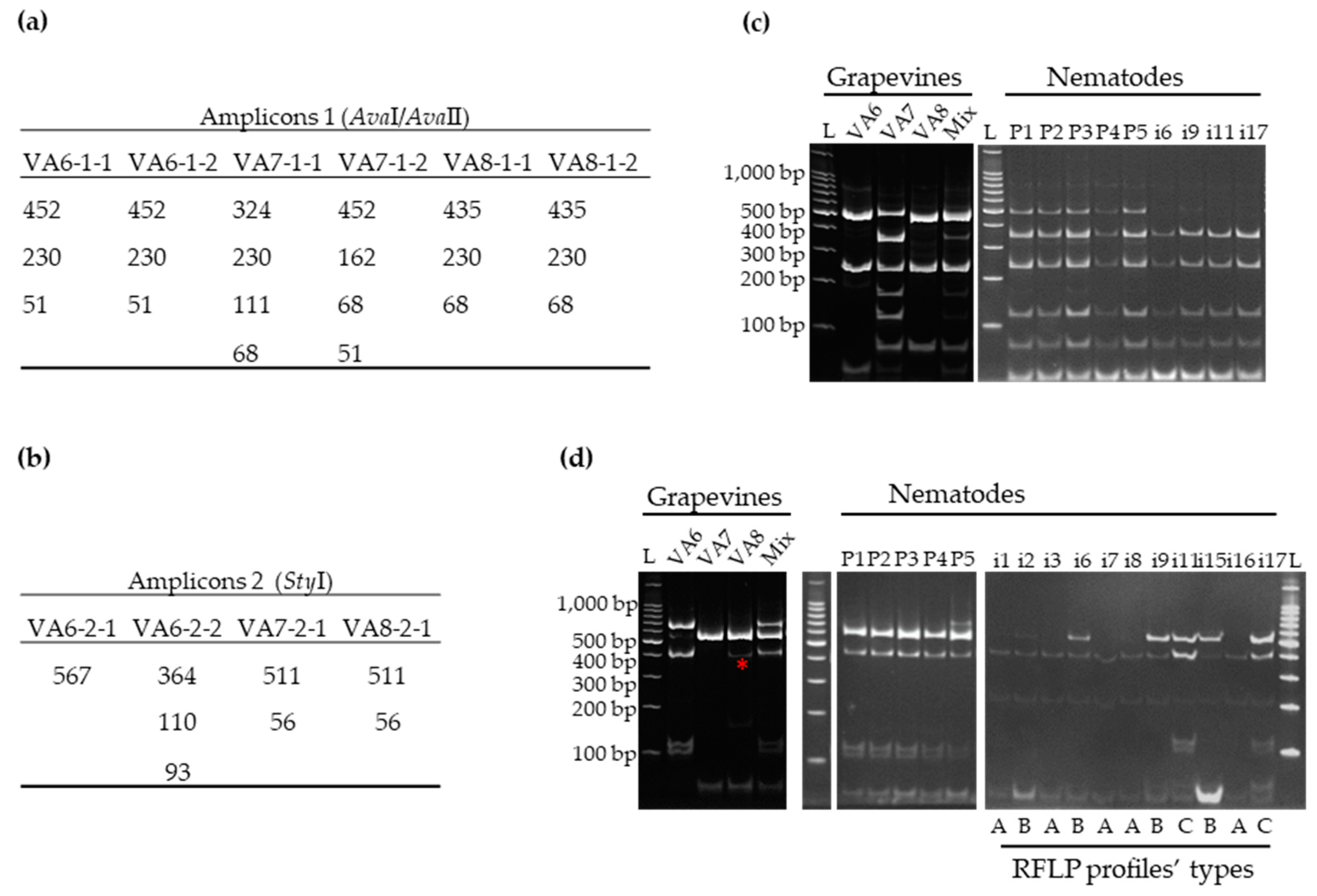
3.2. Distribution of GFLV Variants in Grapevine Plants by RT-PCR-RFLP
3.2.1. Validation by RT-PCR-RFLP of Mixed Infections by GFLV Variants in Grapevines VA6, VA7 and VA8
3.2.2. Distribution of GFLV Variants in Different Organs of Two VA6 Grapevine Cuttings
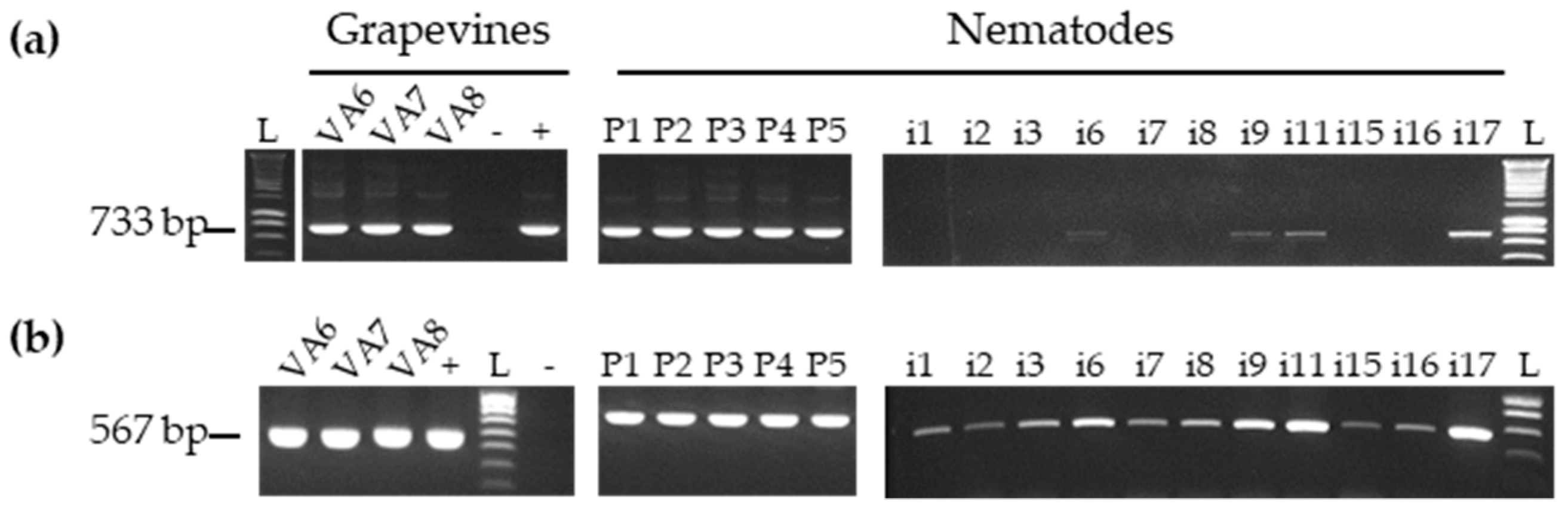
3.3. Detection of GFLV Variants in Pooled and Single Nematodes
3.4. Detection of Other Viruses in Pool and Single Nematodes
4. Discussion
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Steinhauer, D.A.; Domingo, E.; Holland, J.J. Lack of evidence for proofreading mechanisms associated with an RNA virus polymerase. Gene 1992, 122, 281–288. [Google Scholar] [CrossRef]
- Domingo, E.; Sheldon, J.; Perales, C. Viral Quasispecies Evolution. Microbiol. Mol. Biol. Rev. 2012, 76, 159–216. [Google Scholar] [CrossRef] [PubMed]
- Roossinck, M.J. Mechanisms of plant virus evolution. Annu. Rev. Phytopathol. 1997, 35, 191–209. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Roossinck, M.J. Genetic Bottlenecks Reduce Population Variation in an Experimental RNA Virus Population. J. Virol. 2004, 78, 10582–10587. [Google Scholar] [CrossRef]
- Ali, A.; Li, H.; Schneider, W.L.; Sherman, D.J.; Gray, S.; Smith, D.; Roossinck, M.J. Analysis of genetic bottlenecks during horizontal transmission of Cucumber mosaic virus. J. Virol. 2006, 80, 8345–8350. [Google Scholar] [CrossRef]
- Dader, B.; Then, C.; Berthelot, E.; Ducousso, M.; Ng, J.C.K.; Drucker, M. Insect transmission of plant viruses: Multilayered interactions optimize viral propagation. Insect Sci. 2017, 24, 929–946. [Google Scholar] [CrossRef]
- Abel, S.; Abel zur Wiesch, P.; Davis, B.M.; Waldor, M.K. Analysis of Bottlenecks in Experimental Models of Infection. PLoS Pathog. 2015, 11, e1004823. [Google Scholar] [CrossRef]
- Monsion, B.; Froissart, R.; Michalakis, Y.; Blanc, S. Large Bottleneck Size in Cauliflower Mosaic Virus Populations during Host Plant Colonization. PLoS Pathog. 2008, 4, e1000174. [Google Scholar] [CrossRef]
- Zwart, M.P.; Daròs, J.-A.; Elena, S.F. One Is Enough: In Vivo Effective Population Size Is Dose-Dependent for a Plant RNA Virus. PLoS Pathog. 2011, 7, e1002122. [Google Scholar] [CrossRef]
- Zwart, M.P.; Elena, S.F. Matters of Size: Genetic Bottlenecks in Virus Infection and Their Potential Impact on Evolution. Annu. Rev. Virol. 2015, 2, 161–179. [Google Scholar] [CrossRef]
- Visser, P.B.; Brown, D.J.F.; Brederode, F.T.; Bol, J.F. Nematode Transmission of Tobacco Rattle Virus Serves as a Bottleneck to Clear the Virus Population from Defective Interfering RNAs. Virology 1999, 263, 155–165. [Google Scholar] [CrossRef]
- Reynolds, A.G. The Grapevine, Viticulture, and Winemaking: A Brief Introduction. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Meng, B., Martelli, G.P., Golino, D.A., Fuchs, M., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 3–29. [Google Scholar] [CrossRef]
- Martelli, G.P. An Overview on Grapevine Viruses, Viroids, and the Diseases They Cause. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Meng, B., Martelli, G.P., Golino, D.A., Fuchs, M., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 31–46. [Google Scholar] [CrossRef]
- Digiaro, M.; Elbeaino, T.; Martelli, G.P. Grapevine fanleaf virus and Other Old World Nepoviruses. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Meng, B., Martelli, G.P., Golino, D.A., Fuchs, M., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 47–82. [Google Scholar] [CrossRef]
- Vigne, E.; Komar, V.; Tannières, M.; Demangeat, G.; Duchêne, E.; Steyer, D.; Lemarquis, G.; Ritzenthaler, C.; Lemaire, O. Comparative pathogenic effects of distinct Grapevine fanleaf virus strains on Vitis vinifera cvs Gewurztraminer and Chardonnay. In Proceedings of the 18th Congress of the International Council for the Study of Virus and Virus-Like Diseases of the Grapevine (ICVG), Ankara, Turkey, 7–11 September 2015; pp. 236–237. [Google Scholar]
- Andret-Link, P.; Laporte, C.; Valat, L.; Ritzenthaler, C.; Demangeat, G.; Vigne, E.; Laval, V.; Pfeiffer, P.; Stussi-Garaud, C.; Fuchs, M. Grapevine fanleaf virus: Still a major threat to the grapevine industry. J. Plant Pathol. 2004, 86, 183–195. [Google Scholar]
- Schmitt-Keichinger, C.; Hemmer, C.; Berthold, F.; Ritzenthaler, C. Molecular, Cellular, and Structural Biology of Grapevine fanleaf virus. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Meng, B., Martelli, G.P., Golino, D.A., Fuchs, M., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 83–107. [Google Scholar] [CrossRef]
- Quacquarelli, A.; Gallitelli, D.; Savino, V.; Martelli, G.P. Properties of Grapevine Fanleaf Virus. J. Gen. Virol. 1976, 32, 349–360. [Google Scholar] [CrossRef]
- Hewitt, W.B.; Raski, D.J.; Goheen, A.C. Nematode vector of soil-borne fanleaf virus of grapevines. Phytopathology 1958, 48, 586–595. [Google Scholar]
- Andret-Link, P.; Marmonier, A.; Belval, L.; Hleibieh, K.; Ritzenthaler, C.; Demangeat, G. Ectoparasitic Nematode Vectors of Grapevine Viruses. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Meng, B., Martelli, G.P., Golino, D.A., Fuchs, M., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 505–529. [Google Scholar] [CrossRef]
- Demangeat, G.; Voisin, R.; Minot, J.-C.; Bosselut, N.; Fuchs, M.; Esmenjaud, D. Survival of Xiphinema index in Vineyard Soil and Retention of Grapevine fanleaf virus Over Extended Time in the Absence of Host Plants. Phytopathology 2005, 95, 1151–1156. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Taylor, C.E.; Raski, D.J. On the Transmission of Grape Fanleaf By Xiphinema Index. Nematologica 1964, 10, 489–495. [Google Scholar] [CrossRef]
- Das, S.; Raski, D.J. Effect of Grapevine Fanleaf Virus on the Reproduction and Survival of its Nematode Vector, Xiphinema index Thorne &Allen. J. Nematol. 1969, 1, 107–110. [Google Scholar]
- Andret-Link, P.; Schmitt-Keichinger, C.; Demangeat, G.; Komar, V.; Fuchs, M. The specific transmission of Grapevine fanleaf virus by its nematode vector Xiphinema index is solely determined by the viral coat protein. Virology 2004, 320, 12–22. [Google Scholar] [CrossRef]
- Schellenberger, P.; Andret-Link, P.; Schmitt-Keichinger, C.; Bergdoll, M.; Marmonier, A.; Vigne, E.; Lemaire, O.; Fuchs, M.; Demangeat, G.; Ritzenthaler, C. A stretch of 11 amino acids in the betaB-betaC loop of the coat protein of grapevine fanleaf virus is essential for transmission by the nematode Xiphinema index. J. Virol. 2010, 84, 7924–7933. [Google Scholar] [CrossRef]
- Schellenberger, P.; Sauter, C.; Lorber, B.; Bron, P.; Trapani, S.; Bergdoll, M.; Marmonier, A.; Schmitt-Keichinger, C.; Lemaire, O.; Demangeat, G.; et al. Structural Insights into Viral Determinants of Nematode Mediated Grapevine fanleaf virus Transmission. PLoS Pathog. 2011, 7, e1002034. [Google Scholar] [CrossRef]
- Vigne, E.; Bergdoll, M.; Guyader, S.; Fuchs, M. Population structure and genetic variability within isolates of Grapevine fanleaf virus from a naturally infected vineyard in France: Evidence for mixed infection and recombination. J. Gen. Virol. 2004, 85, 2435–2445. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Fan, X.; Dong, Y.; Zhang, Z.; Ren, F.; Hu, G.; Li, Z. Complete nucleotide sequence of a new variant of grapevine fanleaf virus from northeastern China. Arch. Virol. 2017, 162, 577–579. [Google Scholar] [CrossRef] [PubMed]
- Hily, J.-M.; Demanèche, S.; Poulicard, N.; Tannières, M.; Djennane, S.; Beuve, M.; Vigne, E.; Demangeat, G.; Komar, V.; Gertz, C.; et al. Metagenomic-based impact study of transgenic grapevine rootstock on its associated virome and soil bacteriome. Plant Biotechnol. J. 2018, 16, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Vigne, E.; Demangeat, G.; Komar, V.; Fuchs, M. Characterization of a naturally occurring recombinant isolate of Grapevine fanleaf virus. Arch. Virol. 2005, 150, 2241–2255. [Google Scholar] [CrossRef] [PubMed]
- Vigne, E.; Marmonier, A.; Fuchs, M. Multiple interspecies recombination events within RNA2 of Grapevine fanleaf virus and Arabis mosaic virus. Arch. Virol. 2008, 153, 1771–1776. [Google Scholar] [CrossRef] [PubMed]
- Vigne, E.; Garcia, S.; Komar, V.; Lemaire, O.; Hily, J.-M. Comparison of Serological and Molecular Methods With High-Throughput Sequencing for the Detection and Quantification of Grapevine Fanleaf Virus in Vineyard Samples. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Syller, J. Biological and molecular events associated with simultaneous transmission of plant viruses by invertebrate and fungal vectors. Mol. Plant Pathol. 2014, 15, 417–426. [Google Scholar] [CrossRef]
- Demangeat, G.; Komar, V.; Van-Ghelder, C.; Voisin, R.; Lemaire, O.; Esmenjaud, D.; Fuchs, M. Transmission Competency of Single-Female Xiphinema index Lines for Grapevine fanleaf virus. Phytopathology 2010, 100, 384–389. [Google Scholar] [CrossRef]
- Demangeat, G.; Komar, V.; Cornuet, P.; Esmenjaud, D.; Fuchs, M. Sensitive and reliable detection of grapevine fanleaf virus in a single Xiphinema index nematode vector. J. Virol. Methods 2004, 122, 79–86. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Hily, J.-M.; Candresse, T.; Garcia, S.; Vigne, E.; Tannière, M.; Komar, V.; Barnabé, G.; Alliaume, A.; Gilg, S.; Hommay, G.; et al. High-Throughput Sequencing and the Viromic Study of Grapevine Leaves: From the Detection of Grapevine-Infecting Viruses to the Description of a New Environmental Tymovirales Member. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Czotter, N.; Molnar, J.; Szabó, E.; Demian, E.; Kontra, L.; Baksa, I.; Szittya, G.; Kocsis, L.; Deak, T.; Bisztray, G.; et al. NGS of Virus-Derived Small RNAs as a Diagnostic Method Used to Determine Viromes of Hungarian Vineyards. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Beuve, M.; Hily, J.M.; Alliaume, A.; Reinbold, C.; Le Maguet, J.; Candresse, T.; Herrbach, E.; Lemaire, O. A complex virome unveiled by deep sequencing analysis of RNAs from a French Pinot Noir grapevine exhibiting strong leafroll symptoms. Arch. Virol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Huogen, X.; Caihong, L.; Maher, A.R.; Valerian, D.; Baozhong, M. Metagenomic Analysis of Riesling Grapevine Reveals a Complex Virome Including Two New and Divergent Variants of Grapevine leafroll-associated virus 3. Plant Dis. 2019, 103. [Google Scholar] [CrossRef]
- Jo, Y.; Choi, H.; Cho, J.K.; Yoon, J.Y.; Choi, S.K.; Cho, W.K. In silico approach to reveal viral populations in grapevine cultivar Tannat using transcriptome data. Sci. Rep. 2015, 5, 15841. [Google Scholar] [CrossRef]
- Lunden, S.; Meng, B.; Avery, J.; Qiu, W. Association of Grapevine fanleaf virus, Tomato ringspot virus and Grapevine rupestris stem pitting-associated virus with a grapevine vein-clearing complex on var. Chardonnay. Eur. J. Plant Pathol. 2009, 126, 135. [Google Scholar] [CrossRef]
- Wright, S.M.; Kawaoka, Y.; Sharp, G.B.; Senne, D.A.; Webster, R.G. Interspecies Transmission and Reassortment of Influenza a Viruses in Pigs and Turkeys in the United States. Am. J. Epidemiol. 1992, 136, 488–497. [Google Scholar] [CrossRef]
- Schumann, T.; Hotzel, H.; Otto, P.; Johne, R. Evidence of interspecies transmission and reassortment among avian group A rotaviruses. Virology 2009, 386, 334–343. [Google Scholar] [CrossRef]
- Gutierrez, S.; Michalakis, Y.; Blanc, S. Virus population bottlenecks during within-host progression and host-to-host transmission. Curr. Opin. Virol. 2012, 2, 546–555. [Google Scholar] [CrossRef]
- Krebelj, A.; Cepin, U.; Ravnikar, M.; Pompe Novak, M. Spatio-temporal distribution of Grapevine fanleaf virus (GFLV) in grapevine. Eur. J. Plant Pathol. 2015, 142, 159–171. [Google Scholar] [CrossRef]
- Betancourt, M.; Fereres, A.; Fraile, A.; García-Arenal, F. Estimation of the effective number of founders that initiate an infection after aphid transmission of a multipartite plant virus. J. Virol. 2008, 82, 12416–12421. [Google Scholar] [CrossRef] [PubMed]
- Moury, B.; Fabre, F.; Senoussi, R. Estimation of the number of virus particles transmitted by an insect vector. Proc. Natl. Acad. Sci. USA 2007, 104, 17891–17896. [Google Scholar] [CrossRef] [PubMed]
- Gallet, R.; Fabre, F.; Thébaud, G.; Sofonea, M.T.; Sicard, A.; Blanc, S.; Michalakis, Y. Small Bottleneck Size in a Highly Multipartite Virus during a Complete Infection Cycle. J. Virol. 2018, 92, e00139-18. [Google Scholar] [CrossRef] [PubMed]
- Bragard, C.; Caciagli, P.; Lemaire, O.; Lopez-Moya, J.J.; MacFarlane, S.; Peters, D.; Susi, P.; Torrance, L. Status and Prospects of Plant Virus Control Through Interference with Vector Transmission. Annu. Rev. Phytopathol. 2013, 51, 177–201. [Google Scholar] [CrossRef] [PubMed]
- Legavre, T.; Maia, I.G.; Casse-Delbart, F.; Bernardi, F.; Robaglia, C. Switches in the mode of transmission select for or against a poorly aphid-transmissible strain of potato virus Y with reduced helper component and virus accumulation. J. Gen. Virol. 1996, 77, 1343–1347. [Google Scholar] [CrossRef]
- Nolasco, G.; Fonseca, F.; Silva, G. Occurrence of genetic bottlenecks during citrus tristeza virus acquisition by Toxoptera citricida under field conditions. Arch. Virol. 2008, 153, 259–271. [Google Scholar] [CrossRef]
- Albiach-Martí, M.R.; Guerri, J.; de Mendoza, A.H.; Laigret, F.; Ballester-Olmos, J.F.; Moreno, P. Aphid Transmission Alters the Genomic and Defective RNA Populations of Citrus tristeza virus Isolates. Phytopathology 2000, 90, 134–138. [Google Scholar] [CrossRef]
- Keele, B.F.; Giorgi, E.E.; Salazar-Gonzalez, J.F.; Decker, J.M.; Pham, K.T.; Salazar, M.G.; Sun, C.; Grayson, T.; Wang, S.; Li, H.; et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. USA 2008, 105, 7552–7557. [Google Scholar] [CrossRef]
- Smith, D.R.; Adams, A.P.; Kenney, J.L.; Wang, E.; Weaver, S.C. Venezuelan equine encephalitis virus in the mosquito vector Aedes taeniorhynchus: Infection initiated by a small number of susceptible epithelial cells and a population bottleneck. Virology 2008, 372, 176–186. [Google Scholar] [CrossRef]
- Forrester, N.L.; Guerbois, M.; Seymour, R.L.; Spratt, H.; Weaver, S.C. Vector-borne transmission imposes a severe bottleneck on an RNA virus population. PLoS Pathog. 2012, 8, e1002897. [Google Scholar] [CrossRef] [PubMed]
- Lequime, S.; Fontaine, A.; Ar Gouilh, M.; Moltini-Conclois, I.; Lambrechts, L. Genetic Drift, Purifying Selection and Vector Genotype Shape Dengue Virus Intra-host Genetic Diversity in Mosquitoes. PLoS Genet. 2016, 12, e1006111. [Google Scholar] [CrossRef] [PubMed]
- Grubaugh, N.D.; Weger-Lucarelli, J.; Murrieta, R.A.; Fauver, J.R.; Garcia-Luna, S.M.; Prasad, A.N.; Black, W.C.; Ebel, G.D. Genetic Drift during Systemic Arbovirus Infection of Mosquito Vectors Leads to Decreased Relative Fitness during Host Switching. Cell Host Microbe 2016, 19, 481–492. [Google Scholar] [CrossRef] [PubMed]
- Simmons, H.E.; Dunham, J.P.; Stack, J.C.; Dickins, B.J.A.; Pagán, I.; Holmes, E.C.; Stephenson, A.G. Deep sequencing reveals persistence of intra-and inter-host genetic diversity in natural and greenhouse populations of zucchini yellow mosaic virus. J. Gen. Virol. 2012, 93, 1831–1840. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.C.; Villate, L.; Gutierrez-Gutierrez, C.; Castillo, P.; Van Ghelder, C.; Plantard, O.; Esmenjaud, D. Phylogeography of the soil-borne vector nematode Xiphinema index highly suggests Eastern origin and dissemination with domesticated grapevine. Sci. Rep. 2019, 9, 7313. [Google Scholar] [CrossRef] [PubMed]

| ID | VA6 | VA7 | VA8 | |
|---|---|---|---|---|
| Total clean reads | 32,435,733 | 30,338,401 | 34,907,736 | |
| GFLV | RNA1-1 | 772 (VA6-1-1, clade Ic) | 680 (VA7-1-1, clade Ia) | 670 (VA8-1-1, clade Ia) |
| RNA1-2 | 235 (VA6-1-2, clade Id) | 194 (VA7-1-2, clade Ib) | 577 (VA8-1-2, clade Ia) | |
| RNA2-1 | 904 (VA6-2-1, clade IIc) | 2037 (VA7-2-1, clade IIa) | 2923 (VA8-2-1, clade IIa) | |
| RNA2-2 | 899 (VA6-2-2, clade IIb) | - | - | |
| ArMV | RNA1 | 13 | - | - |
| RNA2 | 26 | - | - | |
| Other viruses | GVB | - | 55 | 105 |
| GRVFV-1 | 15 | - | - | |
| GRVFV-2 | 13 | - | - | |
| GRVFV-3 | 11 | - | - | |
| GRGV | - | ✓ | - | |
| “background” Virome (viruses And viroids) | GRSPaV-1 | 37 | 41 | 20 |
| GRSPaV-2 | 16 | 28 | 19 | |
| GRSPaV-3 | 3 | 26 | 11 | |
| GRSPaV-4 | - | 23 | 10 | |
| GRSPaV-5 | - | 7 | 5 | |
| GFkV | ✓ | ✓ | ✓ | |
| GYSVd1-1 | 91 | 103 | 43 | |
| GYSVd1-2 | - | 49 | - | |
| HSVd | 112 | 25 | 181 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garcia, S.; Hily, J.-M.; Komar, V.; Gertz, C.; Demangeat, G.; Lemaire, O.; Vigne, E. Detection of Multiple Variants of Grapevine Fanleaf Virus in Single Xiphinema index Nematodes. Viruses 2019, 11, 1139. https://doi.org/10.3390/v11121139
Garcia S, Hily J-M, Komar V, Gertz C, Demangeat G, Lemaire O, Vigne E. Detection of Multiple Variants of Grapevine Fanleaf Virus in Single Xiphinema index Nematodes. Viruses. 2019; 11(12):1139. https://doi.org/10.3390/v11121139
Chicago/Turabian StyleGarcia, Shahinez, Jean-Michel Hily, Véronique Komar, Claude Gertz, Gérard Demangeat, Olivier Lemaire, and Emmanuelle Vigne. 2019. "Detection of Multiple Variants of Grapevine Fanleaf Virus in Single Xiphinema index Nematodes" Viruses 11, no. 12: 1139. https://doi.org/10.3390/v11121139
APA StyleGarcia, S., Hily, J.-M., Komar, V., Gertz, C., Demangeat, G., Lemaire, O., & Vigne, E. (2019). Detection of Multiple Variants of Grapevine Fanleaf Virus in Single Xiphinema index Nematodes. Viruses, 11(12), 1139. https://doi.org/10.3390/v11121139





