Spatiotemporal Dynamics of Hantavirus Cardiopulmonary Syndrome Transmission Risk in Brazil
Abstract
:1. Introduction
2. Materials and Methods
2.1. Hantavirus Disease Cases and Population Data
2.2. Potential Host Richness
2.3. Land Use and Climate
2.4. Spatiotemporal Models
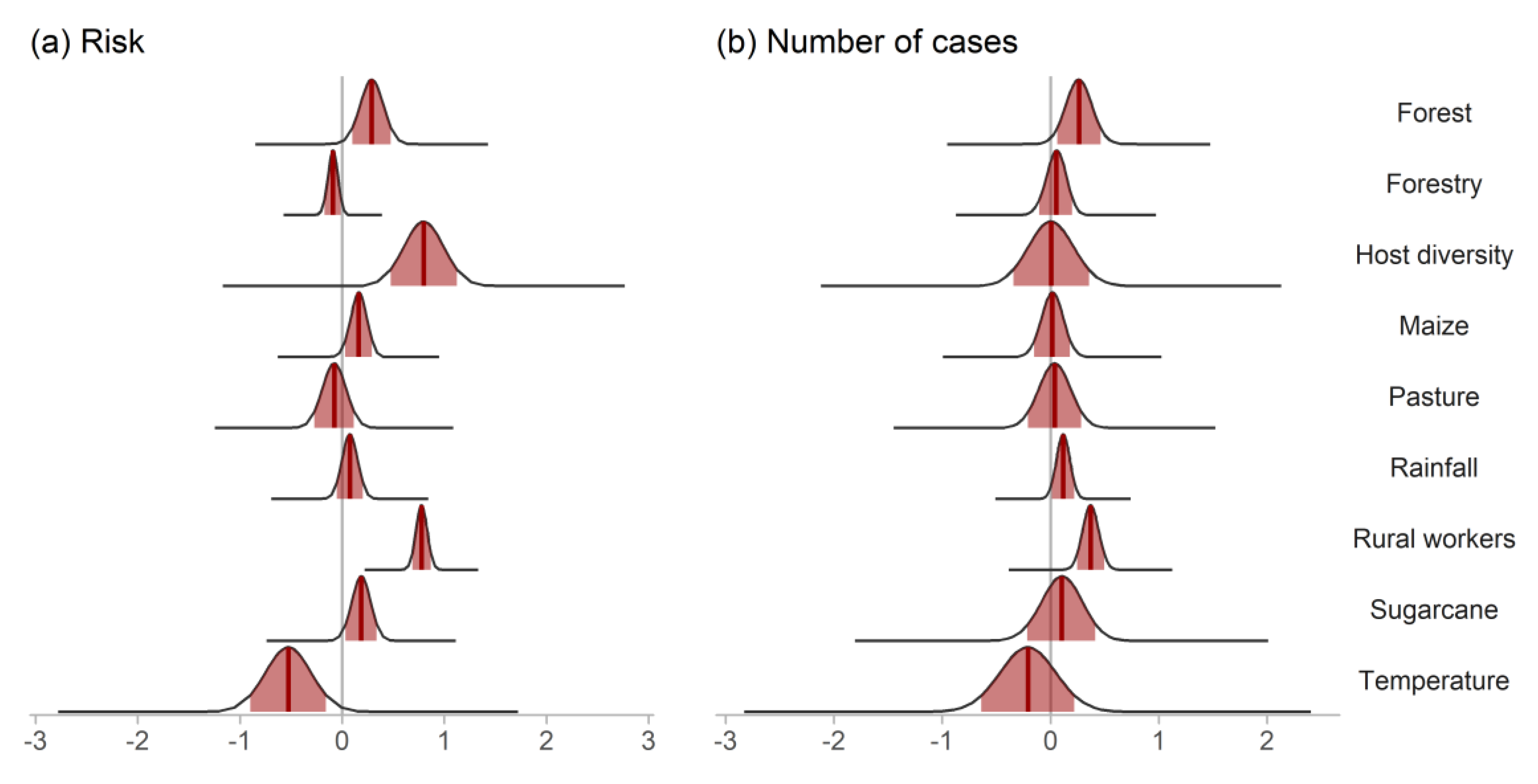
3. Results
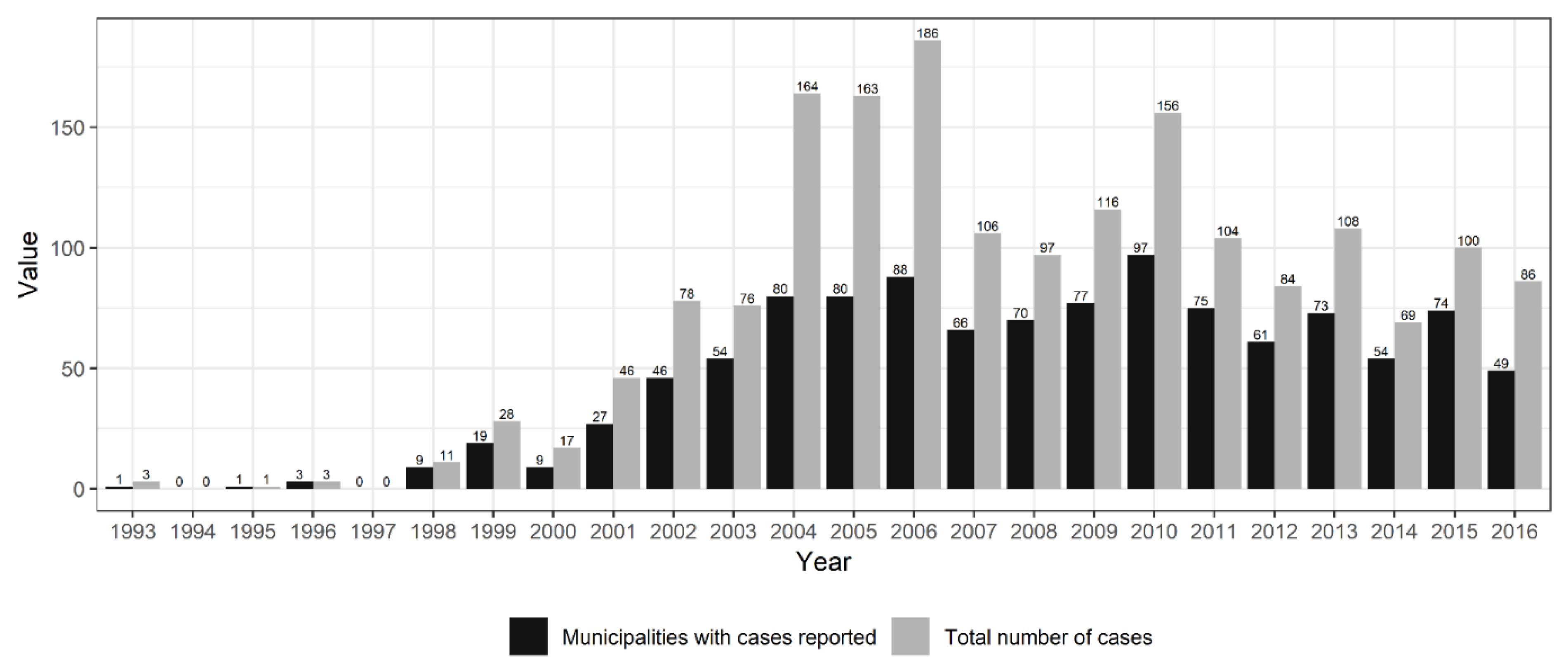
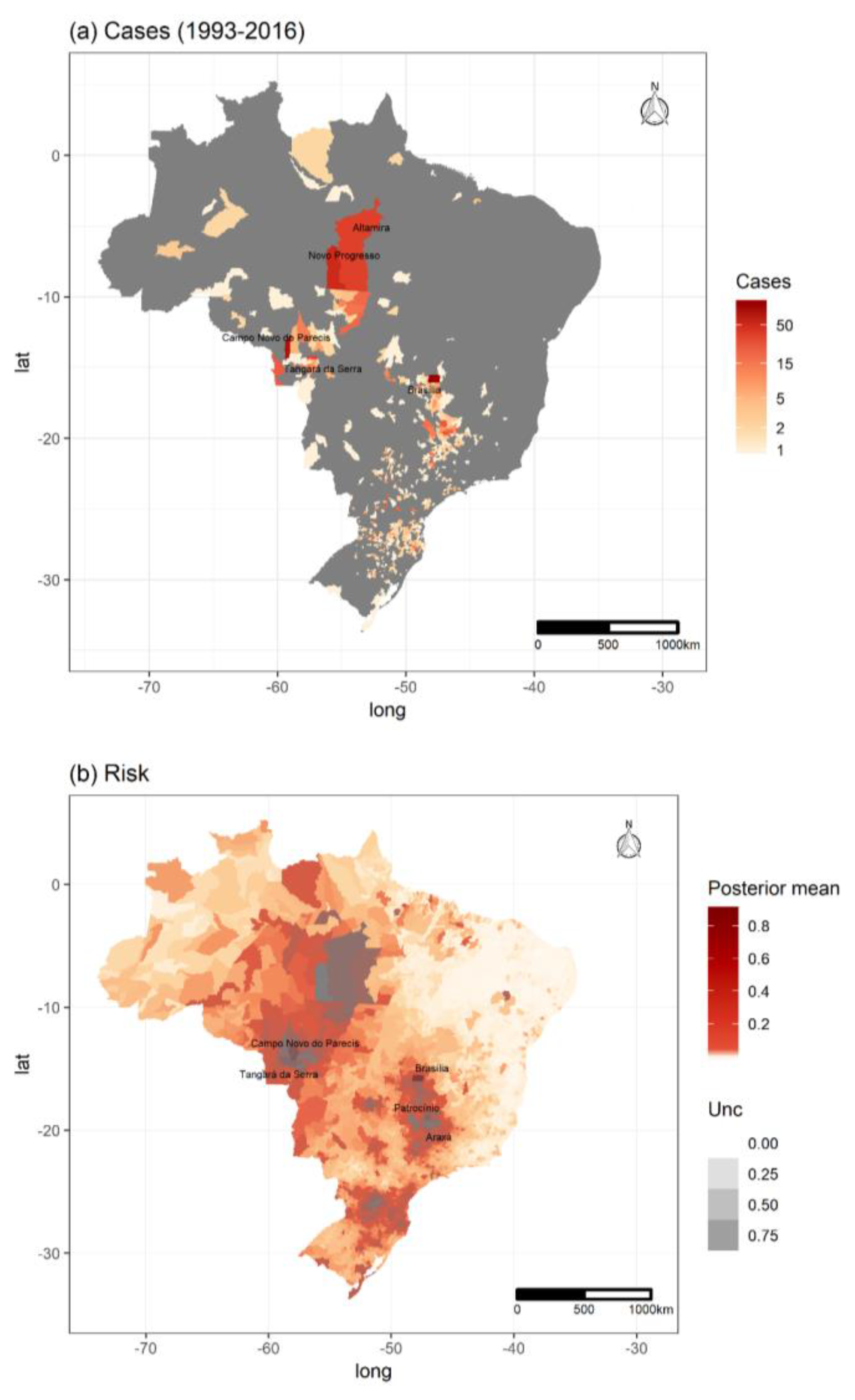
3.1. Hantavirus Disease Cases and Population Data
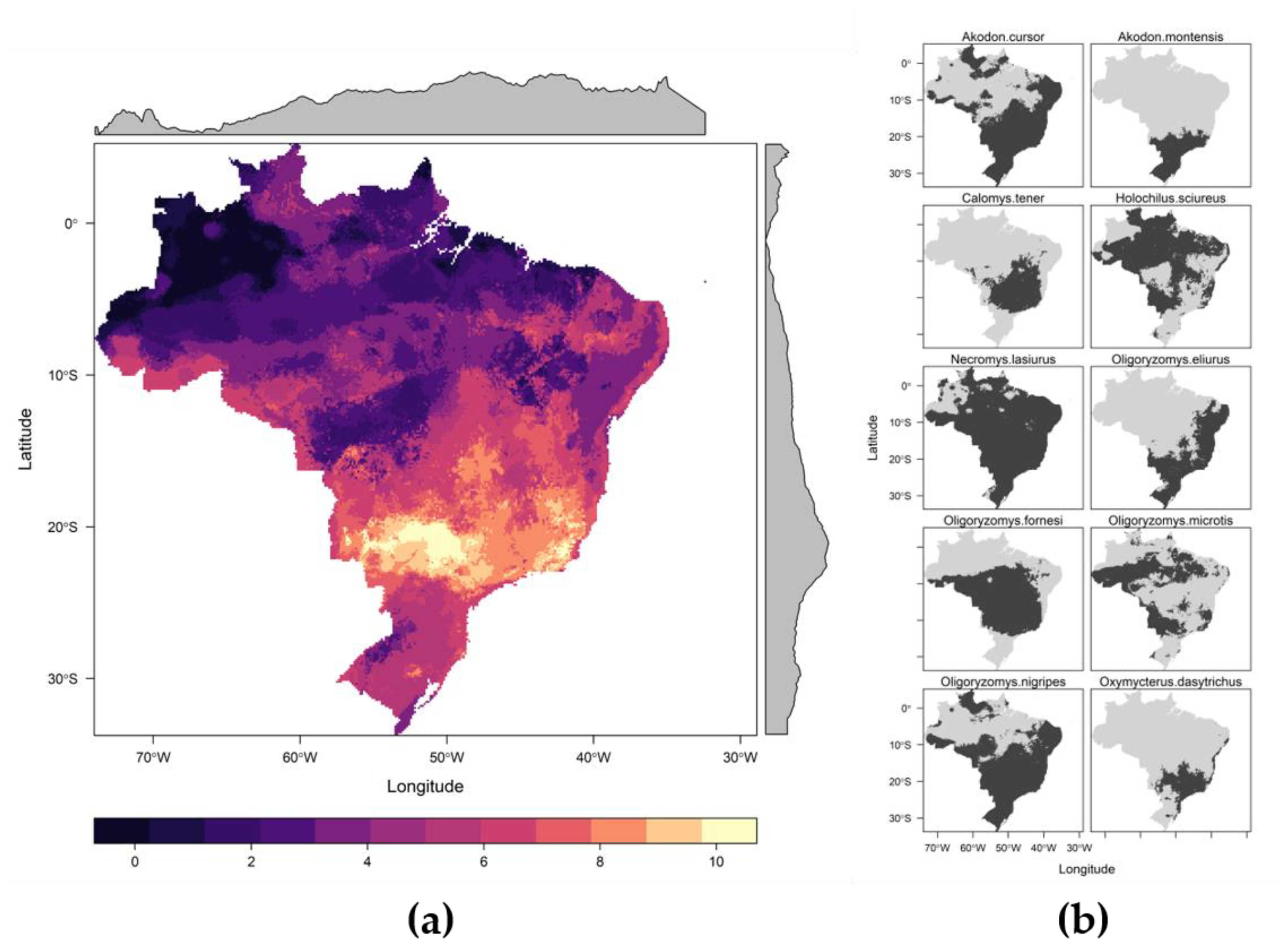
3.2. Potential Host Richness
3.3. Land Use, Population and Climate
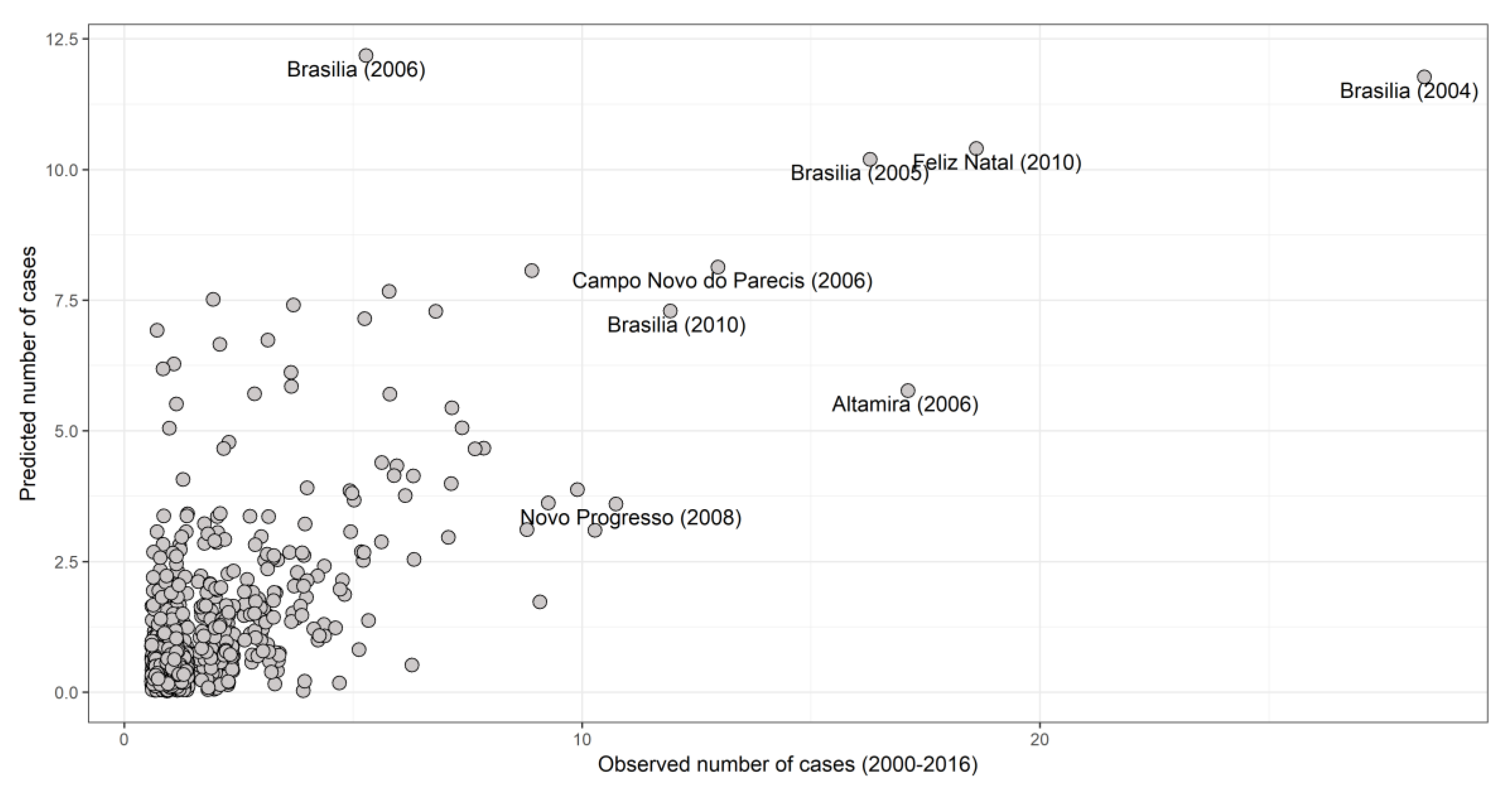
3.4. Spatiotemporal Models
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Allen, T.; Murray, K.A.; Zambrana-Torrelio, C.; Morse, S.S.; Rondinini, C.; Di Marco, M.; Breit, N.; Olival, K.J.; Daszak, P. Global hotspots and correlates of emerging zoonotic diseases. Nat. Commun. 2017, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.P.; Lin, X.D.; Wang, W.; Tian, J.H.; Cong, M.L.; Zhang, H.L.; Wang, M.R.; Zhou, R.H.; Wang, J.B.; Li, M.H.; et al. Phylogeny and Origins of Hantaviruses Harbored by Bats, Insectivores, and Rodents. PLoS Pathog. 2013, 9. [Google Scholar] [CrossRef] [PubMed]
- Maes, P.; Alkhovsky, S.V.; Bào, Y.; Beer, M.; Birkhead, M.; Briese, T.; Buchmeier, M.J.; Calisher, C.H.; Charrel, R.N.; Choi, I.R.; et al. Taxonomy of the family Arenaviridae and the order Bunyavirales: Update 2018. Arch. Virol. 2018, 163, 2295–2310. [Google Scholar] [CrossRef] [PubMed]
- Laenen, L.; Vergote, V.; Calisher, C.H.; Klempa, B.; Klingström, J.; Kuhn, J.H.; Maes, P. Hantaviridae: Current Classification and Future Perspectives. Viruses 2019, 11, 788. [Google Scholar] [CrossRef]
- Saggioro, F.P.; Rossi, M.A.; Duarte, M.I.S.; Martin, C.C.S.; Alves, V.A.F.; Moreli, M.L.; Figueiredo, L.T.M.; Moreira, J.E.; Borges, A.A.; Neder, L. Hantavirus Infection Induces a Typical Myocarditis That May Be Responsible for Myocardial Depression and Shock in Hantavirus Pulmonary Syndrome. J. Infect. Dis. 2007, 195, 1541–1549. [Google Scholar] [CrossRef]
- De Oliveira, S.V.; Fonseca, L.X.; de Araújo Vilges, K.M.; Maniglia, F.V.P.; Pereira, S.V.C.; de Caldas, E.P.; Tauil, P.L.; Gurgel-Gonçalves, R. Vulnerability of Brazilian municipalities to hantavirus infections based on multi-criteria decision analysis. Emerg. Themes Epidemiol. 2015, 12, 15. [Google Scholar] [CrossRef]
- Ermonval, M.; Baychelier, F.; Tordo, N. What Do We Know about How Hantaviruses Interact with Their Different Hosts? Viruses 2016, 8, 223. [Google Scholar] [CrossRef]
- Schmaljohn, C.; Hjelle, B. Hantaviruses: A Global Disease Problem. Emerg. Infect. Dis. 1997, 3, 95–104. [Google Scholar] [CrossRef]
- Mills, J.N.; Yates, T.L.; Childs, J.E.; Parmenter, R.R.; Ksiazek, T.G.; Rollin, P.E.; Peters, C.J. Guidelines for working with rodents potentially infected with hantavirus. J. Mammal. 1995, 76, 716–722. [Google Scholar] [CrossRef]
- Prist, P.R.; Uriarte, M.; Tambosi, L.R.; Prado, A.; Pardini, R.; D’Andrea, P.S.; Metzger, J.P. Landscape, environmental and social predictors of Hantavirus risk in São Paulo, Brazil. PLoS ONE 2016, 11, e0163459. [Google Scholar] [CrossRef]
- Prist, P.R.; D´Andrea, P.S.; Metzger, J.P. Landscape, Climate and Hantavirus Cardiopulmonary Syndrome Outbreaks. Ecohealth 2017, 14, 614–629. [Google Scholar] [CrossRef] [PubMed]
- Brazilian Ministério da Saúde, Secretaria de Vigilância em Saúde, Departamento de Vigilância Epidemiológica. Manual de Vigilância, Prevenção e Controle das Hantaviroses; Brazilian Ministério da Saúde, Secretaria de Vigilância em Saúde, Departamento de Vigilância Epidemiológica: Brasilia, Brazil, 2014; ISBN 9788533420939. [Google Scholar]
- Dias, L.C.P.; Pimenta, F.M.; Santos, A.B.; Costa, M.H.; Ladle, R.J. Patterns of land use, extensification, and intensification of Brazilian agriculture. Glob. Chang. Biol. 2016, 22, 2887–2903. [Google Scholar] [CrossRef] [PubMed]
- Rubio, A.V.; Ávila-Flores, R.; Suzán, G. Responses of Small Mammals to Habitat Fragmentation: Epidemiological Considerations for Rodent-Borne Hantaviruses in the Americas. Ecohealth 2014, 11, 526–533. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, D.A.; Marshall, J.C.; French, N.P.; Hayman, D.T.S. Habitat fragmentation, biodiversity loss and the risk of novel infectious disease emergence. J. R. Soc. Interface 2018, 15, 20180403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pardini, R. Effects of forest fragmentation on small mammals in an Atlantic Forest landscape. Biodivers. Conserv. 2004, 13, 2567–2586. [Google Scholar] [CrossRef]
- Milholland, M.T.; Castro-Arellano, I.; Suzan, G.; Garcia-Pena, G.E.; Lee, T.E., Jr.; Rohde, R.E.; Alonso Aguirre, A.; Mills, J.N. Global Diversity and Distribution of Hantaviruses and Their Hosts. Ecohealth 2018, 15, 163–208. [Google Scholar] [CrossRef]
- De Oliveira, S.V.; Escobar, L.E.; Peterson, A.T.; Gurgel-Gonçalves, R. Potential geographic distribution of hantavirus reservoirs in Brazil. PLoS ONE 2013, 8, e85137. [Google Scholar] [CrossRef]
- Prist, P.R.; Muylaert, R.L.; Prado, A.; Umetsu, F.; Ribeiro, M.C.M.C.; Pardini, R.; Metzger, J.P.J.P. Using Different Proxies To Predict Hantavirus Disease Risk in São Paulo State, Brazil. Oecol. Aust. 2017, 21, 42–53. [Google Scholar] [CrossRef]
- Muylaert, R.L.; Bovendorp, R.S.; Sabino-Santos, G., Jr.; Prist, P.R.; Melo, G.L.; de Fátima Priante, C.; Wilkinson, D.A.; Ribeiro, M.C.; Hayman, D.T.S. Hantavirus host assemblages and human disease in the Atlantic Forest. PLoS Negl. Trop. Dis. 2019. (accepted). [Google Scholar] [CrossRef]
- Kruger, D.H.; Figueiredo, L.T.M.; Song, J.W.; Klempa, B. Hantaviruses-Globally emerging pathogens. J. Clin. Virol. 2015, 64, 128–136. [Google Scholar] [CrossRef]
- Zeier, M.; Handermann, M.; Bahr, U.; Rensch, B.; Muller, S.; Kehm, R.; Muranyi, W.; Darai, G. New Ecological Aspects of Hantavirus Infection: A Change of A Paradigm and a Challenge of Prevention—A Review. Virus Genes 2005, 30, 157–180. [Google Scholar] [CrossRef] [PubMed]
- Kallio, E.R.; Klingström, J.; Gustafsson, E.; Manni, T.; Vaheri, A.; Henttonen, H.; Vapalahti, O.; Lundkvist, Å. Prolonged survival of Puumala hantavirus outside the host: Evidence for indirect transmission via the environment. J. Gen. Virol. 2006, 87, 2127–2134. [Google Scholar] [CrossRef] [PubMed]
- Firth, C.; Tokarz, R.; Simith, D.B.; Nunes, M.R.T.; Bhat, M.; Rosa, E.S.T.; Medeiros, D.B.A.; Palacios, G.; Vasconcelos, P.F.C.; Lipkin, W.I. Diversity and Distribution of Hantaviruses in South America. J. Virol. 2012, 86, 13756–13766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, K.A.; Olivero, J.; Roche, B.; Tiedt, S.; Guégan, J.F. Pathogeography: Leveraging the biogeography of human infectious diseases for global health management. Ecography (Cop.) 2018, 1411–1427. [Google Scholar] [CrossRef]
- Mendonça, A.; Percequillo, A.R.; Camargo, N.F.; Ribeiro, J.F.; Palma, A.R.T.; Oliveira, L.C.; Câmara, E.M.V.C.; Vieira, E.M. Cerrado Small Mammals: abundance and distribution of marsupials, lagomorphs, and rodents in a Neotropical savanna. Ecology 2018, 99, 1900. [Google Scholar] [CrossRef] [PubMed]
- Bovendorp, R.S.; Villar, N.; de Abreu-Junior, E.F.; Bello, C.; Regolin, A.L.; Percequillo, A.R.; Galetti, M. Atlantic small-mammal: a dataset of communities of rodents and marsupials of the Atlantic forests of South America. Ecology 2017, 98, 2226. [Google Scholar] [CrossRef] [PubMed]
- Astorga, F.; Escobar, L.E.; Muñoz, D.P.; Dodero, J.E.; Hucks, S.R.; Rybak, M.A.; Duclos, M.; Alvarez, D.R.; Burgos, B.E.M.; Ricaurte, A.P.; et al. Distributional ecology of Andes hantavirus: a macroecological approach. Int. J. Health Geogr. 2018, 17. [Google Scholar] [CrossRef] [PubMed]
- Qiao, H.; Soberón, J.; Peterson, A.T. No silver bullets in correlative ecological niche modelling: Insights from testing among many potential algorithms for niche estimation. Methods Ecol. Evol. 2015, 6, 1126–1136. [Google Scholar] [CrossRef]
- Tax, D.M.J.; Duin, R.P.W. Support Vector Data Description. J. Dyn. Syst. Meas. Control 2004, 54, 091006. [Google Scholar] [CrossRef]
- Booth, T.H.; Nix, H.A.; Busby, J.R.; Hutchinson, M.F. Bioclim: The first species distribution modelling package, its early applications and relevance to most current MaxEnt studies. Divers. Distrib. 2014, 20, 1–9. [Google Scholar] [CrossRef]
- Phillips, S.J.; Anderson, R.P.; Schapire, R.E. Maximum entropy modeling of species geographic distributions. Ecol. Modell. 2006, 190, 231–259. [Google Scholar] [CrossRef] [Green Version]
- Carpenter, G.; Gillison, A.N.; Winter, J. Domain: A flexible modelling procedure for mapping potential distributions of plants and animals. Biodivers. Conserv. 1993, 2, 667–680. [Google Scholar] [CrossRef]
- Allouche, O.; Tsoar, A.; Kadmon, R. Assessing the accuracy of species distribution models: Prevalence, kappa and the true skill statistic (TSS). J. Appl. Ecol. 2006, 43, 1223–1232. [Google Scholar] [CrossRef]
- Funk, C.; Peterson, P.; Landsfeld, M.; Pedreros, D.; Verdin, J.; Shukla, S.; Husak, G.; Rowland, J.; Harrison, L.; Hoell, A.; et al. The climate hazards infrared precipitation with stations—A new environmental record for monitoring extremes. Sci. Data 2015, 2, 1–21. [Google Scholar] [CrossRef]
- Fan, Y.; van den Dool, H. A global monthly land surface air temperature analysis for 1948-present. J. Geophys. Res. Atmos. 2008, 113, 1–18. [Google Scholar] [CrossRef]
- Meyer, B.J.; Schmaljohn, C.S. Persistent hantavirus infections: Characteristics and mechanisms. Trends Microbiol. 2000, 8, 61–67. [Google Scholar] [CrossRef]
- Blangiardo, M.; Cameletti, M. Spatial and Spatiotemporal Bayesian Models with R-INLA; Wiley: Hoboken, NJ, USA, 2015; ISBN 9781118950203. [Google Scholar]
- Zuur, A.F.; Ieno, E.N.; Saveliev, A.A. Beginner’s Guide to Spatial, Temporal, and Spatial-Temporal Ecological Data Analysis with R-INLA; Highland Satistics LTd: Newburgh, NY, USA, 2017; Volume 1, ISBN 0957174195. [Google Scholar]
- Illian, J.B.; Sørbye, S.H.; Rue, H.; Hendrichsen, D. Using INLA To Fit A Complex Point Process Model With Temporally Varying Effects—A Case Study. J. Environ. Stat. 2012, 3, 1–29. [Google Scholar]
- Zambelli, P.; Gebbert, S.; Ciolli, M. Pygrass: An Object Oriented Python Application Programming Interface (API) for Geographic Resources Analysis Support System (GRASS) Geographic Information System (GIS). ISPRS Int. J. Geo-Information 2013, 2, 201–219. [Google Scholar] [CrossRef] [Green Version]
- Neteler, M.; Bowman, M.H.; Landa, M.; Metz, M. GRASS GIS: A multi-purpose open source GIS. Environ. Model. Softw. 2012, 31, 124–130. [Google Scholar] [CrossRef] [Green Version]
- QGIS Development Team. QGIS Geographic Information System. Open Source Geospatial Found. Proj. Available online: https://qgis.org/en/site/ (accessed on 4 November 2015).
- Hijmans, R.J.; Phillips, S.; Leathwick, J.R.; Elith, J. Species Distribution Modeling. Package ‘dismo’. dismo: Species Distribution Modeling. Cran 2017, 1, 55. [Google Scholar]
- Lindgren, F.; Rue, H. Bayesian Spatial Modelling with R-INLA. J. Stat. Softw. 2015, 63, 1–25. [Google Scholar] [CrossRef]
- Rue, H.; Martino, S.; Chopin, N. Approximate Bayesian inference for latent Gaussian models by using integrated nested Laplace approximations. J. R. Stat. Soc. B 2009, 71, 319–392. [Google Scholar] [CrossRef]
- R Core Team R Core Team. R: A language and Environment for Statistical Computing; R Foundation Statistic Computing: Vienna, Austria, 2014. [Google Scholar]
- De Oliveira, R.C.; Guterres, A.; Fernandes, J.; D’Andrea, P.S.; Bonvicino, C.R.; de Lemos, E.R.S. Hantavirus reservoirs: Current status with an emphasis on data from Brazil. Viruses 2014, 6, 1929–1973. [Google Scholar] [CrossRef] [PubMed]
- Goodin, D.G.; Paige, R.; Owen, R.D.; Ghimire, K.; Koch, D.E.; Chu, Y.-K.; Jonsson, C.B. Microhabitat characteristics of Akodon montensis, a reservoir for hantavirus, and hantaviral seroprevalence in an Atlantic forest site in eastern Paraguay. J. Vector Ecol. 2009, 34, 104–113. [Google Scholar] [CrossRef]
- Guterres, A.; de Oliveira, R.C.; Fernandes, J.; Strecht, L.; Casado, F.; de Oliveira, F.C.G.; D’Andrea, P.S.; Bonvicino, C.R.; Schrago, C.G.; de Lemos, E.R.S. Characterization of Juquitiba virus in Oligoryzomys fornesi from Brazilian Cerrado. Viruses 2014, 6, 1473–1482. [Google Scholar] [CrossRef]
- González-Ittig, R.E.; Rivera, P.C.; Levis, S.C.; Calderón, G.E.; Gardenal, C.N. The molecular phylogenetics of the genus Oligoryzomys (Rodentia: Cricetidae) clarifies rodent host-hantavirus associations. Zool. J. Linn. Soc. 2014, 171, 457–474. [Google Scholar] [CrossRef]
- Sabino-Santos, G., Jr. Detecção de Hantavírus em roedores silvestres e estudo de sua dinâmica populacional na região Nordeste do Estado de São Paulo; Universidade de São Paulo: Ribeirão Preto, Brazil, 2010. [Google Scholar]
- Gheler-Costa, C.; Sabino-Santos, G.; Amorim, L.S.; Rosalino, L.M.; Figueiredo, L.T.M.; Verdade, L.M. The effect of pre-harvest fire on the small mammal assemblage in sugarcane fields. Agric. Ecosyst. Environ. 2013, 171, 85–89. [Google Scholar] [CrossRef]
- Salkeld, D.J.; Padgett, K.A.; Jones, J.H. A meta-analysis suggesting that the relationship between biodiversity and risk of zoonotic pathogen transmission is idiosyncratic. Ecol. Lett. 2013, 16, 679–686. [Google Scholar] [CrossRef]
- Luis, A.D.; O’Shea, T.J.; Hayman, D.T.S.; Wood, J.L.N.; Cunningham, A.A.; Gilbert, A.T.; Mills, J.N.; Webb, C.T. Network analysis of host—Virus communities in bats and rodents reveals determinants of cross-species transmission. Ecol. Lett. 2015, 18, 1153–1162. [Google Scholar] [CrossRef]
- Badra, S.J.; Maia, F.G.M.; Figueiredo, G.G.; dos Santos, S., Jr.; Campos, G.M.; Figueiredo, L.T.M.; Passos, A.D.C. A retrospective serologic survey of hantavirus infections in the county of Cássia dos Coqueiros, State of São Paulo, Brazil. Rev. Soc. Bras. Med. Trop. 2012, 45, 468–470. [Google Scholar] [CrossRef]
- Capaz, R.S.; Carvalho, V.S.B.; Nogueira, L.A.H. Impact of mechanization and previous burning reduction on GHG emissions of sugarcane harvesting operations in Brazil. Appl. Energy 2013, 102, 220–228. [Google Scholar] [CrossRef]
- Leite, M.R.; Zanetta, D.M.T.; Trevisan, I.B.; de Almeida Burdmann, E.; de Santos, U.P. O trabalho no corte de cana-de-açúcar, riscos e efeitos na saúde: revisão da literatura. Rev. Saúde Públ. 2018, 52, 80. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muylaert, R.L.; Sabino-Santos, G.; Prist, P.R.; Oshima, J.E.F.; Niebuhr, B.B.; Sobral-Souza, T.; Oliveira, S.V.d.; Bovendorp, R.S.; Marshall, J.C.; Hayman, D.T.S.; et al. Spatiotemporal Dynamics of Hantavirus Cardiopulmonary Syndrome Transmission Risk in Brazil. Viruses 2019, 11, 1008. https://doi.org/10.3390/v11111008
Muylaert RL, Sabino-Santos G, Prist PR, Oshima JEF, Niebuhr BB, Sobral-Souza T, Oliveira SVd, Bovendorp RS, Marshall JC, Hayman DTS, et al. Spatiotemporal Dynamics of Hantavirus Cardiopulmonary Syndrome Transmission Risk in Brazil. Viruses. 2019; 11(11):1008. https://doi.org/10.3390/v11111008
Chicago/Turabian StyleMuylaert, Renata L., Gilberto Sabino-Santos, Paula R. Prist, Júlia E. F. Oshima, Bernardo Brandão Niebuhr, Thadeu Sobral-Souza, Stefan Vilges de Oliveira, Ricardo Siqueira Bovendorp, Jonathan C. Marshall, David T. S. Hayman, and et al. 2019. "Spatiotemporal Dynamics of Hantavirus Cardiopulmonary Syndrome Transmission Risk in Brazil" Viruses 11, no. 11: 1008. https://doi.org/10.3390/v11111008
APA StyleMuylaert, R. L., Sabino-Santos, G., Prist, P. R., Oshima, J. E. F., Niebuhr, B. B., Sobral-Souza, T., Oliveira, S. V. d., Bovendorp, R. S., Marshall, J. C., Hayman, D. T. S., & Ribeiro, M. C. (2019). Spatiotemporal Dynamics of Hantavirus Cardiopulmonary Syndrome Transmission Risk in Brazil. Viruses, 11(11), 1008. https://doi.org/10.3390/v11111008






