Molecular Piracy: Redirection of Bacteriophage Capsid Assembly by Mobile Genetic Elements
Abstract
1. Introduction
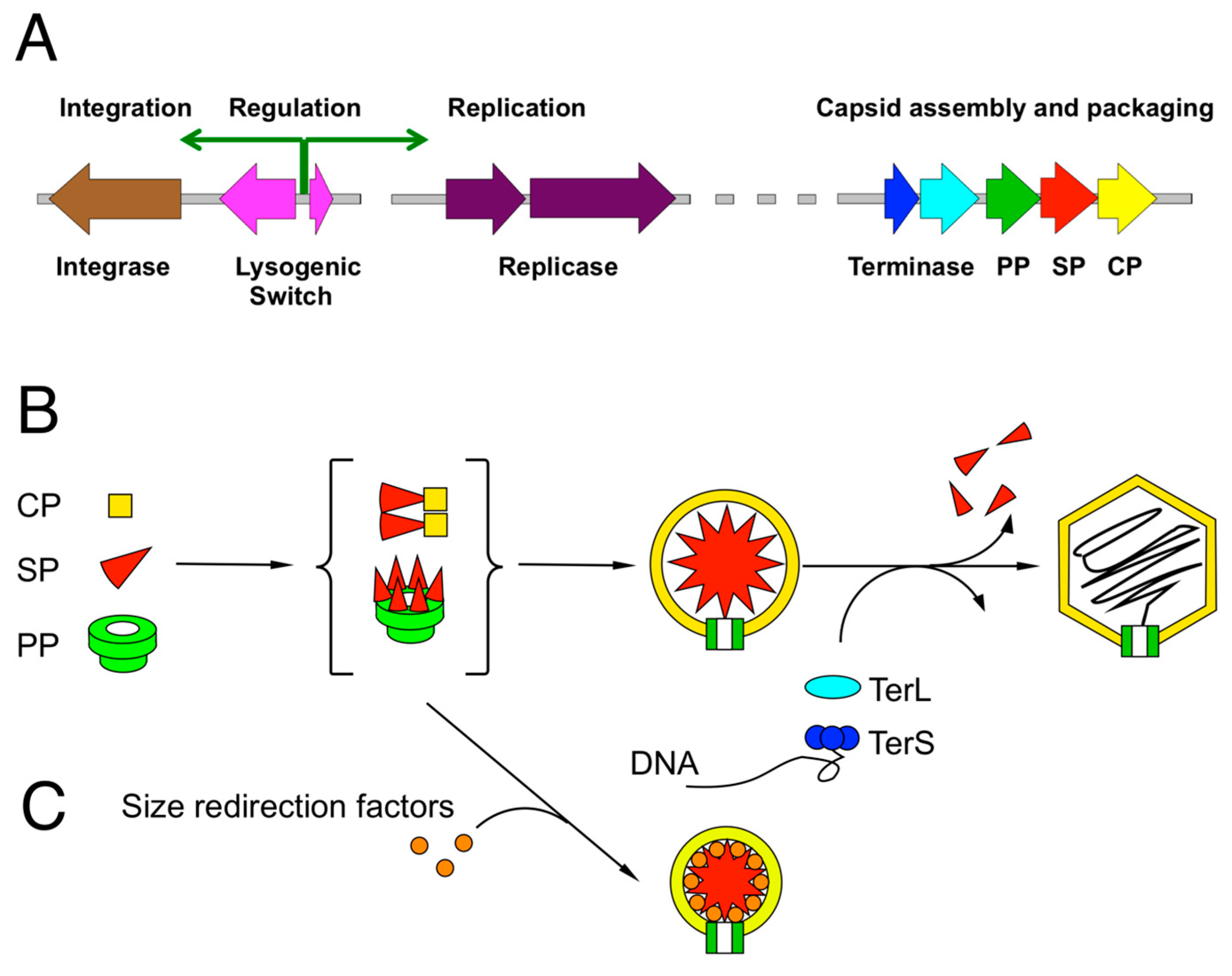
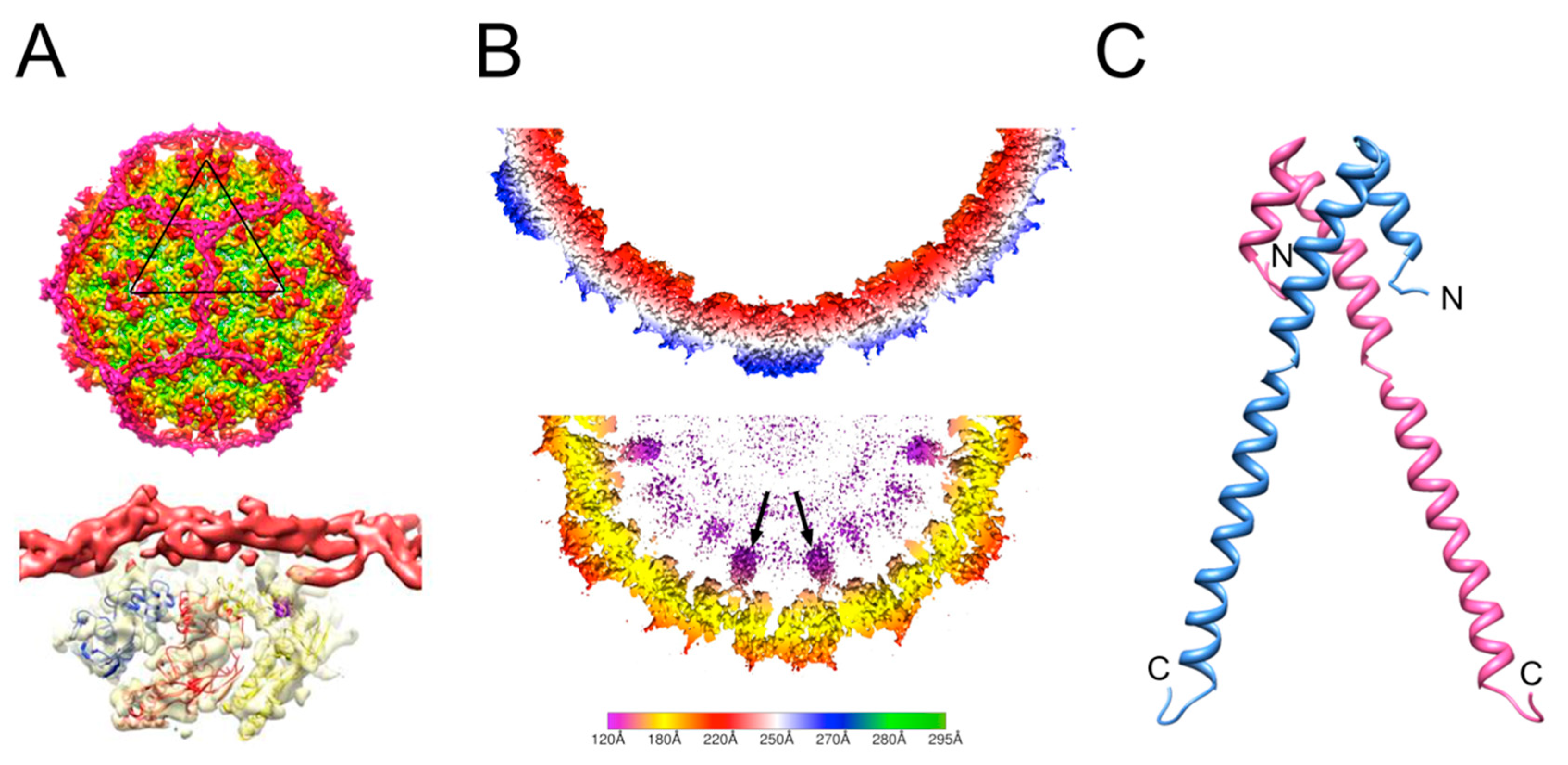
2. Review of Capsid Assembly and DNA Packaging in the Caudovirales
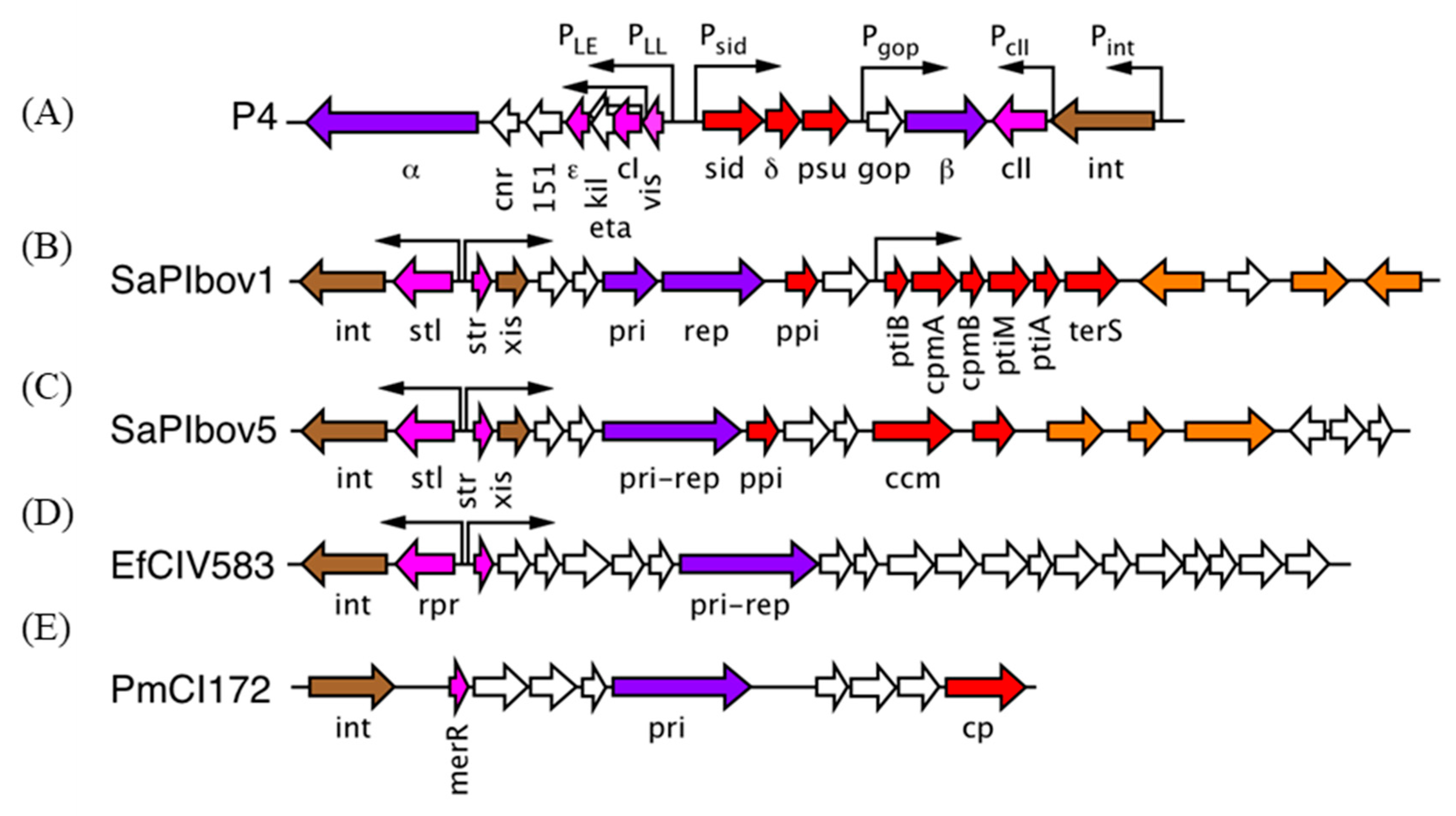
3. The P2/P4 Paradigm
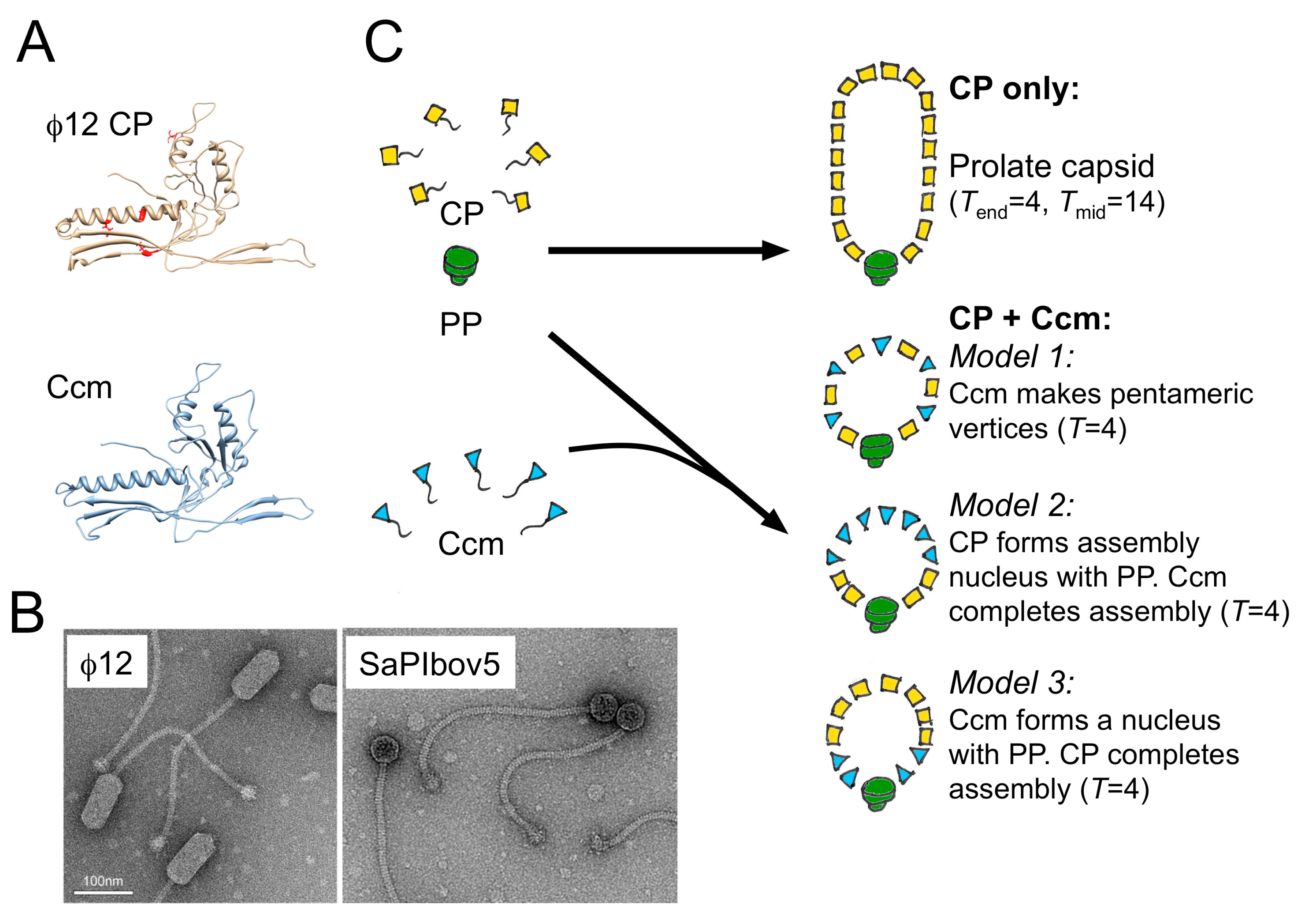
4. The Staphylococcus aureus Pathogenicity Islands (SaPIs)
5. A Different Type of SaPI
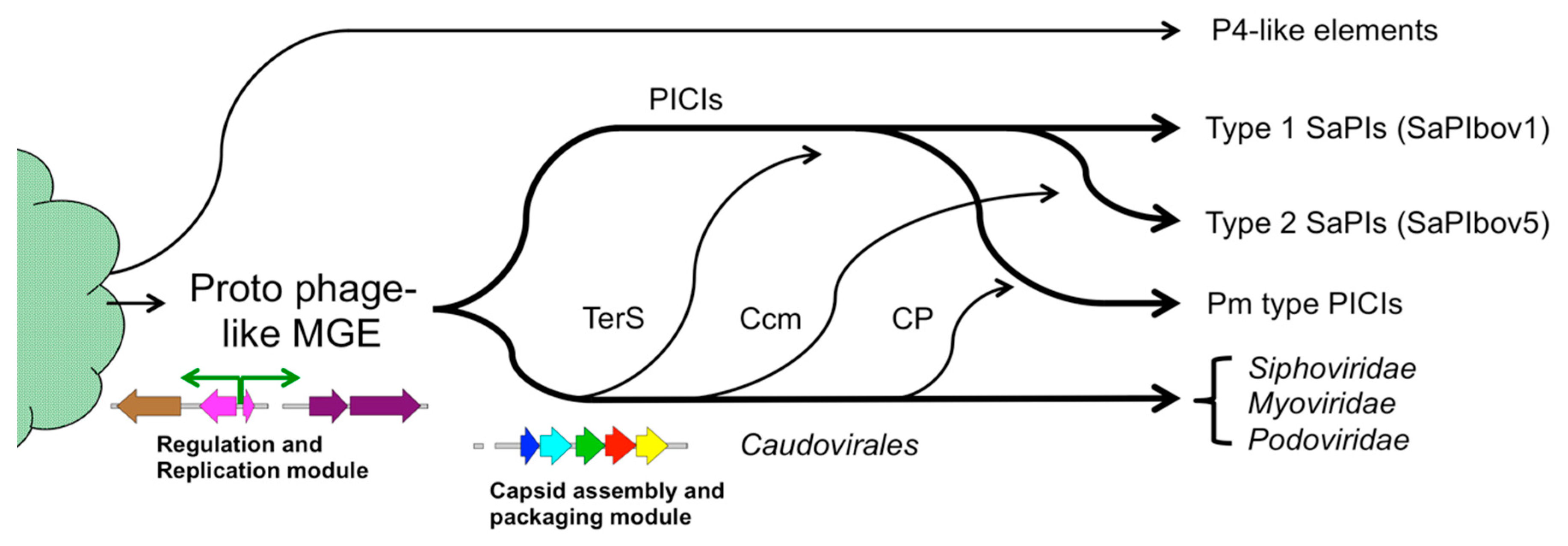
6. The Families of Phage-Induced Chromosomal Islands (PICIs)
7. Evolution of Molecular Piracy
Funding
Acknowledgments
Conflicts of Interest
References
- Bobay, L.M.; Ochman, H. The Evolution of Bacterial Genome Architecture. Front. Genet. 2017, 8, 72. [Google Scholar] [CrossRef] [PubMed]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, J.A. Staphylococcus aureus genomics and the impact of horizontal gene transfer. Int. J. Med. Microbiol. 2014, 304, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Xia, G.; Wolz, C. Phages of Staphylococcus aureus and their impact on host evolution. Infect. Genet. Evol. 2014, 21, 593–601. [Google Scholar] [CrossRef]
- Tolstoy, I.; Kropinski, A.M.; Brister, J.R. Bacteriophage Taxonomy: An Evolving Discipline. Methods Mol. Biol. 2018, 1693, 57–71. [Google Scholar]
- Hatfull, G.F.; Hendrix, R.W. Bacteriophages and their genomes. Curr. Opin. Virol. 2011, 1, 298–303. [Google Scholar] [CrossRef]
- Chiang, Y.N.; Penades, J.R.; Chen, J. Genetic transduction by phages and chromosomal islands: The new and noncanonical. PLoS Pathog. 2019, 15, e1007878. [Google Scholar] [CrossRef]
- Bobay, L.M.; Touchon, M.; Rocha, E.P. Pervasive domestication of defective prophages by bacteria. Proc. Natl. Acad. Sci. USA 2014, 111, 12127–12132. [Google Scholar] [CrossRef]
- Christie, G.E.; Dokland, T. Pirates of the Caudovirales. Virology 2012, 434, 210–221. [Google Scholar] [CrossRef]
- Rao, V.B.; Feiss, M. Mechanisms of DNA Packaging by Large Double-Stranded DNA Viruses. Annu. Rev. Virol. 2015, 2, 351–378. [Google Scholar] [CrossRef]
- Wikoff, W.R.; Liljas, L.; Duda, R.L.; Tsuruta, H.; Hendrix, R.W.; Johnson, J.E. Topologically linked protein rings in the bacteriophage HK97 capsid. Science 2000, 289, 2129–2133. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Sun, L.; Zhang, Z.; Fokine, A.; Padilla-Sanchez, V.; Hanein, D.; Jiang, W.; Rossmann, M.G.; Rao, V.B. Cryo-EM structure of the bacteriophage T4 isometric head at 3.3-Å resolution and its relevance to the assembly of icosahedral viruses. Proc. Natl. Acad. Sci. USA 2017, 114, E8184–E8193. [Google Scholar] [CrossRef]
- Chen, D.H.; Baker, M.L.; Hryc, C.F.; Dimaio, F.; Jakana, J.; Wu, W.; Dougherty, M.; Haase-Pettingell, C.; Schmid, M.F.; Jiang, W.; et al. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc. Natl. Acad. Sci. USA 2011, 108, 1355–1360. [Google Scholar] [CrossRef] [PubMed]
- Hua, J.; Huet, A.; Lopez, C.A.; Toropova, K.; Pope, W.H.; Duda, R.L.; Hendrix, R.W.; Conway, J.F. Capsids and Genomes of Jumbo-Sized Bacteriophages Reveal the Evolutionary Reach of the HK97 Fold. MBio 2017, 8, e015791-7. [Google Scholar] [CrossRef] [PubMed]
- Duda, R.L.; Teschke, C.M. The amazing HK97 fold: Versatile results of modest differences. Curr. Opin. Virol. 2019, 36, 9–16. [Google Scholar] [CrossRef]
- Tao, Y.; Olson, N.H.; Xu, W.; Anderson, D.L.; Rossmann, M.G.; Baker, T.S. Assembly of a tailed bacterial virus and its genome release studied in three dimensions. Cell 1998, 95, 431–437. [Google Scholar] [CrossRef]
- Guo, P.; Erickson, S.; Xu, W.; Olson, N.; Baker, T.S.; Anderson, D. Regulation of phage phi29 prohead shape and size by the portal vertex. Virology 1991, 183, 366–373. [Google Scholar] [CrossRef]
- Katsura, I. Structure and inherent properties of the bacteriophage lambda head shell. IV. Small-head mutants. J. Mol. Biol. 1983, 171, 297–317. [Google Scholar] [CrossRef]
- Doermann, A.H.; Eiserling, F.A.; Boehner, L. Genetic control. of capsid length in bacteriophage T4. I. Isolation and preliminary description of four new mutants. J. Virol. 1973, 12, 374–385. [Google Scholar]
- Dokland, T. Freedom and restraint: Themes in virus capsid assembly. Structure 2000, 8, R157–R167. [Google Scholar] [CrossRef]
- Dokland, T. Scaffolding proteins and their role in viral assembly. Cell. Mol. Life Sci. 1999, 56, 580–603. [Google Scholar] [CrossRef]
- Black, L.W.; Thomas, J.A. Dealing with the whole head: Diversity and function of ejection proteins in tailed phages. In Encyclopedia of Virology, 4th ed.; Academic Press: San Diego, CA, USA, 2019; in press. [Google Scholar]
- Walker, D.H., Jr.; Anderson, T.F. Morphological variants of coliphage P1. J. Virol. 1970, 5, 765–782. [Google Scholar] [PubMed]
- Piya, D.; Vara, L.; Russell, W.K.; Young, R.; Gill, J.J. The multicomponent antirestriction system of phage P1 is linked to capsid morphogenesis. Mol. Microbiol. 2017, 105, 399–412. [Google Scholar] [CrossRef] [PubMed]
- Ang, D.; Keppel, F.; Klein, G.; Richardson, A.; Georgopoulos, C. Genetic analysis of bacteriophage-encoded cochaperonins. Annu. Rev. Genet. 2000, 34, 439–456. [Google Scholar] [CrossRef] [PubMed]
- Kizziah, J.L.; Manning, K.A.; Dearborn, A.D.; Wall, E.A.; Klenow, L.; Hill, R.L.L.; Spilman, M.S.; Stagg, S.M.; Christie, G.E.; Dokland, T. Cleavage and Structural Transitions during Maturation of Staphylococcus aureus Bacteriophage 80α and SaPI1 Capsids. Viruses 2017, 9, 384. [Google Scholar] [CrossRef]
- Wall, E.A.; Caufield, J.H.; Lyons, C.E.; Manning, K.A.; Dokland, T.; Christie, G.E. Specific N-terminal cleavage of ribosomal protein L27 in Staphylococcus aureus and related bacteria. Mol. Microbiol. 2015, 95, 258–269. [Google Scholar] [CrossRef]
- Morais, M.C.; Kanamaru, S.; Badasso, M.O.; Koti, J.S.; Owen, B.A.L.; McMurray, C.T.; Anderson, D.L.; Rossmann, M.G. Bacteriophage phi29 scaffolding protein gp7 before and after prohead assembly. Nat. Struct. Biol. 2003, 10, 572–576. [Google Scholar] [CrossRef]
- Sun, Y.; Parker, M.H.; Weigele, P.; Casjens, S.; Prevelige, P.E.; Krishna, N.R. Structure of the coat protein-binding domain of the scaffolding protein from a double-stranded DNA virus. J. Mol. Biol. 2000, 297, 1195–1202. [Google Scholar] [CrossRef]
- Dearborn, A.D.; Wall, E.A.; Kizziah, J.L.; Klenow, L.; Parker, L.K.; Manning, K.A.; Spilman, M.S.; Spear, J.M.; Christie, G.E.; Dokland, T. Competing scaffolding proteins determine capsid size during mobilization of Staphylococcus aureus pathogenicity islands. eLife 2017, 6, e30822. [Google Scholar] [CrossRef]
- Bertani, G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 1951, 62, 293–300. [Google Scholar]
- Bertani, L.E.; Six, E. The P2-like phages and their parasite, P4. In The Bacteriophages, 2nd ed.; Calendar, R., Ed.; Plenum Press: New York, NY, USA, 1988; pp. 731–743. [Google Scholar]
- Nilsson, A.S.; Karlsson, J.L.; Haggård-Ljungquist, E. Site-specific recombination links the evolution of P2-like coliphages and pathogenic enterobacteria. Mol. Biol. Evol. 2004, 21, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Six, E.W.; Klug, C.A. Bacteriophage P4: A satellite virus depending on a helper such as prophage P2. Virology 1973, 51, 327–344. [Google Scholar] [CrossRef]
- Briani, F.; Dehò, G.; Forti, F.; Ghisotti, D. The plasmid status of satellite bacteriophage P4. Plasmid 2001, 45, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Dehò, G.; Ghisotti, D. The satellite phage P4. In The Bacteriophages; Calendar, R., Ed.; Oxford University Press: New York, NY, USA, 2006; pp. 391–408. [Google Scholar]
- Six, E.W. The helper dependence of satellite bacteriophage P4: Which gene functions of bacteriophage P2 are needed by P4? Virology 1975, 67, 249–263. [Google Scholar] [CrossRef]
- Dokland, T.; Lindqvist, B.H.; Fuller, S.D. Image reconstruction from cryo-electron micrographs reveals the morphopoietic mechanism in the P2-P4 bacteriophage system. EMBO J. 1992, 11, 839–846. [Google Scholar] [CrossRef]
- Dearborn, A.D.; Laurinmäki, P.; Chandramouli, P.; Rodenburg, C.M.; Wang, S.; Butcher, S.J.; Dokland, T. Structure and size determination of bacteriophage P2 and P4 procapsids: Function of size responsiveness mutations. J. Struct Biol. 2012, 178, 215–224. [Google Scholar] [CrossRef]
- Shore, D.; Dehò, G.; Tsipis, J.; Goldstein, R. Determination of capsid size by satellite bacteriophage P4. Proc. Natl. Acad. Sci. USA 1978, 75, 400–404. [Google Scholar] [CrossRef]
- Marvik, O.J.; Sharma, P.; Dokland, T.; Lindqvist, B.H. Bacteriophage P2 and P4 assembly: Alternative scaffolding proteins regulate capsid size. Virology 1994, 200, 702–714. [Google Scholar] [CrossRef]
- Marvik, O.J.; Dokland, T.; Nøkling, R.H.; Jacobsen, E.; Larsen, T.; Lindqvist, B.H. The capsid size-determining protein Sid forms an external scaffold on phage P4 procapsids. J. Mol. Biol. 1995, 251, 59–75. [Google Scholar] [CrossRef]
- Chang, J.R.; Spilman, M.S.; Rodenburg, C.M.; Dokland, T. Functional domains of the bacteriophage P2 scaffolding protein: Identification of residues involved in assembly and protease activity. Virology 2009, 384, 144–150. [Google Scholar] [CrossRef]
- Novick, R.P. Mobile genetic elements and bacterial toxinoses: The superantigen-encoding pathogenicity islands of Staphylococcus aureus. Plasmid 2003, 49, 93–105. [Google Scholar] [CrossRef]
- Novick, R.P.; Christie, G.E.; Penadés, J.R. The phage-related chromosomal islands of Gram-positive bacteria. Nat. Rev. Microbiol. 2010, 8, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Penadés, J.R.; Christie, G.E. The Phage-Inducible Chromosomal Islands: A Family of Highly Evolved Molecular Parasites. Annu. Rev. Virol. 2015, 2, 181–201. [Google Scholar] [CrossRef] [PubMed]
- Novick, R.P.; Ram, G. Staphylococcal pathogenicity islands-movers and shakers in the genomic firmament. Curr. Opin. Microbiol. 2017, 38, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, J.A.; Ruzin, A.; Ross, H.F.; Kurepina, N.; Novick, R.P. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol. Microbiol. 1998, 29, 527–543. [Google Scholar] [CrossRef] [PubMed]
- Novick, R.P.; Ram, G. The Floating (Pathogenicity) Island: A Genomic Dessert. Trends Genet. 2016, 32, 114–126. [Google Scholar] [CrossRef]
- Damle, P.K.; Wall, E.A.; Spilman, M.S.; Dearborn, A.D.; Ram, G.; Novick, R.P.; Dokland, T.; Christie, G.E. The roles of SaPI1 proteins gp7 (CpmA) and gp6 (CpmB) in capsid size determination and helper phage interference. Virology 2012, 432, 277–282. [Google Scholar] [CrossRef]
- Dearborn, A.D.; Spilman, M.S.; Damle, P.K.; Chang, J.R.; Monroe, E.B.; Saad, J.S.; Christie, G.E.; Dokland, T. The Staphylococcus aureus pathogenicity island protein gp6 functions as an internal scaffold during capsid size determination. J. Mol. Biol. 2011, 412, 710–722. [Google Scholar] [CrossRef]
- Spilman, M.S.; Damle, P.K.; Dearborn, A.D.; Rodenburg, C.M.; Chang, J.R.; Wall, E.A.; Christie, G.E.; Dokland, T. Assembly of bacteriophage 80α capsids in a Staphylococcus aureus expression system. Virology 2012, 434, 242–250. [Google Scholar] [CrossRef]
- Ubeda, C.; Olivarez, N.P.; Barry, P.; Wang, H.; Kong, X.; Matthews, A.; Tallent, S.M.; Christie, G.E.; Novick, R.P. Specificity of staphylococcal phage and SaPI DNA packaging as revealed by integrase and terminase mutations. Mol. Microbiol. 2009, 72, 98–108. [Google Scholar] [CrossRef]
- Ram, G.; Chen, J.; Kumar, K.; Ross, H.F.; Ubeda, C.; Damle, P.K.; Lane, K.D.; Penadés, J.R.; Christie, G.E.; Novick, R.P. Staphylococcal pathogenicity island interference with helper phage reproduction is a paradigm of molecular parasitism. Proc. Natl. Acad. Sci. USA 2012, 109, 16300–16305. [Google Scholar] [CrossRef] [PubMed]
- Viana, D.; Blanco, J.; Tormo-Mas, M.A.; Selva, L.; Guinane, C.M.; Baselga, R.; Corpa, J.M.; Lasa, I.; Novick, R.P.; Fitzgerald, J.R.; et al. Adaptation of Staphylococcus aureus to ruminant and equine hosts involves SaPI-carried variants of von Willebrand factor-binding protein. Mol. Microbiol. 2010, 77, 1583–1594. [Google Scholar] [CrossRef] [PubMed]
- Quiles-Puchalt, N.; Carpena, N.; Alonso, J.C.; Novick, R.P.; Marina, A.; Penadés, J.R. Staphylococcal pathogenicity island DNA packaging system involving cos-site packaging and phage-encoded HNH endonucleases. Proc. Natl. Acad. Sci. USA 2014, 111, 6016–6021. [Google Scholar] [CrossRef] [PubMed]
- Carpena, N.; Manning, K.A.; Dokland, T.; Marina, A.; Penadés, J.R. Convergent evolution of pathogenicity islands in helper cos phage interference. Phil. Trans. Roy. Soc. B 2016, 371, 20150505. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Rubio, R.; Quiles-Puchalt, N.; Martí, M.; Humphrey, S.; Ram, G.; Smyth, D.; Chen, J.; Novick, R.P.; Penadés, J.R. Phage-inducible islands in the Gram-positive cocci. ISME J. 2017, 11, 1029–1042. [Google Scholar] [CrossRef]
- Fillol-Salom, A.; Martínez-Rubio, R.; Abdulrahman, R.F.; Chen, J.; Davies, R.; Penadés, J.R. Phage-inducible chromosomal islands are ubiquitous within the bacterial universe. ISME J. 2018, 12, 2114–2128. [Google Scholar] [CrossRef] [PubMed]
- Fillol-Salom, A.; Bacarizo, J.; Alqasmi, M.; Ciges-Tomas, J.R.; Martínez-Rubio, R.; Roszak, A.W.; Cogdell, R.J.; Chen, J.; Marina, A.; Penadés, J.R. Hijacking the Hijackers: Escherichia coli Pathogenicity Islands Redirect Helper Phage Packaging for Their Own Benefit. Mol. Cell 2019, 75, 1020–1030.e4. [Google Scholar] [CrossRef]
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dokland, T. Molecular Piracy: Redirection of Bacteriophage Capsid Assembly by Mobile Genetic Elements. Viruses 2019, 11, 1003. https://doi.org/10.3390/v11111003
Dokland T. Molecular Piracy: Redirection of Bacteriophage Capsid Assembly by Mobile Genetic Elements. Viruses. 2019; 11(11):1003. https://doi.org/10.3390/v11111003
Chicago/Turabian StyleDokland, Terje. 2019. "Molecular Piracy: Redirection of Bacteriophage Capsid Assembly by Mobile Genetic Elements" Viruses 11, no. 11: 1003. https://doi.org/10.3390/v11111003
APA StyleDokland, T. (2019). Molecular Piracy: Redirection of Bacteriophage Capsid Assembly by Mobile Genetic Elements. Viruses, 11(11), 1003. https://doi.org/10.3390/v11111003




