Root Transcriptomic Analysis Reveals Global Changes Induced by Systemic Infection of Solanum lycopersicum with Mild and Severe Variants of Potato Spindle Tuber Viroid
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Growth and PSTVd Inoculation
2.2. Lignin Quantification
2.3. RNA Extraction
2.4. Northern Blot Hybridization
2.5. PSTVd Sequencing
2.6. Transcriptome Profiling Using Microarrays
2.7. Transcriptome Profiling by RNA-seq
2.8. Functional Annotation of Genes and Pathway Analysis
2.9. NanoString nCounter Analysis
2.10. Statistical Analysis
3. Results
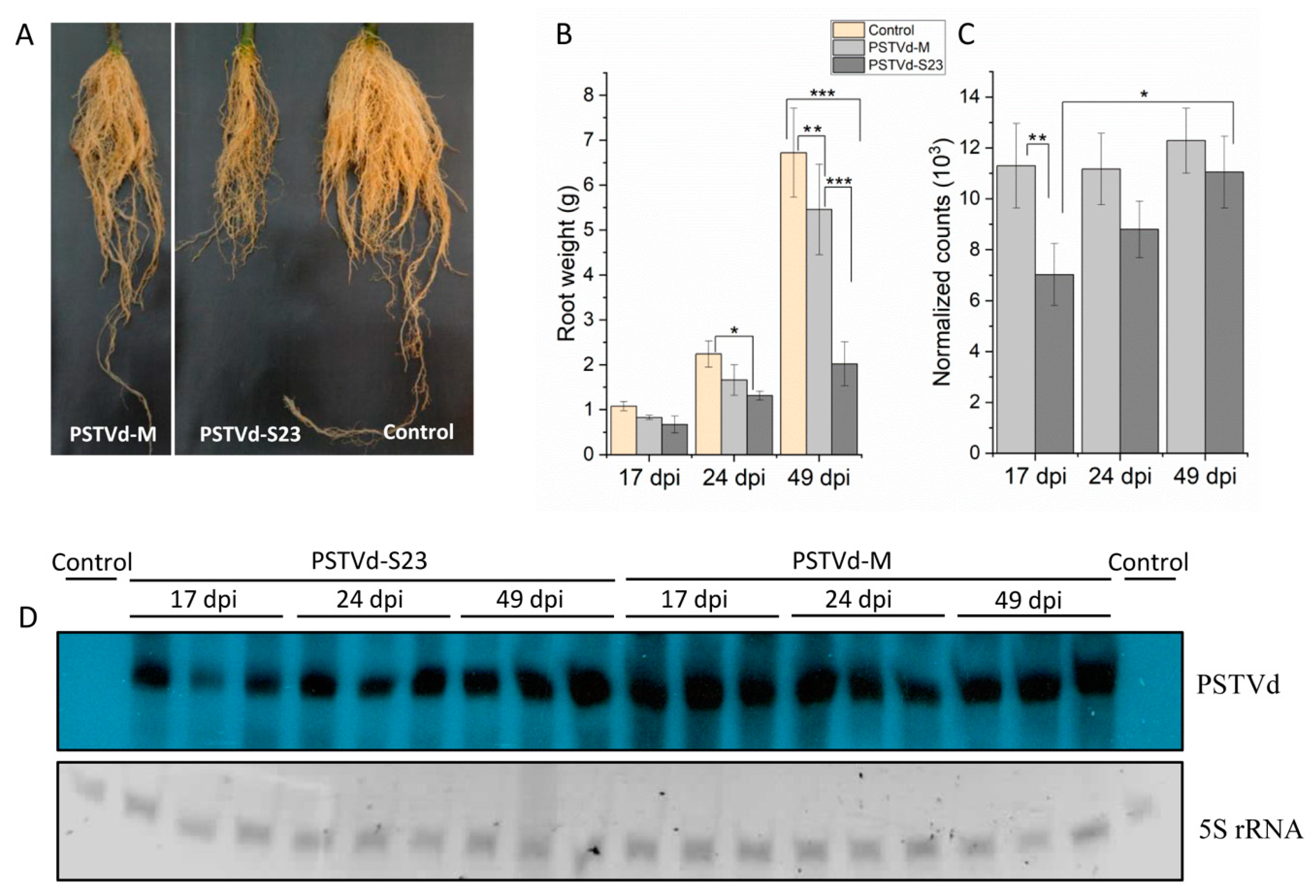
3.1. Mild and Severe PSTVd Infection in Tomato Roots
3.2. Overview of Gene Expression in Roots during PSTVd Infection
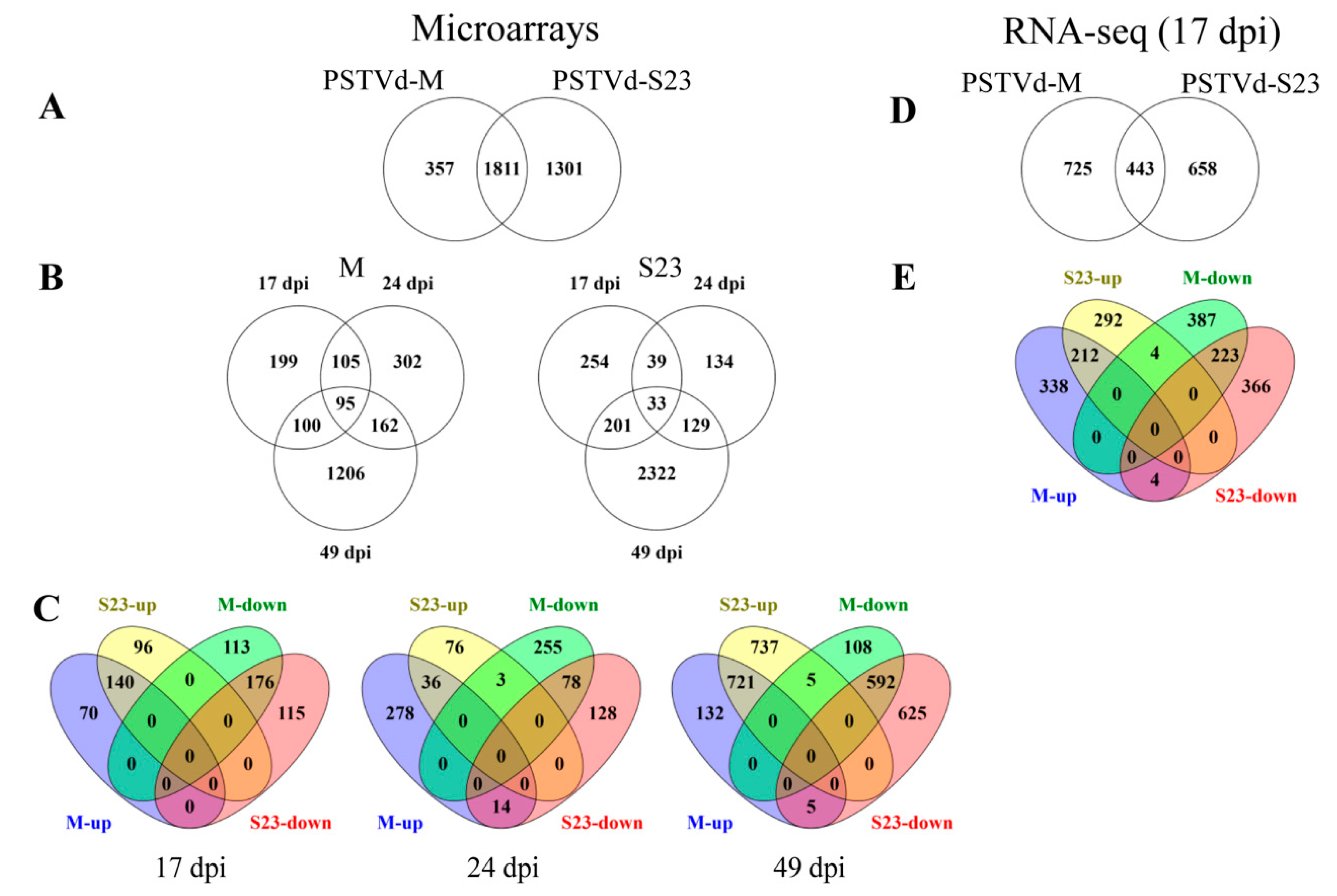
3.2.1. Microarray
3.2.2. RNA-seq
3.3. KEGG Pathways Influenced by the Infection Time Course
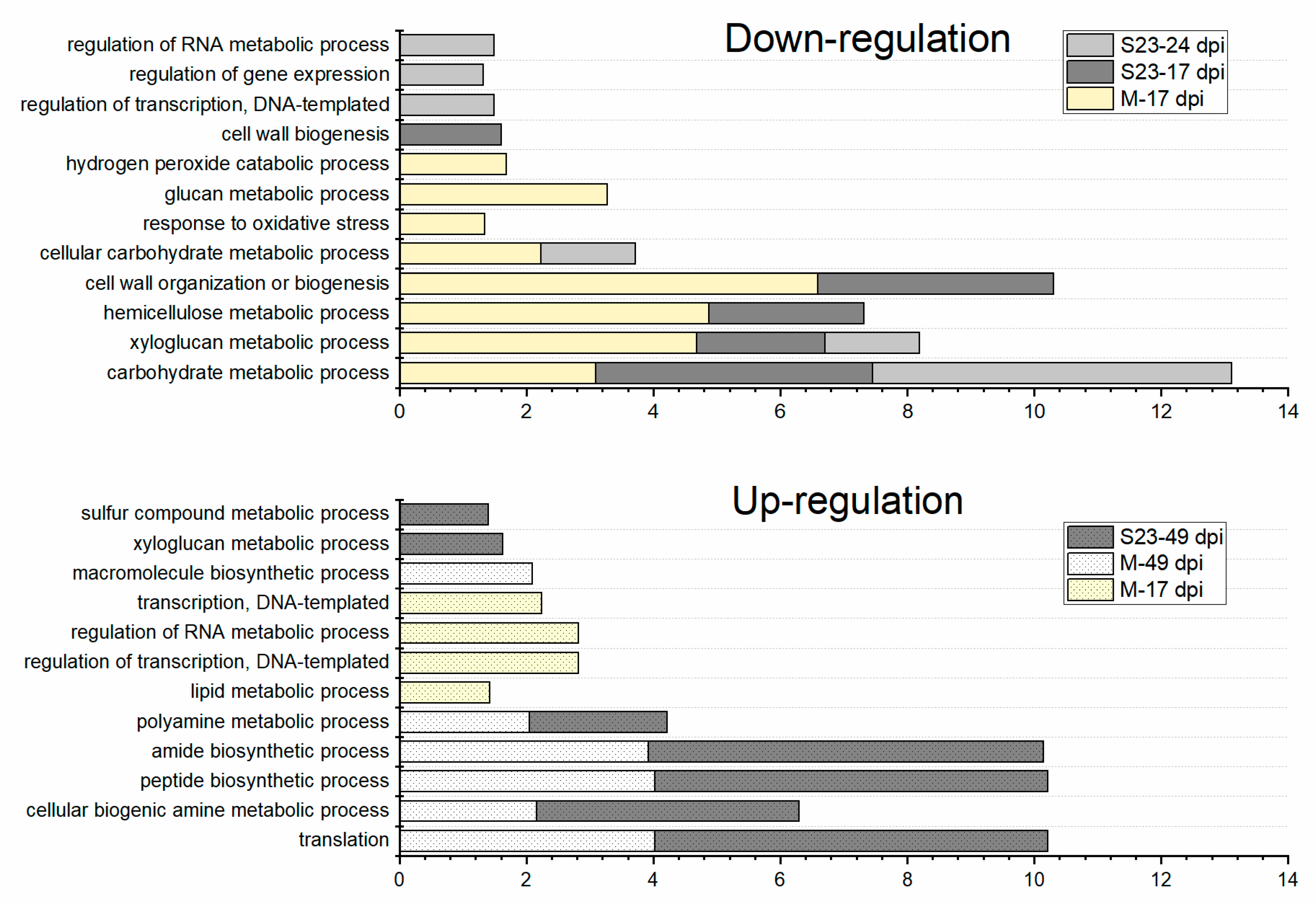
3.4. Enriched GO Terms Related to Biological Process over the Infection Time Course
3.5. Analysis of DEGs Identified by RNA-seq Analysis
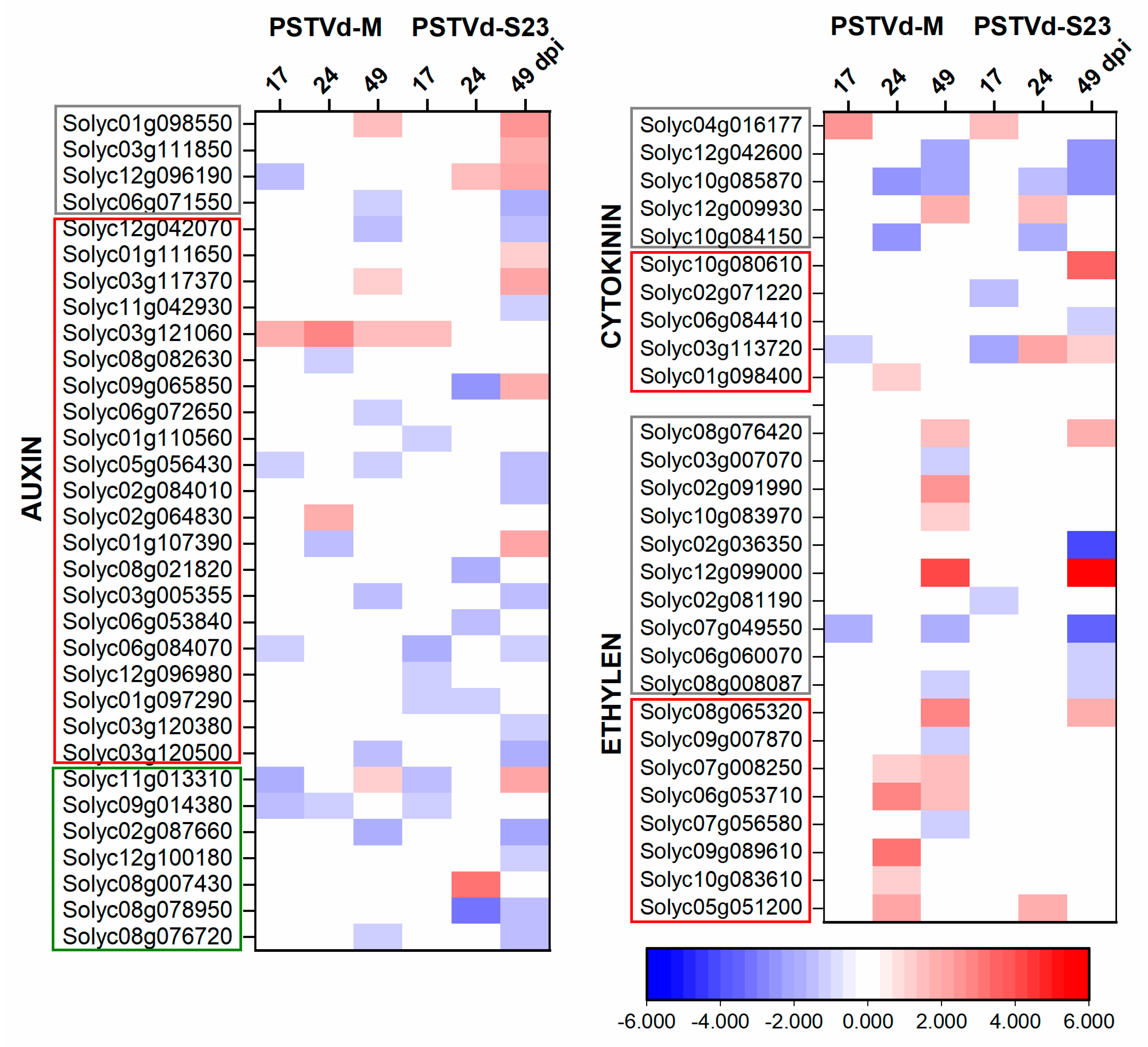
3.6. Expression of Hormone-Related DEGs
3.7. DEGs That Encode TFs and PKs
3.8. PSTVd Infection Repressed the Expression of Cell-Wall-Related Genes during Early Infection
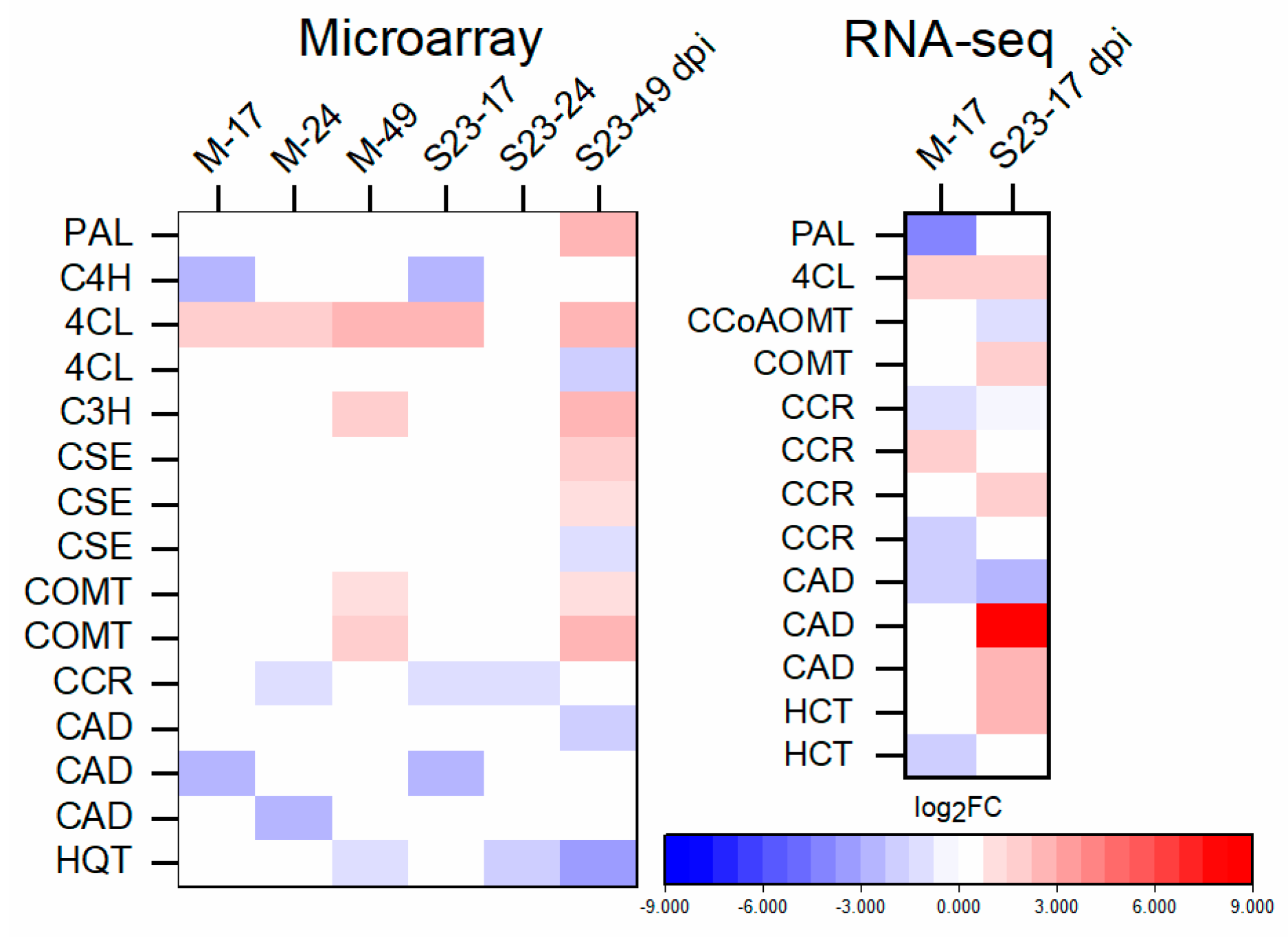
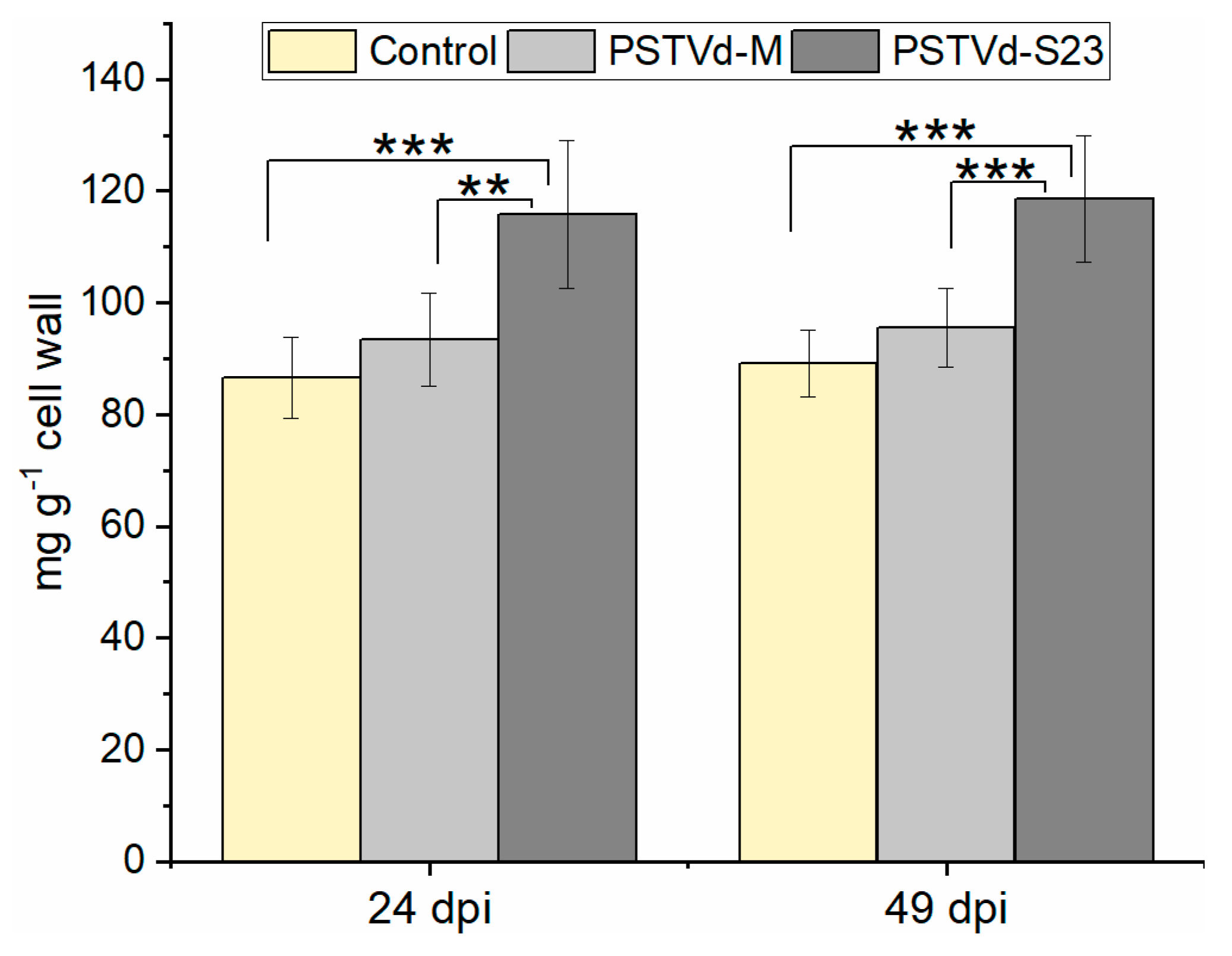
3.9. Expression of Lignin-Biosynthesis-Specific Genes and Lignin Quantification
3.10. Up-Regulation of Proteasome-Related Genes
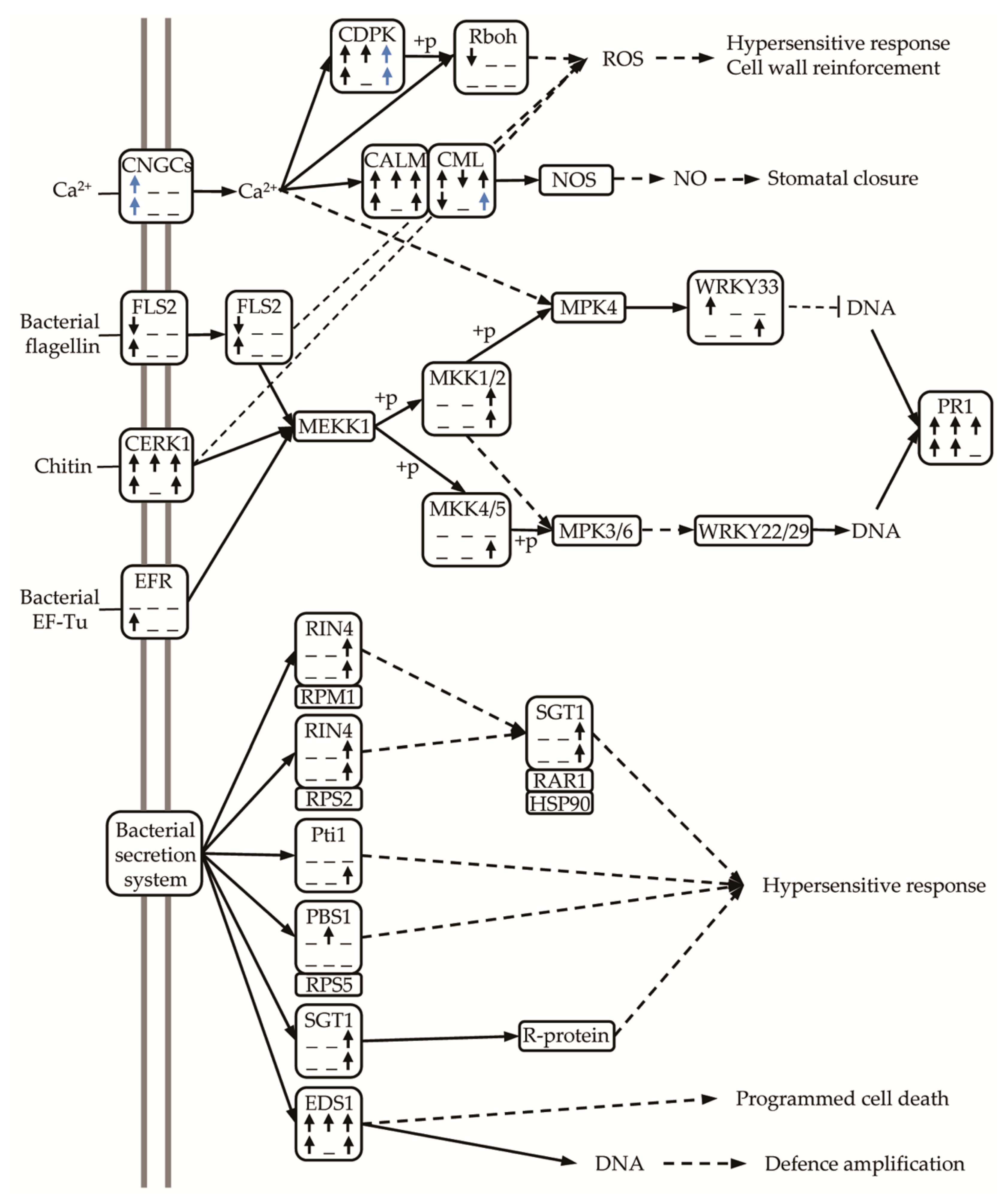
3.11. Up-Regulation of DEGs Related to Plant-Pathogen Interaction
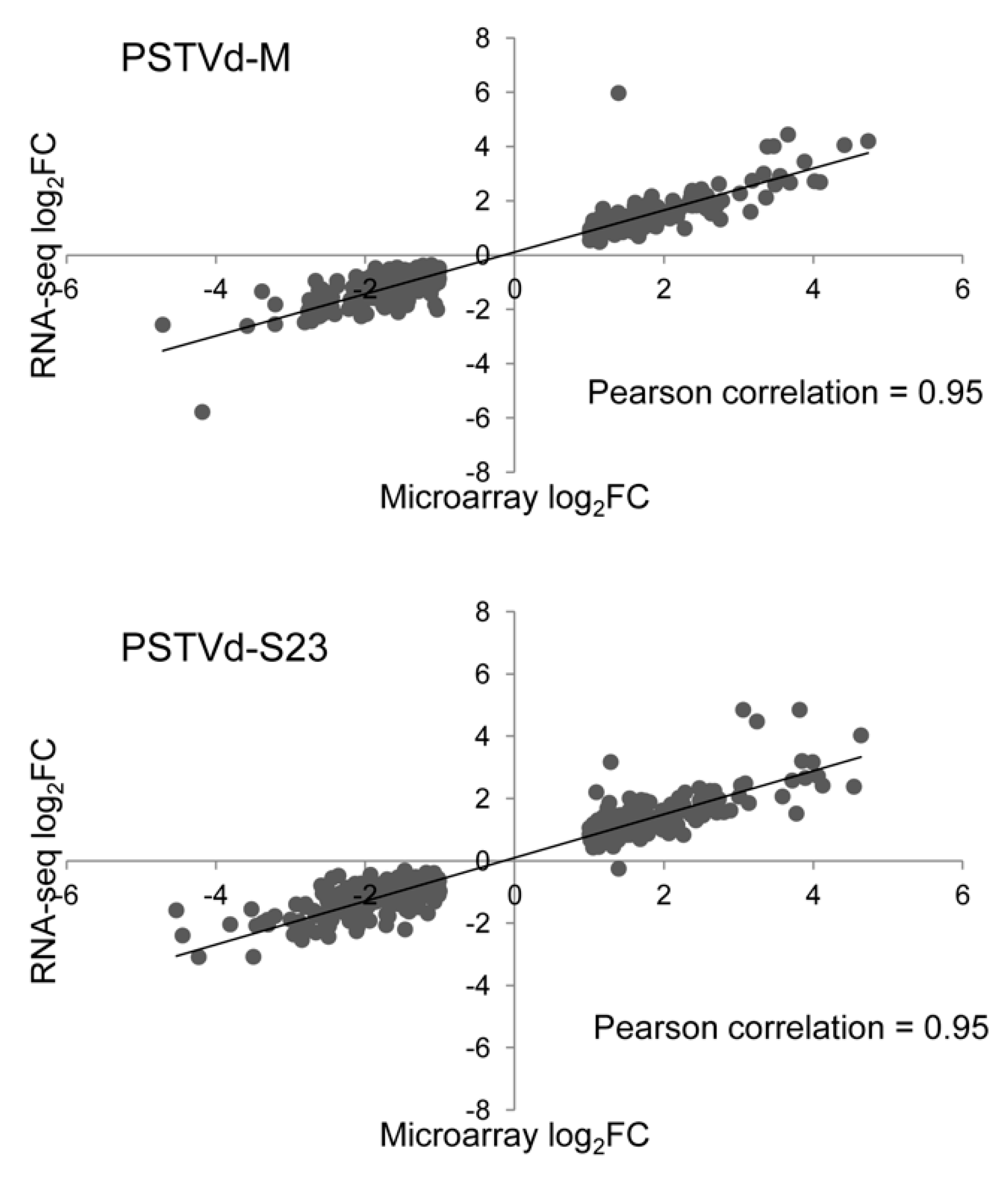
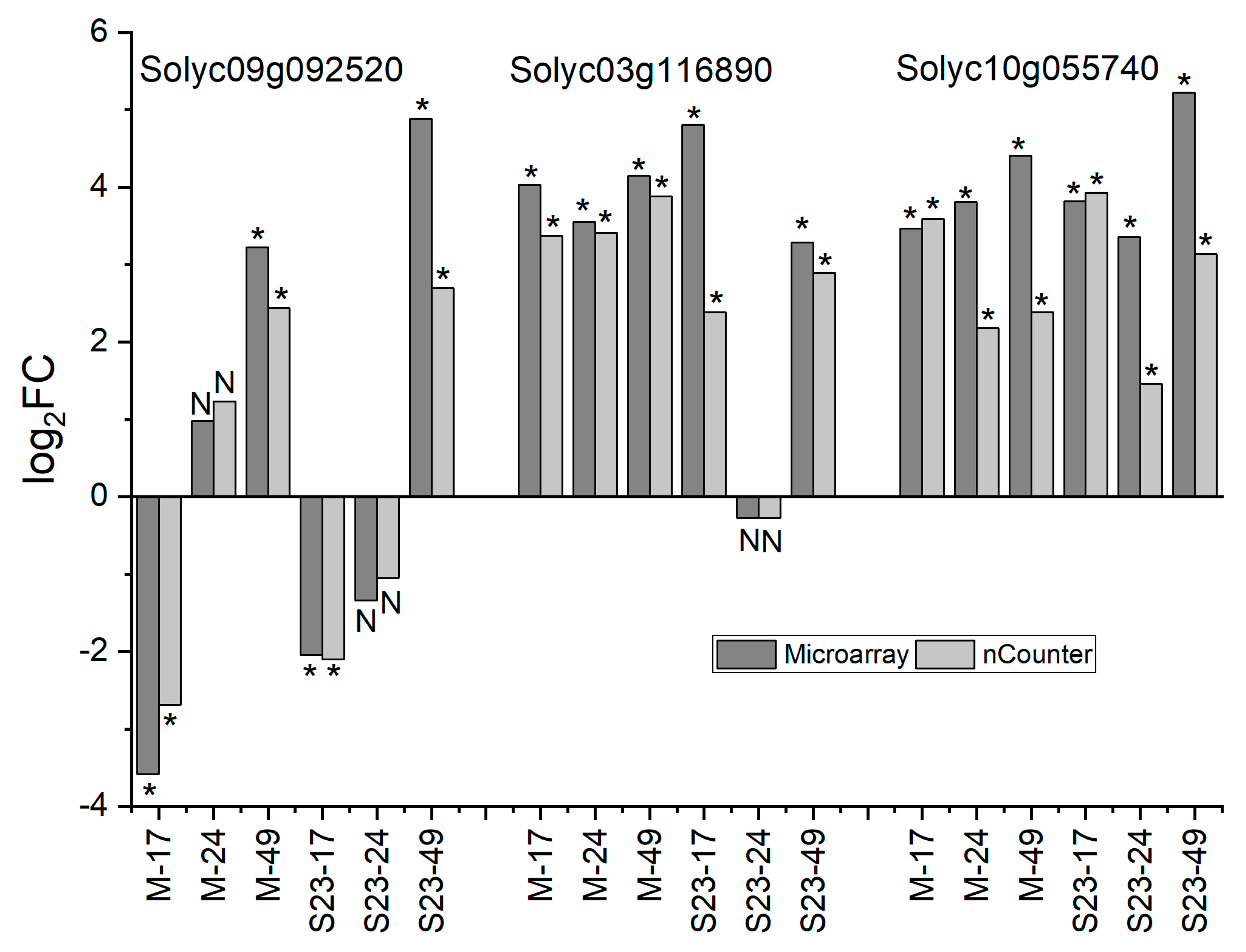
3.12. Validation of Microarray and RNA-seq
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Flores, R.; Minoia, S.; Carbonell, A.; Gisel, A.; Delgado, S.; López-Carrasco, A.; Navarro, B.; Di Serio, F. Viroids, the simplest RNA replicons: How they manipulate their hosts for being propagated and how their hosts react for containing the infection. Virus Res. 2015, 209, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Gago-Zachert, S. Viroids, infectious long non-coding RNAs with autonomous replication. Virus Res. 2016, 212, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Palukaitis, P. What has been happening with viroids? Virus Genes 2014, 49, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Hadidi, A.; Flores, R.; Randles, J.; Palukaitis, P. Viroids and Satellies; Academic Press: Cambridge, MA, USA; Elsevier: Oxford, UK, 2017; ISBN 978-0-12-801498-1. [Google Scholar]
- Mühlbach, H.P.; Sänger, H.L. Viroid replication is inhibited by alpha-amanitin. Nature 1979, 278, 185–188. [Google Scholar] [CrossRef]
- Bojić, T.; Beeharry, Y.; Zhang, D.J.; Pelchat, M. Tomato RNA polymerase II interacts with the rod-like conformation of the left terminal domain of the potato spindle tuber viroid positive RNA genome. J. Gen. Virol. 2012, 93, 1591–1600. [Google Scholar] [CrossRef] [Green Version]
- Gas, M.E.; Hernández, C.; Flores, R.; Daròs, J.A. Processing of nuclear viroids in vivo: an interplay between RNA conformations. PLoS Pathog. 2007, 3, e182. [Google Scholar] [CrossRef]
- Nohales, M.; Flores, R.; Daròs, J.A. Viroid RNA redirects host DNA ligase 1 to act as an RNA ligase. Proc. Natl. Acad. Sci. USA 2012, 109, 13805–13810. [Google Scholar] [CrossRef] [Green Version]
- Kovalskaya, N.; Hammond, R.W. Molecular biology of viroid-host interactions and disease control strategies. Plant Sci. 2014, 228, 48–60. [Google Scholar] [CrossRef]
- Itaya, A.; Matsuda, Y.; Gonzales, R.A.; Nelson, R.S.; Ding, B. Potato spindle tuber viroid strains of different pathogenicity induces and suppresses expression of common and unique genes in infected tomato. Mol. Plant Microbe Interact. 2002, 15, 990–999. [Google Scholar] [CrossRef]
- Wang, Y.; Shibuya, M.; Taneda, A.; Kurauchi, T.; Senda, M.; Owens, R.A.; Sano, T. Accumulation of Potato spindle tuber viroid-specific small RNAs is accompanied by specific changes in gene expression in two tomato cultivars. Virology 2011, 413, 72–83. [Google Scholar] [CrossRef] [Green Version]
- Owens, R.A.; Tech, K.B.; Shao, J.Y.; Sano, T.; Baker, C.J. Global analysis of tomato gene expression during Potato spindle tuber viroid infection reveals a complex array of changes affecting hormone signaling. Mol. Plant Microbe Interact. 2012, 25, 582–598. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wang, Y.; Ding, B.; Fei, Z. Comprehensive Transcriptome Analyses Reveal that Potato Spindle Tuber Viroid Triggers Genome-Wide Changes in Alternative Splicing, Inducible trans-Acting Activity of Phased Secondary Small Interfering RNAs, and Immune Responses. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Więsyk, A.; Iwanicka-Nowicka, R.; Fogtman, A.; Zagórski-Ostoja, W.; Góra-Sochacka, A. Time-Course Microarray Analysis Reveals Differences between Transcriptional Changes in Tomato Leaves Triggered by Mild and Severe Variants of Potato Spindle Tuber Viroid. Viruses 2018, 10, 257. [Google Scholar] [CrossRef] [PubMed]
- Rizza, S.; Conesa, A.; Juarez, J.; Catara, A.; Navarro, L.; Duran-Vila, N.; Ancillo, G. Microarray analysis of Etrog citron (Citrus medica L.) reveals changes in chloroplast, cell wall, peroxidase and symporter activities in response to viroid infection. Mol. Plant Pathol. 2012, 13, 852–864. [Google Scholar] [CrossRef]
- Thibaut, O.; Claude, B. Innate Immunity Activation and RNAi Interplay in Citrus Exocortis Viroid-Tomato Pathosystem. Viruses 2018, 10, 587. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, J.; Qiu, Y.; Atta, S.; Zhou, C.; Cao, M. Global Transcriptomic Analysis Reveals Insights into the Response of ‘Etrog’ Citron (Citrus medica L.) to Citrus Exocortis Viroid Infection. Viruses 2019, 11, 453. [Google Scholar] [CrossRef]
- Tessitori, M.; Maria, G.; Capasso, C.; Catara, G.; Rizza, S.; De Luca, V.; Catara, A.; Capasso, A.; Carginale, V. Differential display analysis of gene expression in Etrog citron leaves infected by Citrus viroid III. Biochim. Biophys. Acta. 2007, 1769, 228–235. [Google Scholar] [CrossRef]
- Herranz, M.C.; Niehl, A.; Rosales, M.; Fiore, N.; Zamorano, A.; Granell, A.; Pallas, V. A remarkable synergistic effect at the transcriptomic level in peach fruits doubly infected by prunus necrotic ringspot virus and peach latent mosaic viroid. Virol. J. 2013, 10, 164. [Google Scholar] [CrossRef]
- Kappagantu, M.; Bullock, J.M.; Nelson, M.E.; Eastwell, K.C. Hop stunt viroid: Effect on Host (Humulus lupulus) Transcriptome and Its Interactions With Hop Powdery Mildew (Podospheara macularis). Mol. Plant Microbe Interact. 2017, 30, 842–851. [Google Scholar] [CrossRef]
- Xia, C.; Li, S.; Hou, W.; Fan, Z.; Xiao, H.; Lu, M.; Sano, T.; Zhang, Z. Global Transcriptomic Changes Induced by Infection of Cucumber (Cucumis sativum L.) with mild and severe variants of hop stunt viroid. Front. Microbiol. 2017, 8, 2427. [Google Scholar] [CrossRef]
- Takino, H.; Kitajima, S.; Hirano, S.; Oka, M.; Matsuura, T.; Ikeda, Y.; Kojima, M.; Takebayashi, Y.; Sakakibara, H.; Mino, M. Global transcriptome analyses reveal that infection with chrysanthemum stunt viroid (CSVd) affects gene expression profile of chrysanthemum plants, but the genes involved in plant hormone metabolism and signaling may not be silencing target of CSVd-siRNAs. Plant Gene 2019, 18, 100181. [Google Scholar] [CrossRef]
- Pokorn, T.; Radišek, S.; Javornik, B.; Štajner, N.; Jakše, J. Development of hop transcriptome to support research into host-viroid interactions. PLoS One 2017, 12, e0184528. [Google Scholar] [CrossRef] [PubMed]
- Štajner, N.; Radišek, S.; Mishra, A.K.; Nath, V.S.; Matoušek, J.; Jakše, J. Evaluation of Disease Severity and Global Transcriptome Response Induced by Citrus bark cracking viroid, Hop latent viroid, and Their Co-Infection in Hop (Humulus lupulus L.). Int. J. Mol. Sci. 2019, 20, 3154. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.K.; Kumar, A.; Mishra, D.; Nath, V.S.; Jakše, J.; Kocábek, T.; Killi, U.K.; Morina, F.; Matoušek, J. Genome-Wide Transcriptomic Analysis Reveals Insights into the Response to Citrus bark cracking viroid (CBCVd) in Hop (Humulus lupulus L.). Viruses 2018, 10, 570. [Google Scholar] [CrossRef] [PubMed]
- Katsarou, K.; Wu, Y.; Zhang, R.; Bonar, N.; Morris, J.; Hedley, P.E.; Bryan, G.J.; Kalantidis, K.; Hornyik, C. Insight on Genes Affecting Tuber Development in Potato upon Potato spindle tuber viroid (PSTVd) Infection. PLoS One 2016, 11, e0150711. [Google Scholar] [CrossRef] [PubMed]
- Calil, I.P.; Fontes, E.P.B. Plant immunity against viruses: antiviral immune receptors in focus. Ann. Bot 2017, 119, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Withers, J.; Dong, X. Post-translational regulation of plant immunity. Curr. Opin. Plant Biol. 2017, 38, 124–132. [Google Scholar] [CrossRef]
- Muthamilarasan, M.; Prasad, M. Plant innate immunity: an updated insight into defense mechanism. J. Biosci. 2013, 38, 433–449. [Google Scholar] [CrossRef]
- Millet, Y.A.; Danna, C.H.; Clay, N.K.; Songnuan, W.; Simon, M.D.; Werck-Reichhart, D.; Ausubel, F.M. Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell 2010, 22, 973–990. [Google Scholar] [CrossRef]
- Teixeira, M.A.; Wei, L.; Kaloshian, I. Root-knot nematodes induce pattern-triggered immunity in Arabidopsis thaliana roots. New Phytol. 2016, 211, 276–287. [Google Scholar] [CrossRef]
- French, E.; Kim, B.S.; Rivera-Zuluaga, K.; Iyer-Pascuzzi, A.S. Whole Root Transcriptomic Analysis Suggests a Role for Auxin Pathways in Resistance to Ralstonia solanacearum in Tomato. Mol. Plant Microbe Interact. 2018, 31, 432–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- dos Santos, W.D.; Ferrarese, M.e.L.; Finger, A.; Teixeira, A.C.; Ferrarese-Filho, O. Lignification and related enzymes in Glycine max root growth-inhibition by ferulic acid. J. Chem. Ecol. 2004, 30, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Moreira-Vilar, F.C.; Siqueira-Soares, R.e.C.; Finger-Teixeira, A.; de Oliveira, D.M.; Ferro, A.P.; da Rocha, G.J.; Ferrarese, M.e.L.; dos Santos, W.D.; Ferrarese-Filho, O. The acetyl bromide method is faster, simpler and presents best recovery of lignin in different herbaceous tissues than Klason and thioglycolic acid methods. PLoS One 2014, 9, e110000. [Google Scholar] [CrossRef] [PubMed]
- Parizotto, A.V.; Bubna, G.A.; Marchiosi, R.; Soares, A.R.; Ferrarese, M.e.L.; Ferrarese-Filho, O. Benzoxazolin-2(3H)-one inhibits soybean growth and alters the monomeric composition of lignin. Plant Signal. Behav. 2015, 10, e989059. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef] [Green Version]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [Green Version]
- Usadel, B.; Poree, F.; Nagel, A.; Lohse, M.; Czedik-Eysenberg, A.; Stitt, M. A guide to using MapMan to visualize and compare Omics data in plants: a case study in the crop species, Maize. Plant Cell Environ. 2009, 32, 1211–1229. [Google Scholar] [CrossRef]
- Zheng, Y.; Jiao, C.; Sun, H.; Rosli, H.G.; Pombo, M.A.; Zhang, P.; Banf, M.; Dai, X.; Martin, G.B.; Giovannoni, J.J.; et al. iTAK: A Program for Genome-wide Prediction and Classification of Plant Transcription Factors, Transcriptional Regulators, and Protein Kinases. Mol. Plant 2016, 9, 1667–1670. [Google Scholar] [CrossRef] [Green Version]
- Lehti-Shiu, M.D.; Shiu, S.H. Diversity, classification and function of the plant protein kinase superfamily. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2012, 367, 2619–2639. [Google Scholar] [CrossRef] [Green Version]
- Oliveros, J.C.; Blaschke, C.; Herrero, J.; Dopazo, J.; Valencia, A. Expression profiles and biological function. Genome Inform. Ser. Workshop Genome Inform. 2000, 11, 106–117. [Google Scholar] [PubMed]
- Geiss, G.K.; Bumgarner, R.E.; Birditt, B.; Dahl, T.; Dowidar, N.; Dunaway, D.L.; Fell, H.P.; Ferree, S.; George, R.D.; Grogan, T.; et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 2008, 26, 317–325. [Google Scholar] [PubMed]
- Malkov, V.A.; Serikawa, K.A.; Balantac, N.; Watters, J.; Geiss, G.; Mashadi-Hossein, A.; Fare, T. Multiplexed measurements of gene signatures in different analytes using the Nanostring nCounter Assay System. BMC Res. Notes 2009, 2, 80. [Google Scholar]
- Overvoorde, P.; Fukaki, H.; Beeckman, T. Auxin control of root development. Cold Spring Harb Perspect Biol. 2010, 2, a001537. [Google Scholar] [PubMed]
- Ferreira, F.J.; Kieber, J.J. Cytokinin signaling. Curr. Opin. Plant Biol. 2005, 8, 518–525. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Chiang, Y.H.; Kieber, J.J.; Schaller, G.E. SCF(KMD) controls cytokinin signaling by regulating the degradation of type-B response regulators. Proc. Natl. Acad. Sci. USA 2013, 110, 10028–10033. [Google Scholar] [CrossRef]
- Zhang, Y.; Lubberstedt, T.; Xu, M. The genetic and molecular basis of plant resistance to pathogens. J. Genet. Genomics 2013, 40, 23–35. [Google Scholar] [CrossRef]
- Gapper, N.E.; Giovannoni, J.J.; Watkins, C.B. Understanding development and ripening of fruit crops in an ‘omics’ era. Hortic. Res. 2014, 1, 14034. [Google Scholar]
- Mickelbart, M.V.; Hasegawa, P.M.; Bailey-Serres, J. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat. Rev. Genet. 2015, 16, 237–251. [Google Scholar]
- Xu, J.; Zhang, S. Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci. 2015, 20, 56–64. [Google Scholar]
- Xie, M.; Zhang, J.; Tschaplinski, T.J.; Tuskan, G.A.; Chen, J.G.; Muchero, W. Regulation of Lignin Biosynthesis and Its Role in Growth-Defense Tradeoffs. Front. Plant Sci. 2018, 9, 1427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tronchet, M.; Balagué, C.; Kroj, T.; Jouanin, L.; Roby, D. Cinnamyl alcohol dehydrogenases-C and D, key enzymes in lignin biosynthesis, play an essential role in disease resistance in Arabidopsis. Mol. Plant Pathol. 2010, 11, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Dreher, K.; Callis, J. Ubiquitin, hormones and biotic stress in plants. Ann. Bot 2007, 99, 787–822. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.; Joshi, D.; Yadav, P.K.; Gupta, A.K.; Bhatt, T.K. Role of Ubiquitin-Mediated Degradation System in Plant Biology. Front. Plant Sci. 2016, 7, 806. [Google Scholar] [CrossRef] [PubMed]
- Toepel, J.; Illmer-Kephalides, M.; Jaenicke, S.; Straube, J.; May, P.; Goesmann, A.; Kruse, O. New insights into Chlamydomonas reinhardtii hydrogen production processes by combined microarray/RNA-seq transcriptomics. Plant Biotechnol. J. 2013, 11, 717–733. [Google Scholar] [CrossRef] [PubMed]
- Mukaka, M.M. Statistics corner: A guide to appropriate use of correlation coefficient in medical research. Malawi. Med. J. 2012, 24, 69–71. [Google Scholar]
- Kogenaru, S.; Qing, Y.; Guo, Y.; Wang, N. RNA-seq and microarray complement each other in transcriptome profiling. BMC Genomics 2012, 13, 629. [Google Scholar] [CrossRef]
- Peng, J.; Hao, B.; Liu, L.; Wang, S.; Ma, B.; Yang, Y.; Xie, F.; Li, Y. RNA-Seq and microarrays analyses reveal global differential transcriptomes of Mesorhizobium huakuii 7653R between bacteroids and free-living cells. PLoS One 2014, 9, e93626. [Google Scholar] [CrossRef]
- Krishnamurthy, A.; Ferl, R.J.; Paul, A.L. Comparing RNA-Seq and microarray gene expression data in two zones of the. Appl. Plant Sci. 2018, 6, e01197. [Google Scholar] [CrossRef]
- Wang, Y.; Qu, J.; Ji, S.; Wallace, A.J.; Wu, J.; Li, Y.; Gopalan, V.; Ding, B. A Land Plant-Specific Transcription Factor Directly Enhances Transcription of a Pathogenic Noncoding RNA Template by DNA-Dependent RNA Polymerase II. Plant Cell 2016, 28, 1094–1107. [Google Scholar] [CrossRef]
- Catoni, M.; Miozzi, L.; Fiorilli, V.; Lanfranco, L.; Accotto, G.P. Comparative analysis of expression profiles in shoots and roots of tomato systemically infected by Tomato spotted wilt virus reveals organ-specific transcriptional responses. Mol. Plant Microbe Interact. 2009, 22, 1504–1513. [Google Scholar] [CrossRef] [PubMed]
- Milanović, J.; Oklestkova, J.; Majdandžić, A.; Novák, O.; Mihaljević, S. Organ-specific differences in endogenous phytohormone and antioxidative responses in potato upon PSTVd infection. J. Plant Physiol. 2019, 232, 107–114. [Google Scholar] [CrossRef] [PubMed]
- López-Gresa, M.P.; Lisón, P.; Yenush, L.; Conejero, V.; Rodrigo, I.; Bellés, J.M. Salicylic Acid Is Involved in the Basal Resistance of Tomato Plants to Citrus Exocortis Viroid and Tomato Spotted Wilt Virus. PLoS ONE 2016, 11, e0166938. [Google Scholar] [CrossRef] [PubMed]
- Smakowska, E.; Kong, J.; Busch, W.; Belkhadir, Y. Organ-specific regulation of growth-defense tradeoffs by plants. Curr. Opin. Plant Biol. 2016, 29, 129–137. [Google Scholar] [CrossRef]
- Vera, P.; Tornero, P.; Conejero, V. Cloning and expression analysis of a viroid-induced peroxidase from tomato plants. Mol. Plant Microbe Interact. 1993, 6, 790–794. [Google Scholar]
- Wang, M.C.; Lin, J.J.; Duranvila, N.; Semancik, J.S. Alteration in cell-wall composition and structure in viroid-infected cells. Physiol. Mol. Plant Pathol. 1986, 28, 107–124. [Google Scholar] [CrossRef]
- Di Serio, F.; De Stradis, A.; Delgado, S.; Flores, R.; Navarro, B. Cytopathic Effects Incited by Viroid RNAs and Putative Underlying Mechanisms. Front. Plant Sci. 2012, 3, 288. [Google Scholar] [CrossRef]
- Sampedro, J.; Cosgrove, D.J. The expansin superfamily. Genome Biol. 2005, 6, 242. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Plant expansins: diversity and interactions with plant cell walls. Curr. Opin. Plant Biol. 2015, 25, 162–172. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Liu, L.; Wang, X.; Han, Z.; Ouyang, B.; Zhang, J.; Li, H. Genome-wide identification and expression analysis of the expansin gene family in tomato. Mol. Genet. Genomics 2016, 291, 597–608. [Google Scholar] [CrossRef]
- Lee, H.W.; Kim, J. EXPANSINA17 up-regulated by LBD18/ASL20 promotes lateral root formation during the auxin response. Plant Cell Physiol. 2013, 54, 1600–1611. [Google Scholar] [CrossRef]
- Soltys, D.; Rudzińska-Langwald, A.; Gniazdowska, A.; Wiśniewska, A.; Bogatek, R. Inhibition of tomato (Solanum lycopersicum L.) root growth by cyanamide is due to altered cell division, phytohormone balance and expansin gene expression. Planta 2012, 236, 1629–1638. [Google Scholar] [CrossRef] [Green Version]
- Reinhardt, D.; Wittwer, F.; Mandel, T.; Kuhlemeier, C. Localized upregulation of a new expansin gene predicts the site of leaf formation in the tomato meristem. Plant. Cell 1998, 10, 1427–1437. [Google Scholar] [CrossRef]
- Vogler, H.; Caderas, D.; Mandel, T.; Kuhlemeier, C. Domains of expansin gene expression define growth regions in the shoot apex of tomato. Plant Mol. Biol. 2003, 53, 267–272. [Google Scholar] [CrossRef]
- Qi, Y.; Ding, B. Inhibition of cell growth and shoot development by a specific nucleotide sequence in a noncoding viroid RNA. Plant Cell 2003, 15, 1360–1374. [Google Scholar] [CrossRef]
- Tsabary, G.; Shani, Z.; Roiz, L.; Levy, I.; Riov, J.; Shoseyov, O. Abnormal ‘wrinkled’ cell walls and retarded development of transgenic Arabidopsis thaliana plants expressing endo-1,4-beta-glucanase (cell) antisense. Plant Mol. Biol. 2003, 51, 213–224. [Google Scholar] [CrossRef]
- Koka, C.V.; Cerny, R.E.; Gardner, R.G.; Noguchi, T.; Fujioka, S.; Takatsuto, S.; Yoshida, S.; Clouse, S.D. A putative role for the tomato genes DUMPY and CURL-3 in brassinosteroid biosynthesis and response. Plant Physiol. 2000, 122, 85–98. [Google Scholar] [CrossRef]
- Zurek, D.M.; Rayle, D.L.; McMorris, T.C.; Clouse, S.D. Investigation of Gene Expression, Growth Kinetics, and Wall Extensibility during Brassinosteroid-Regulated Stem Elongation. Plant Physiol. 1994, 104, 505–513. [Google Scholar] [CrossRef] [Green Version]
- Gu, C.; Guo, Z.H.; Hao, P.P.; Wang, G.M.; Jin, Z.M.; Zhang, S.L. Multiple regulatory roles of AP2/ERF transcription factor in angiosperm. Bot. Stud. 2017, 58, 6. [Google Scholar] [CrossRef]
- Guo, J.; Wang, M.-H. Expression Profiling of the DREB2 Type Gene from Tomato (Solanum lycopersicum L.) under Various Abiotic Stresses. Hortic. Environ. Biotechnol. 2011, 52, 105–111. [Google Scholar] [CrossRef]
- Nie, J.; Wen, C.; Xi, L.; Lv, S.; Zhao, Q.; Kou, Y.; Ma, N.; Zhao, L.; Zhou, X. The AP2/ERF transcription factor CmERF053 of chrysanthemum positively regulates shoot branching, lateral root, and drought tolerance. Plant Cell Rep. 2018, 37, 1049–1060. [Google Scholar] [CrossRef]
- Miedes, E.; Vanholme, R.; Boerjan, W.; Molina, A. The role of the secondary cell wall in plant resistance to pathogens. Front. Plant Sci. 2014, 5, 358. [Google Scholar] [CrossRef] [Green Version]
- Matoušek, J.; Stehlík, J.; Procházková, J.; Orctová, L.; Wullenweber, J.; Füssy, Z.; Kováčik, J.; Duraisamy, G.S.; Ziegler, A.; Schubert, J.; et al. Biological and molecular analysis of the pathogenic variant C3 of potato spindle tuber viroid (PSTVd) evolved during adaptation to chamomile (Matricaria chamomilla). Biol. Chem. 2012, 393, 605–615. [Google Scholar] [CrossRef]
- Moura, J.C.; Bonine, C.A.; de Oliveira Fernandes Viana, J.; Dornelas, M.C.; Mazzafera, P. Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 2010, 52, 360–376. [Google Scholar] [CrossRef]
- Lauvergeat, V.; Lacomme, C.; Lacombe, E.; Lasserre, E.; Roby, D.; Grima-Pettenati, J. Two cinnamoyl-CoA reductase (CCR) genes from Arabidopsis thaliana are differentially expressed during development and in response to infection with pathogenic bacteria. Phytochemistry 2001, 57, 1187–1195. [Google Scholar] [CrossRef]
- Lima, R.B.; Salvador, V.H.; dos Santos, W.D.; Bubna, G.A.; Finger-Teixeira, A.; Soares, A.R.; Marchiosi, R.; Ferrarese, M.e.L.; Ferrarese-Filho, O. Enhanced lignin monomer production caused by cinnamic Acid and its hydroxylated derivatives inhibits soybean root growth. PLoS ONE 2013, 8, e80542. [Google Scholar] [CrossRef]
- Chabannes, M.; Barakate, A.; Lapierre, C.; Marita, J.M.; Ralph, J.; Pean, M.; Danoun, S.; Halpin, C.; Grima-Pettenati, J.; Boudet, A.M. Strong decrease in lignin content without significant alteration of plant development is induced by simultaneous down-regulation of cinnamoyl CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD) in tobacco plants. Plant J. 2001, 28, 257–270. [Google Scholar] [CrossRef]
- Thévenin, J.; Pollet, B.; Letarnec, B.; Saulnier, L.; Gissot, L.; Maia-Grondard, A.; Lapierre, C.; Jouanin, L. The simultaneous repression of CCR and CAD, two enzymes of the lignin biosynthetic pathway, results in sterility and dwarfism in Arabidopsis thaliana. Mol. Plant 2011, 4, 70–82. [Google Scholar] [CrossRef]
- Deng, X.W.; Gruissem, W. Constitutive transcription and regulation of gene expression in non-photosynthetic plastids of higher plants. EMBO J. 1988, 7, 3301–3308. [Google Scholar] [CrossRef]
- Kang, J.; Yu, H.; Tian, C.; Zhou, W.; Li, C.; Jiao, Y.; Liu, D. Suppression of Photosynthetic Gene Expression in Roots Is Required for Sustained Root Growth under Phosphate Deficiency. Plant Physiol. 2014, 165, 1156–1170. [Google Scholar] [CrossRef] [Green Version]
- Heydarian, Z.; Yu, M.; Gruber, M.; Coutu, C.; Robinson, S.J.; Hegedus, D.D. Changes in gene expression in Camelina sativa roots and vegetative tissues in response to salinity stress. Sci. Rep. 2018, 8, 9804. [Google Scholar] [CrossRef]
- Kobayashi, K.; Ohnishi, A.; Sasaki, D.; Fujii, S.; Iwase, A.; Sugimoto, K.; Masuda, T.; Wada, H. Shoot Removal Induces Chloroplast Development in Roots via Cytokinin Signaling. Plant Physiol. 2017, 173, 2340–2355. [Google Scholar] [CrossRef]
| Pathway | PSTVd-M | PSTVd-S23 | ||||
|---|---|---|---|---|---|---|
| 17 dpi | 24 dpi | 49 dpi | 17 dpi | 24 dpi | 49 dpi | |
| Down-regulated | ||||||
| Biosynthesis of secondary metabolites | 2.50 × 10−4 | 4.06 × 10−2 | 2.03 × 10−2 | 2.25 × 10−4 | 4.66 × 10−4 | |
| Phenylpropanoid biosynthesis | 4.27 × 10−4 | 4.60 × 10−3 | ||||
| Plant hormone signal transduction | 1.77 × 10−2 | 1.37 × 10−3 | ||||
| Biosynthesis of unsaturated fatty acids | 2.20 × 10−2 | |||||
| Lysine degradation | 2.20 × 10−2 | |||||
| Carotenoid biosynthesis | 2.31 × 10−5 | |||||
| Regulation of autophagy | 9.90 × 10−3 | |||||
| Up-regulated | ||||||
| alpha-Linolenic acid metabolism | 1.04 × 10−2 | 7.72 × 10−7 | 9.53 × 10−3 | 1.65 × 10−3 | ||
| Plant hormone signal transduction | 1.04 × 10−2 | 1.19 × 10−5 | 3.08 × 10−2 | 1.22 × 10−2 | ||
| Biosynth of secondary metabolites | 2.52 × 10−4 | 6.62 × 10−7 | 3.23 × 10−2 | 6.40 × 10−5 | ||
| Endocytosis | 2.26 × 10−2 | 1.01 × 10−3 | ||||
| Ribosome | 7.99 × 10−13 | 3.55 × 10−12 | ||||
| Steroid biosynthesis | 1.14 × 10−4 | 8.13 × 10−3 | ||||
| Cysteine and methionine metabolism | 1.60 × 10−4 | 1.24 × 10−4 | ||||
| Plant-pathogen interaction | 2.53 × 10−2 | 4.99 × 10−2 | ||||
| Biosynthesis of amino acids | 4.54 × 10−3 | 1.12 × 10−6 | ||||
| Protein processing in ER | 9.19 × 10−3 | 1.56 × 10−2 | ||||
| Phenylalanine, tyrosine and tryptophan biosynthesis | 3.10 × 10−2 | 9.77 × 10−4 | ||||
| Proteasome | 3.16 × 10−2 | 7.98 × 10−6 | ||||
| Carbon metabolism | 4.43 × 10−5 | |||||
| Carbon fixation in photosynthetic organisms | 1.65 × 10−3 | 4.35 × 10−2 | ||||
| Carotenoid biosynthesis | 2.26 × 10−2 | 3.23 × 10−2 | ||||
| Glyoxylate and dicarboxylate metabolism | 4.86 × 10−2 | |||||
| Spliceosome | 1.24 × 10−4 | |||||
| Peroxisome | 3.88 × 10−3 | |||||
| Oxidative phosphorylation | 8.13 × 10−3 | |||||
| Phagosome | 2.49 × 10−2 | |||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Góra-Sochacka, A.; Więsyk, A.; Fogtman, A.; Lirski, M.; Zagórski-Ostoja, W. Root Transcriptomic Analysis Reveals Global Changes Induced by Systemic Infection of Solanum lycopersicum with Mild and Severe Variants of Potato Spindle Tuber Viroid. Viruses 2019, 11, 992. https://doi.org/10.3390/v11110992
Góra-Sochacka A, Więsyk A, Fogtman A, Lirski M, Zagórski-Ostoja W. Root Transcriptomic Analysis Reveals Global Changes Induced by Systemic Infection of Solanum lycopersicum with Mild and Severe Variants of Potato Spindle Tuber Viroid. Viruses. 2019; 11(11):992. https://doi.org/10.3390/v11110992
Chicago/Turabian StyleGóra-Sochacka, Anna, Aneta Więsyk, Anna Fogtman, Maciej Lirski, and Włodzimierz Zagórski-Ostoja. 2019. "Root Transcriptomic Analysis Reveals Global Changes Induced by Systemic Infection of Solanum lycopersicum with Mild and Severe Variants of Potato Spindle Tuber Viroid" Viruses 11, no. 11: 992. https://doi.org/10.3390/v11110992
APA StyleGóra-Sochacka, A., Więsyk, A., Fogtman, A., Lirski, M., & Zagórski-Ostoja, W. (2019). Root Transcriptomic Analysis Reveals Global Changes Induced by Systemic Infection of Solanum lycopersicum with Mild and Severe Variants of Potato Spindle Tuber Viroid. Viruses, 11(11), 992. https://doi.org/10.3390/v11110992





