Abstract
Despite significant improvements in therapy, the HIV/AIDS pandemic remains an important threat to public health. Current treatments fail to eradicate HIV as proviral DNA persists in long-living cellular reservoirs, leading to viral rebound whenever treatment is discontinued. Hence, a better understanding of viral reservoir establishment and maintenance is required to develop novel strategies to destroy latently infected cells, and/or to durably silence the latent provirus in infected cells. Whereas the mechanism of integration has been well studied from a catalytic point of view, it remains unknown how integration site selection and transcription are linked. In recent years, evidence has grown that lens epithelium-derived growth factor p75 (LEDGF/p75) is the main determinant of HIV integration site selection and that the integration site affects the transcriptional state of the provirus. LEDGINs have been developed as small molecule inhibitors of the interaction between LEDGF/p75 and integrase. Recently, it was shown that LEDGIN treatment in cell culture shifts the residual integrated provirus towards the inner nuclear compartment and out of transcription units in a dose dependent manner. This LEDGIN-mediated retargeting increased the proportion of provirus with a transcriptionally silent phenotype and the residual reservoir proved refractory to reactivation in vitro. LEDGINs provide us with a research tool to study the link between integration and transcription, a quintessential question in retrovirology. LEDGIN-mediated retargeting of the residual reservoirs provides a novel potential “block-and-lock” strategy as a functional cure of HIV infection.
1. Introduction
Despite significant improvements in therapy, the HIV/AIDS pandemic remains an important threat to public health. According to the World Health Organization (WHO), 36.9 million people are infected worldwide and 1.8 million new infections occurred in 2017 [1]. In 2017, 21.7 million people worldwide were receiving antiretroviral treatment. Expanded access to combination antiretroviral therapy (cART), in particular in economically disfavored regions, has reduced the overall incidence of HIV-related deaths to about 940,000 in 2017 compared to 1.9 million in 2004. However, the 90–90–90 HIV treatment targets of UNAIDS remain unmet, as in 2017, only 75% of people living with HIV in the world knew their HIV status; among those only 79% were receiving treatment and among the patients receiving treatment only 81% were virally suppressed [1]. Still, current treatments fail to eradicate HIV, as proviral DNA persists in long-living cellular reservoirs, leading to viral rebound whenever cART is discontinued. As a result, the current treatment for HIV is life-long and requires strict adherence to prevent selection of antiviral resistance, which inevitably occurs under suboptimal treatment conditions. In addition, increasing treatment coverage in resource-limited settings means facing enormous infrastructural, logistic, and financial challenges. Therefore, a continuous academic effort to increase the basic understanding of HIV molecular virology, and in particular its mechanisms to persist in infected cells, is of paramount importance to validate novel therapeutic strategies and targets to cure HIV infection [2].
2. The Hurdles on the Road Towards an HIV-1 Cure
Even in patients on effective cART, a residual low-level viremia can be detected. In fact, the existence of a pool of persistent, replication competent HIV in patients on cART was demonstrated as early as 1997 [3,4]. However, studying HIV persistence has proven quite difficult. HIV DNA has been found in many anatomical sites, including blood circulating cells, the lymphoid system, the gut-associated lymphoid tissues, the brain, and other tissues [5,6], but it is not clear if replication competent virus can arise from all these sites. HIV can infect both myeloid and lymphoid cells and the relative contributions of different cell types to the reservoir remains a hot topic (reviewed in [6]). Additionally, two mechanisms can explain the persistent viremia in cART-treated subjects: ongoing low-grade viral replication owing to suboptimal tissue drug concentrations in so called sanctuary sites [7,8,9,10], and/or reactivation of HIV expression from pools of latently-infected cells [11,12,13]. In the literature, both sanctuary sites and latently infected cells are sometimes called reservoirs, since they both contain replication competent virus that can contribute to viral persistence and viral resurgence when treatment is interrupted. However, on the molecular level, latently infected cells contain intact, replication competent HIV DNA but do not produce new virus particles until reactivation occurs. Pre-integration latency refers to unintegrated HIV DNA that can still lead to expression of some viral proteins or integration upon stimulation, but its contribution to long-term persistence in patients treated with cART is considered to be limited [14,15]. In this review, we will focus on post-integration latency, where cells contain an integrated provirus that produces little to no viral mRNA, proteins, and/or progeny virus until reactivated. These latently infected cells are therefore not destroyed by viral cytopathic effects nor recognized by the immune system. However, upon reactivation of the provirus, transcription resumes, resulting in the completion of the viral replication cycle [15]. Since the focus of our review is on post-integration latency, we will thus define a reservoir as a certain pool of latently infected cells. To date, memory CD4+ T cells [16], including long-lived CD4+ T central memory stem cells (TCMs) [17] as well as T follicular helper cells (Tfh) [18], are considered to be the major subsets harboring the viral reservoirs. In addition to their long half-life, all memory CD4+ T cells are maintained through homeostatic proliferation [16] and they can also undergo clonal proliferation depending on their HIV-1 integration site [19,20,21]. Since expression of viral proteins is low or absent, these cells are not cleared by the immune system and can persist indefinitely. As such, memory CD4+ T cells provide a permanent source for virus reactivation and are likely responsible for the rapid rebound of plasma viral load observed after cART interruption. Therefore, the existence of latent reservoirs carrying replication competent provirus is the major obstacle towards finding a cure for HIV-1 infection [2]. This implies that a better understanding of viral reservoir establishment and maintenance is required to develop novel therapeutic strategies to target these latent reservoirs. Multiple strategies are being pursued [2,22]. So far, most efforts have been focused on the so-called ”shock-and-kill” strategy, where latency reversing agents (LRA), such as histone deacetylase (HDAC) inhibitors, are given to deliberately reactivate proviral transcription in latently infected cells [23] (Figure 1A). The hypothesis behind this approach is that reactivated cells will express viral proteins, allowing them to be recognized and destroyed by the host immune system. This strategy has entered exploratory clinical trials, but early results show limited evidence of efficacy [24,25,26,27,28,29]. Alternatively, autologous T- or stem cells can be modified to provide an HIV-1-resistant pool, e.g., by disrupting the CCR5 gene with zinc-finger or CRISPR/Cas technology [30]. Furthermore, researchers are attempting to remove HIV-1 provirus from latent cells in cell culture using gene editing [31,32]. However, delivery of gene editing constructs to all reservoir cells in vivo remains a formidable hurdle. Moreover, all gene-editing strategies suffer from unknown risks due to off-target effects. A final strategy is to create a cellular reservoir resistant to reactivation, thereby preventing viral rebound [22]. This so-called “block-and-lock” strategy is the focus of this review (Figure 1B).
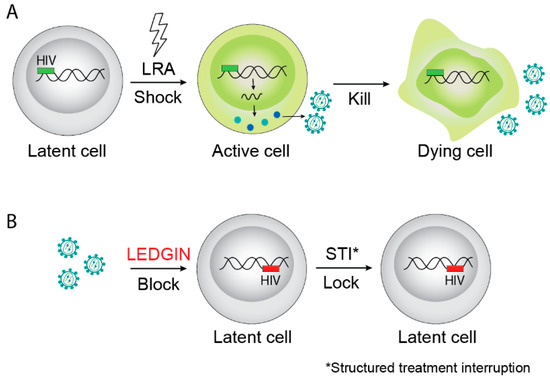
Figure 1.
Examples of HIV cure strategies. (A) The “shock-and-kill” strategy aims to reactivate latent provirus followed by killing of activated cells by viral cytopathic effects on the host immune system. (B) The “block-and-lock” functional cure strategy aims to permanently silence latent reservoirs, for instance by LEDGIN-mediated retargeting of integration to sites that are less susceptible to reactivation after interruption of combination antiretroviral therapy (cART). LRA is latency reversing agents.
3. Molecular Determinants of HIV Latency
Multiple molecular factors determine HIV latency. Basic research on the cellular mechanisms that underlie the rate-limiting steps controlling viral gene expression in resting CD4+ T cells is required. Although latent HIV-1 proviruses are preferentially found integrated into actively transcribed genes [33,34], many HIV-1 proviruses display a transcriptional repression at the level of initiation and elongation, leading to establishment and maintenance of post-integration latency. The state of latency is associated with a block of transcription from the main HIV-1 promoter, the 5’ long terminal repeat (5’LTR). While HIV-1 Tat (trans-activator of transcription) activates viral transcription and limited Tat transactivation correlates with latency, other elements like epigenetic silencing and the absence of transcription factors are involved in the block of viral transcription (reviewed in References [12,22,35]). Moreover, not only HIV transcriptional silencing needs to be overcome to disrupt latent infection effectively. The link between the viral Rev protein, involved in HIV mRNA splicing and export, and latency is not well understood. Additionally, the role of cellular processes as translation, viral antigen expression, and/or processing in HIV latency, especially within resting CD4+ T cells, is not well defined. Furthermore, the potential role of exosomes in HIV-1 latency needs to be elucidated as they influence HIV infection and modulate immune responses by delivering viral proteins [36,37]. Recent evidence even suggests that exosomes from (un)infected cells can reactivate latent HIV [38,39,40]. Finally, a better understanding of host factors that enable or restrict latency is needed as well. In this review, we will focus on our understanding of the HIV-1 integration process and its possible link with transcription and latency.
4. HIV-1 Integration Is Mediated by LEDGF/p75, the “Global Positioning System (GPS)” of HIV
Whereas the integration step has been well studied from a catalytic point of view, it remains unknown how integration site selection and transcription are linked. In recent years, evidence has grown that the site of integration affects the transcriptional state of the provirus. Indeed, HIV integration is not random, but targeted towards the nuclear periphery and active transcription units in gene dense regions [34,41,42,43,44,45,46]. Interestingly, this pattern is highly conserved between all primate lentiviruses (simian immunodeficiency virus (SIV), HIV-1, and HIV-2) [47,48,49]. The observed differences between these viruses are small and involve the preferred orientation of integration (sense or antisense) and some integration hot spots. To achieve such specific integration patterns, lentiviruses use cellular cofactors. Lens epithelium-derived growth factor p75 (LEDGF/p75) in particular is a major determinant of HIV integration site selection (for a review see Reference [50]). In 2003, Cherepanov et al. identified LEDGF/p75 as a cellular binding partner of HIV integrase [51]. However, further research demonstrated that interaction with LEDGF/p75 is a conserved feature amongst all lentiviral integrases (including Feline Immunodeficiency Virus, Equine Infectious Anemia Virus, SIV, and HIV-2), pointing to its evolutionary importance. Additionally, this could explain the similarity between lentiviral integration patterns [52]. LEDGF/p75 is ubiquitously expressed and has been implicated in neuroepithelial stem cell differentiation and neurogenesis [53], lens epithelial cell gene regulation, stress responses [54], and regulation of gene expression during embryogenesis [55]. Next to its functions in cell biology, LEDGF/p75 has been associated with various pathological processes, such as autoimmune diseases [56], prostate cancer [57], mixed lineage leukemia [58], and HIV infection [51]. Both in health and in disease, LEDGF/p75 functions as a molecular tether, linking proteins and protein complexes with chromatin. As a chromatin reader, LEDGF/p75 recognizes nucleosomes associated with actively transcribed genes [59,60]. As such, it provides HIV with a molecular “global positioning system (GPS)”.
LEDGF/p75 is encoded by the PC4 and SFRS-interacting protein 1 (PSIP1) gene on chromosome 9 [54,61] and belongs to the hepatoma-derived growth factor related (HDGF) family [50]. Next to LEDGF/p75 and HDGF, this family contains four additional members (HDGF related proteins 1–4 (HRP1–4)). The HDGF family of proteins shares a conserved N-terminal HATH (homologous to the N-terminus of HDGF) domain with a characteristic structural domain: PWWP (Pro–Trp–Trp–Pro). The PWWP domain is known to bind methylated histones H3K36me2 and 3, DNA, and other negatively charged molecules, such as heparin. Next to the HATH domain, HDGF proteins contain a nuclear localization signal (NLS).
PSIP1 encodes two isoforms, p52 and p75, resulting from alternative splicing [54,61]. Both splice-variants share their N-terminal region (amino acids 1–325), which harbors the PWWP domain, the nuclear localization signal (NLS), two AT-hook like motifs (ATH), and the supercoil recognition domain (SRD) [50]. LEDGF/p52 contains a C-terminal tail of only eight amino acids whilst LEDGF/p75 contains a largely unstructured region of an additional 205 amino acids. The only structured domain within this region is the integrase binding domain (IBD), named for its interaction with HIV integrase (IN) [62], although later work has demonstrated that many other cellular interaction partners also bind LEDGF/p75 through this domain [63]. The IBD domain of LEDGF/p75 interacts with the catalytic core domain (CCD) and the N-terminal domain (NTD) of integrase [62,64,65], and tethers the integration complex to the host chromatin [66]. This facilitates the integration into actively transcribed genes and ensures viral replication [67,68,69,70]. Next to its tethering function, LEDGF/p75 also stimulates the catalytic activity of IN and protects it from proteolytic degradation [51,71,72]. In human cells depleted of LEDGF/p75 by RNAi, in embryonic knockout mice-derived fibroblasts and in stable human LEDGF/p75 knock-down and knock-out cell lines, the absence of LEDGF/p75 significantly inhibits viral replication [67,68,69,70,73]. In the absence of LEDGF/p75, the related HRP-2 can substitute for its function [70,73]. The fact that overexpression of the IBD in cell culture effectively competes with endogenous LEDGF/p75 and potently inhibits HIV replication [68,74] provided proof-of-concept that the LEDGF/p75-integrase interaction indeed is a good target for antiviral therapy.
6. LEDGF/p75 Points the Way to a Block-and-Lock Strategy for a Functional Cure of HIV Infection
Since LEDGF/p75 is the main determinant of HIV integration site selection and since LEDGINs provide an elegant molecular tool to intervene with LEDGF/p75-mediated integration site selection, it was hypothesized that residual provirus after LEDGIN therapy may well be retargeted away from preferential integration sites. If so, this would open studies on the impact of retargeted integration on proviral gene expression and latency, and provide the rationale for a “block-and-lock” functional cure strategy (Figure 1B). In 2016, Vranckx et al. published a paper describing such experiments [83]. LEDGIN treatment clearly shifted residual HIV integration out of transcription units in a dose dependent manner [83]. An increased proportion of provirus integrated in the inverse orientation after LEDGIN treatment. In addition, the 3D localization of the integrated provirus shifted towards the inner nuclear compartment away from the nuclear periphery. Using the double reporter virus designed by the Verdin lab [84], it was shown that LEDGIN-mediated retargeting increased the proportion of provirus with a transcriptionally silent phenotype. Furthermore, this residual reservoir proved less susceptible to HIV reactivation. Of interest, almost simultaneously Chen et al. published a paper wherein a barcoded HIV vector (Barcoded HIV Ensembles (B-HIVE)) was used to experimentally prove that transcription is affected by the proximity of the integration site to genomic enhancers and that the extent of reactivation of the provirus with different LRAs is dependent on the integration site [85,86]. This finding corroborates previous findings whereby different HIV infected clonal cell lines express varying HIV RNA levels [41].
Many research questions remain to be answered. LEDGINs are known to interfere with HIV assembly. Does this so-called late effect also affect the transcriptional state after infection with the crippled virus? Of note, although LEDGIN treatment in cell culture partially shifts integration out of genes, no random integration is obtained. Still, transcription of residual provirus and reactivation of latent provirus can be completely suppressed by higher concentrations of inhibitor. This teaches us that fine tuning of the optimal chromatin environment of the provirus by LEDGF/p75 surpasses mere integration inside or outside of genes. Research is ongoing to elucidate the mechanism whereby LEDGF/p75 mediates optimal integration site selection. New methods, such as B-HIVE [85,86] and branched DNA imaging (bDNA) [87], can be used to study the link between LEDGIN-mediated retargeting and the transcriptional state of the provirus at the single-cell level. Furthermore, DNase or MNase assays can be used to investigate the role of nucleosome positioning [88]. Finally, the interaction between HIV capsid and cellular CPSF6 was recently proposed to bypass heterochromatin in the nuclear periphery to allow integration in the nuclear interior [89]. These data are at odds with reports on preferential integration in the nuclear periphery and thus require further investigation [44,45,46]. Another line of research focuses on the possible role of a complex containing LEDGF/p75, Iws1, and Spt6 in the regulation of HIV latency [90]. In conclusion, HIV may have evolved to use LEDGF/p75 as a molecular tether to ensure both a productive and a latent provirus population. In the absence of LEDGF/p75 the provirus may end up in a third deep latent population refractory to reactivation.
7. Tat Inhibition Provides a Second Block-and-Lock Strategy
As discussed, interference with the GPS of HIV, LEDGF/p75, results in a deep latent state, an approach coined the “block-and-lock” strategy [22]. Next to targeting the integration event, a “block-and-lock” functional cure strategy that aims to inhibit HIV transcription is also being explored. Recent studies have shown that HIV transcription can be abrogated by inhibition of the trans-activator of transcription (Tat) [91,92,93]. Mousseau et al. first showed that the Tat inhibitor didehydro-cortistatin A (dCA), an analog of the natural product cortistatin A, selectively inhibits Tat transactivation of the HIV-1 promoter by binding to the trans-activating response (TAR)-binding domain of Tat [91]. Later, the same group demonstrated that dCA establishes a state of latency with a diminished capacity for reactivation. In a primary cell model of HIV-1 latency, dCA-induced inactivation of viral transcription persists even after drug removal, suggesting that the HIV-1 promoter is epigenetically repressed [92]. dCA mediates epigenetic silencing by increasing nucleosomal occupancy at Nucleosome-1, restricting RNAPolII recruitment to the HIV-1 promoter. The efficacy of dCA was also studied in the bone marrow–liver–thymus (BLT) mouse model of HIV persistence. Adding dCA to cART-suppressed mice systemically reduced viral mRNA in tissues. Moreover, dCA significantly delayed and reduced viral rebound levels upon treatment interruption, proving the validity of a ″block-and-lock″ strategy for obtaining a functional cure.
8. The Debate is Open
Can LEDGINs contribute to a functional cure? The intuitive response to this question is negative, because HIV reservoirs are established early after infection, preventing provirus retargeting strategies. Still, we now know that initiation of cART early after infection is effective in reducing the size of the viral reservoir and early treatment has now become standard care [94,95,96]. If early after infection a therapeutic window exists to reduce the size of the reservoir, addition of an antiviral to initial cART regimens that can modulate the functional reservoir would make perfect sense. Once LEDGINs are tested as antivirals in clinical trials for acute infection, follow-up of their reservoirs with proviral DNA loads and quantitative viral outgrowth assay (qVOA) analysis may be a wise thing to do. LEDGINs may also be beneficial when added to HIV pre-exposure prophylaxis (PrEP), as they might ensure that any residual infection under PrEP treatment results in a non-functional, deeply latent provirus.
With respect to chronic infection, in case no residual replication occurs after establishment of the viral reservoirs, LEDGINs can no longer modulate the functional reservoir. Yet, any residual replication at sanctuary sites with poor drug penetration by current drugs would be retargeted by LEDGIN treatment. Secondly, at present, the proportion of the functional reservoir that is mobilized upon treatment interruption is not known. Possibly, re-initiation of a cART regimen including LEDGINs after treatment interruption may modulate the functional residual reservoir if enough proviruses are mobilized and if not all replication is fully blocked. Although this is currently a speculative model, it should be testable once LEDGINs are evaluated in a clinical setting.
In conclusion, LEDGINs provide us with a formidable research tool to study the link between integration and transcription, a quintessential question in retrovirology. When LEDGINs enter clinical trials, adding diagnostic readouts to monitor their impact on the functional reservoir would address the question on a potential clinical use of the proposed “block-and-lock” strategy.
Funding
G. Vansant and A. Bruggemans are doctoral fellows supported by the Flemish Fund for Scientific Research (FWO; Fonds voor Wetenschappelijk Onderzoek). Research at KU Leuven received financial support from the FWO, the KU Leuven Research Council (C1 C14/17/095-3M170311), HIV-ERA EURECA (IWT-SBO-EURECA, ZL345530), the KU Leuven IDO program (IDO/12/008), the Belgian IAP Belvir (ZKC4893-P7/45-P).
Conflicts of Interest
The authors declare no conflict of interest.
References
- UNAIDS. UNAIDS Data 2017; Joint United Nations Programme on HIV/AIDS (UNAIDS); UNAIDS Resources/UNAIDS, 2017; pp. 1–248. ISBN 978-92-9173-945-5. [Google Scholar]
- Deeks, S.G.; Lewin, S.R.; Ross, A.L.; Ananworanich, J.; Benkirane, M.; Cannon, P.; Chomont, N.; Douek, D.; Lifson, J.D.; Lo, Y.-R.; et al. International AIDS Society Global Scientific Strategy: Towards an HIV Cure 2016. Nat. Med. 2016, 22, 839. [Google Scholar] [CrossRef] [PubMed]
- Chun, T.-W.; Stuyver, L.; Mizell, S.B.; Ehler, L.A.; Mican, J.A.M.; Baseler, M.; Lloyd, A.L.; Nowak, M.A.; Fauci, A.S.; Vandekerckhove, L.; et al. Presence of an Inducible HIV-1 Latent Reservoir during Highly Active Antiretroviral Therapy. Proc. Natl. Acad. Sci. USA 1997, 94, 13193–13197. [Google Scholar] [CrossRef] [PubMed]
- Finzi, D.; Hermankova, M.; Pierson, T.; Carruth, L.M.; Buck, C.; Chaisson, R.E.; Quinn, T.C.; Chadwick, K.; Margolick, J.; Brookmeyer, R.; et al. Identification of a Reservoir for HIV-1 in Patients on Highly Active Antiretroviral Therapy Identification of a Reservoir for HIV-1 in Patients on Highly Active Antiretroviral Therapy. Science 1997, 278, 1295–1300. [Google Scholar] [CrossRef] [PubMed]
- Barton, K.; Winckelmann, A.; Palmer, S. HIV-1 Reservoirs During Suppressive Therapy. Trends Microbiol. 2016, 24, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.K.; Yukl, S.A. Tissue Reservoirs of HIV. Curr. Opin. HIV AIDS 2016, 11, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo-Redondo, R.; Fryer, H.R.; Bedford, T.; Kim, E.-Y.; Archer, J.; Kosakovsky Pond, S.L.; Chung, Y.-S.; Penugonda, S.; Chipman, J.G.; Fletcher, C.V.; et al. Persistent HIV-1 Replication Maintains the Tissue Reservoir during Therapy. Nature 2016, 530, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Buzón, M.J.; Massanella, M.; Llibre, J.M.; Esteve, A.; Dahl, V.; Puertas, M.C.; Gatell, J.M.; Domingo, P.; Paredes, R.; Sharkey, M.; et al. HIV-1 Replication and Immune Dynamics Are Affected by Raltegravir Intensification of HAART-Suppressed Subjects. Nat. Med. 2010, 16, 460. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, C.V.; Staskus, K.; Wietgrefe, S.W.; Rothenberger, M.; Reilly, C.; Chipman, J.G.; Beilman, G.J.; Khoruts, A.; Thorkelson, A.; Schmidt, T.E.; et al. Persistent HIV-1 Replication Is Associated with Lower Antiretroviral Drug Concentrations in Lymphatic Tissues. Proc. Natl. Acad. Sci. USA 2014, 111, 2307–2312. [Google Scholar] [CrossRef]
- Hatano, H.; Strain, M.C.; Scherzer, R.; Bacchetti, P.; Wentworth, D.; Hoh, R.; Martin, J.N.; McCune, J.M.; Neaton, J.D.; Tracy, R.P.; et al. Increase in 2-Long Terminal Repeat Circles and Decrease in D-Dimer after Raltegravir Intensification in Patients with Treated HIV Infection: A Randomized, Placebo-Controlled Trial. J. Infect. Dis. 2013, 208, 1436–1442. [Google Scholar] [CrossRef]
- Ho, Y.C.; Shan, L.; Hosmane, N.N.; Wang, J.; Laskey, S.B.; Rosenbloom, D.I. Replication-Competent Noninduced Proviruses in the Latent Reservoir Increase Barrier to HIV-1 Cure. Cell 2013, 155, 540–551. [Google Scholar] [CrossRef]
- Van Lint, C.; Bouchat, S.; Marcello, A. HIV-1 Transcription and Latency: An Update. Retrovirology 2013, 10, 67. [Google Scholar] [CrossRef] [PubMed]
- Churchill, M.J.; Deeks, S.G.; Margolis, D.M.; Siliciano, R.F.; Swanstrom, R. HIV Reservoirs: What, Where and How to Target Them. Nat. Rev. Microbiol. 2015, 14, 55. [Google Scholar] [CrossRef] [PubMed]
- Lassen, K.; Han, Y.; Zhou, Y.; Siliciano, J.; Siliciano, R.F. The Multifactorial Nature of HIV-1 Latency. Trends Mol. Med. 2004, 10, 525–531. [Google Scholar] [CrossRef] [PubMed]
- Coiras, M.; López-Huertas, M.R.; Pérez-Olmeda, M.; Alcamí, J. Understanding HIV-1 Latency Provides Clues for the Eradication of Long-Term Reservoirs. Nat. Rev. Microbiol. 2009, 7, 798–812. [Google Scholar] [CrossRef]
- Chomont, N.; El-Far, M.; Ancuta, P.; Trautmann, L.; Procopio, F.A.; Yassine-Diab, B.; Boucher, G.; Boulassel, M.-R.; Ghattas, G.; Brenchley, J.M.; et al. HIV Reservoir Size and Persistence Are Driven by T Cell Survival and Homeostatic Proliferation. Nat. Med. 2009, 15, 893. [Google Scholar] [CrossRef] [PubMed]
- Buzon, M.J.; Sun, H.; Li, C.; Shaw, A.; Seiss, K.; Ouyang, Z.; Martin-Gayo, E.; Leng, J.; Henrich, T.J.; Li, J.Z.; et al. HIV-1 Persistence in CD4+ T Cells with Stem Cell–like Properties. Nat. Med. 2014, 20, 139. [Google Scholar] [CrossRef] [PubMed]
- Pallikkuth, S.; Sharkey, M.; Babic, D.Z.; Gupta, S.; Stone, G.W.; Fischl, M.A.; Stevenson, M.; Pahwa, S. Peripheral T Follicular Helper Cells Are the Major HIV Reservoir within Central Memory CD4 T Cells in Peripheral Blood from Chronically HIV-Infected Individuals on Combination Antiretroviral Therapy. J. Virol. 2016, 90, 2718–2728. [Google Scholar] [CrossRef]
- Maldarelli, F.; Wu, X.; Su, L.; Simonetti, F.R.; Shao, W.; Hill, S.; Spindler, J.; Ferris, A.L.; Mellors, J.W.; Kearney, M.F.; et al. Specific HIV Integration Sites Are Linked to Clonal Expansion and Persistence of Infected Cells. Science 2014, 345, 179–183. [Google Scholar] [CrossRef]
- Wagner, T.A.; McLaughlin, S.; Garg, K.; Cheung, C.Y.K.; Larsen, B.B.; Styrchak, S.; Huang, H.C.; Edlefsen, P.T.; Mullins, J.I.; Frenkel, L.M. Proliferation of Cells with HIV Integrated into Cancer Genes Contributes to Persistent Infection. Science 2014, 345, 570–573. [Google Scholar] [CrossRef]
- Cohn, L.B.; Silva, I.T.; Oliveira, T.Y.; Rosales, R.A.; Parrish, E.H.; Learn, G.H.; Hahn, B.H.; Czartoski, J.L.; McElrath, M.J.; Lehmann, C.; et al. HIV-1 Integration Landscape during Latent and Active Infection. Cell 2015, 160, 420–432. [Google Scholar] [CrossRef]
- Darcis, G.; Van Driessche, B.; Van Lint, C. HIV Latency: Should We Shock or Lock? Trends Immunol. 2017, 38, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Darcis, G.; Van Driessche, B.; Van Lint, C. Preclinical Shock Strategies to Reactivate Latent HIV-1: An Update. Curr. Opin. HIV AIDS 2016, 11, 388–393. [Google Scholar] [CrossRef] [PubMed]
- Lehrman, G.; Hogue, I.B.; Palmer, S.; Jennings, C.; Spina, C.A.; Wiegand, A.; Landay, A.L.; Coombs, R.W.; Richman, D.D.; Mellors, J.W.; et al. Depletion of Latent HIV-1 Infection in Vivo: A Proof-of-Concept Study. Lancet (Lond. Engl.) 2005, 366, 549–555. [Google Scholar] [CrossRef]
- Siliciano, J.D.; Lai, J.; Callender, M.; Pitt, E.; Zhang, H.; Margolick, J.B.; Gallant, J.E.; Cofrancesco Joseph, J.; Moore, R.D.; Gange, S.J.; et al. Stability of the Latent Reservoir for HIV-1 in Patients Receiving Valproic Acid. J. Infect. Dis. 2007, 195, 833–836. [Google Scholar] [CrossRef] [PubMed]
- Routy, J.P.; Tremblay, C.L.; Angel, J.B.; Trottier, B.; Rouleau, D.; Baril, J.G.; Harris, M.; Trottier, S.; Singer, J.; Chomont, N.; et al. Valproic Acid in Association with Highly Active Antiretroviral Therapy for Reducing Systemic HIV-1 Reservoirs: Results from a Multicentre Randomized Clinical Study. HIV Med. 2012, 13, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Archin, N.M.; Liberty, A.L.; Kashuba, A.D.; Choudhary, S.K.; Kuruc, J.D.; Crooks, A.M.; Parker, D.C.; Anderson, E.M.; Kearney, M.F.; Strain, M.C.; et al. Administration of Vorinostat Disrupts HIV-1 Latency in Patients on Antiretroviral Therapy. Nature 2012, 487, 482. [Google Scholar] [CrossRef] [PubMed]
- Elliott, J.H.; Wightman, F.; Solomon, A.; Ghneim, K.; Ahlers, J.; Cameron, M.J.; Smith, M.Z.; Spelman, T.; McMahon, J.; Velayudham, P.; et al. Activation of HIV Transcription with Short-Course Vorinostat in HIV-Infected Patients on Suppressive Antiretroviral Therapy. PLoS Pathog. 2014, 10, e1004473. [Google Scholar] [CrossRef] [PubMed]
- Søgaard, O.S.; Graversen, M.E.; Leth, S.; Olesen, R.; Brinkmann, C.R.; Nissen, S.K.; Kjaer, A.S.; Schleimann, M.H.; Denton, P.W.; Hey-Cunningham, W.J.; et al. The Depsipeptide Romidepsin Reverses HIV-1 Latency In Vivo. PLoS Pathog. 2015, 11, e1005142. [Google Scholar] [CrossRef]
- Tebas, P.; Stein, D.; Tang, W.W.; Frank, I.; Wang, S.Q.; Lee, G.; Spratt, S.K.; Surosky, R.T.; Giedlin, M.A.; Nichol, G.; et al. Gene Editing of CCR5 in Autologous CD4 T Cells of Persons Infected with HIV. N. Engl. J. Med. 2014, 370, 901–910. [Google Scholar] [CrossRef]
- Wang, G.; Zhao, N.; Berkhout, B.; Das, A.T. CRISPR-Cas9 Can Inhibit HIV-1 Replication but NHEJ Repair Facilitates Virus Escape. Mol. Ther. 2016, 24, 522–526. [Google Scholar] [CrossRef]
- Liao, H.-K.; Gu, Y.; Diaz, A.; Marlett, J.; Takahashi, Y.; Li, M.; Suzuki, K.; Xu, R.; Hishida, T.; Chang, C.-J.; et al. Use of the CRISPR/Cas9 System as an Intracellular Defense against HIV-1 Infection in Human Cells. Nat. Commun. 2015, 6, 6413. [Google Scholar] [CrossRef]
- Han, Y.; Lin, Y.B.; An, W.; Xu, J.; Yang, H.-C.; O’Connell, K.; Dordai, D.; Boeke, J.D.; Siliciano, J.D.; Siliciano, R.F. Orientation-Dependent Regulation of Integrated HIV-1 Expression by Host Gene Transcriptional Readthrough. Cell Host Microbe 2008, 4, 134–146. [Google Scholar] [CrossRef]
- Schroder, A.R.; Shinn, P.; Chen, H.; Berry, C.; Ecker, J.R.; Bushman, F. HIV-1 Integration in the Human Genome Favors Active Genes and Local Hotspots. Cell 2002, 110, S0092–S8674. [Google Scholar] [CrossRef]
- Khoury, G.; Darcis, G.; Lee, M.Y.; Bouchat, S.; Van Driessche, B.; Purcell, D.F.J.; Van Lint, C. The Molecular Biology of HIV Latency BT—HIV Vaccines and Cure: The Path Towards Finding an Effective Cure and Vaccine; Zhang, L., Lewin, S.R., Eds.; Springer: Singapore, 2018; pp. 187–212. [Google Scholar]
- Madison, M.N.; Okeoma, C.M. Exosomes: Implications in HIV-1 Pathogenesis. Viruses 2015, 7, 4093–4118. [Google Scholar] [CrossRef] [PubMed]
- Sampey, G.C.; Saifuddin, M.; Schwab, A.; Barclay, R.; Punya, S.; Chung, M.-C.; Hakami, R.M.; Zadeh, M.A.; Lepene, B.; Klase, Z.A.; et al. Exosomes from HIV-1-Infected Cells Stimulate Production of Pro-Inflammatory Cytokines through Trans-Activating Response (TAR) RNA. J. Biol. Chem. 2016, 291, 1251–1266. [Google Scholar] [CrossRef] [PubMed]
- Barclay, R.A.; Schwab, A.; DeMarino, C.; Akpamagbo, Y.; Lepene, B.; Kassaye, S.; Iordanskiy, S.; Kashanchi, F. Exosomes from Uninfected Cells Activate Transcription of Latent HIV-1. J. Biol. Chem. 2017, 292, 11682–11701. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Lu, H.; Dooner, M.; Chapman, S.; Quesenberry, P.J.; Ramratnam, B. Exosomal Tat Protein Activates Latent HIV-1 in Primary, Resting CD4+ T Lymphocytes. JCI Insight 2018, 7. [Google Scholar] [CrossRef]
- Arenaccio, C.; Anticoli, S.; Manfredi, F.; Chiozzini, C.; Olivetta, E.; Federico, M. Latent HIV-1 Is Activated by Exosomes from Cells Infected with Either Replication-Competent or Defective HIV-1. Retrovirology 2015, 12, 87. [Google Scholar] [CrossRef] [PubMed]
- Jordan, A.; Defechereux, P.; Verdin, E. The Site of HIV-1 Integration in the Human Genome Determines Basal Transcriptional Activity and Response to Tat Transactivation. EMBO J. 2001, 20, 1726–1738. [Google Scholar] [CrossRef]
- Maxfield, L.F.; Fraize, C.D.; Coffin, J.M. Relationship between Retroviral DNA-Integration-Site Selection and Host Cell Transcription. Proc. Natl. Acad. Sci. USA 2005, 102, 1436–1441. [Google Scholar] [CrossRef]
- Felice, B.; Cattoglio, C.; Cittaro, D.; Testa, A.; Miccio, A.; Ferrari, G.; Luzi, L.; Recchia, A.; Mavilio, F. Transcription Factor Binding Sites Are Genetic Determinants of Retroviral Integration in the Human Genome. PLoS ONE 2009, 4, e4571. [Google Scholar] [CrossRef] [PubMed]
- Marini, B.; Kertesz-Farkas, A.; Ali, H.; Lucic, B.; Lisek, K.; Manganaro, L. Nuclear Architecture Dictates HIV-1 Integration Site Selection. Nature 2015, 521, 227–231. [Google Scholar] [CrossRef] [PubMed]
- Albanese, A.; Arosio, D.; Terreni, M.; Cereseto, A. HIV-1 Pre-Integration Complexes Selectively Target Decondensed Chromatin in the Nuclear Periphery. PLoS ONE 2008, 3, e2413. [Google Scholar] [CrossRef] [PubMed]
- Dieudonné, M.; Maiuri, P.; Biancotto, C.; Knezevich, A.; Kula, A.; Lusic, M.; Marcello, A. Transcriptional Competence of the Integrated HIV-1 Provirus at the Nuclear Periphery. EMBO J. 2009, 28, 2231–2243. [Google Scholar] [CrossRef] [PubMed]
- MacNeil, A.; Sankale, J.-L.; Meloni, S.T.; Sarr, A.D.; Mboup, S.; Kanki, P. Genomic Sites of Human Immunodeficiency Virus Type 2 (HIV-2) Integration: Similarities to HIV-1 In Vitro and Possible Differences In Vivo. J. Virol. 2006, 80, 7316–7321. [Google Scholar] [CrossRef] [PubMed]
- Hematti, P.; Hong, B.-K.; Ferguson, C.; Adler, R.; Hanawa, H.; Sellers, S.; Holt, I.E.; Eckfeldt, C.E.; Sharma, Y.; Schmidt, M.; et al. Distinct Genomic Integration of MLV and SIV Vectors in Primate Hematopoietic Stem and Progenitor Cells. PLoS Biol. 2004, 2, e423. [Google Scholar] [CrossRef] [PubMed]
- Soto, M.J.; Peña, Á.; Vallejo, F.G. A Genomic and Bioinformatics Analysis of the Integration of HIV in Peripheral Blood Mononuclear Cells. AIDS Res. Hum. Retroviruses 2010, 27, 547–555. [Google Scholar] [CrossRef]
- Debyser, Z.; Christ, F.; De Rijck, J.; Gijsbers, R. Host Factors for Retroviral Integration Site Selection. Trends Biochem. Sci. 2015, 40, 108–116. [Google Scholar] [CrossRef]
- Cherepanov, P.; Maertens, G.; Proost, P.; Devreese, B.; Van Beeumen, J.; Engelborghs, Y.; De Clercq, E.; Debyser, Z. HIV-1 Integrase Forms Stable Tetramers and Associates with LEDGF/P75 Protein in Human Cells. J. Biol. Chem. 2003, 278, 372–381. [Google Scholar] [CrossRef]
- Cherepanov, P. LEDGF/P75 Interacts with Divergent Lentiviral Integrases and Modulates Their Enzymatic Activity in Vitro. Nucleic Acids Res. 2007, 35, 113–124. [Google Scholar] [CrossRef]
- Chylack, L.T.; Fu, L.; Mancini, R.; Martin-Rehrmann, M.D.; Saunders, A.J.; Konopka, G.; Tian, D.; Hedley-Whyte, E.T.; Folkerth, R.D.; Goldstein, L.E. Lens Epithelium-Derived Growth Factor (LEDGF/P75) Expression in Fetal and Adult Human Brain. Exp. Eye Res. 2004, 79, 941–948. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.P.; Kimura, A.; Chylack, L.T.; Shinohara, T. Lens Epithelium-Derived Growth Factor (LEDGF/P75) and P52 Are Derived from a Single Gene by Alternative Splicing. Gene 2000, 242, 265–273. [Google Scholar] [CrossRef]
- Sutherland, H.G.; Newton, K.; Brownstein, D.G.; Holmes, M.C.; Kress, C.; Semple, C.A.; Bickmore, W.A. Disruption of Ledgf/Psip1 Results in Perinatal Mortality and Homeotic Skeletal Transformations. Mol. Cell. Biol. 2006, 26, 7201–7210. [Google Scholar] [CrossRef] [PubMed]
- Ochs, R.L.; Muro, Y.; Si, Y.; Ge, H.; Chan, E.K.L.; Tan, E.M. Autoantibodies to DFS 70 Kd/Transcription Coactivator P75 in Atopic Dermatitis and Other Conditions. J. Allergy Clin. Immunol. 2000, 105, 1211–1220. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.; Cajigas-Du Ross, C.K.; Rios-Colon, L.; Mediavilla-Varela, M.; Daniels-Wells, T.R.; Leoh, L.S.; Rojas, H.; Banerjee, H.; Martinez, S.R.; Acevedo-Martinez, S.; et al. LEDGF/P75 Overexpression Attenuates Oxidative Stress-Induced Necrosis and Upregulates the Oxidoreductase ERP57/PDIA3/GRP58 in Prostate Cancer. PLoS ONE 2016, 11, e0146549. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, A.; Cleary, M.L. Menin Critically Links MLL Proteins with LEDGF on Cancer-Associated Target Genes. Cancer Cell 2008, 14, 36–46. [Google Scholar] [CrossRef]
- Eidahl, J.O.; Crowe, B.L.; North, J.A.; McKee, C.J.; Shkriabai, N.; Feng, L.; Plumb, M.; Graham, R.L.; Gorelick, R.J.; Hess, S.; et al. Structural Basis for High-Affinity Binding of LEDGF PWWP to Mononucleosomes. Nucleic Acids Res. 2013, 41, 3924–3936. [Google Scholar] [CrossRef]
- Van Nuland, R.; van Schaik, F.M.; Simonis, M.; van Heesch, S.; Cuppen, E.; Boelens, R.; Timmers, H.M.; van Ingen, H. Nucleosomal DNA Binding Drives the Recognition of H3K36-Methylated Nucleosomes by the PSIP1-PWWP Domain. Epigenetics Chromatin 2013, 6, 12. [Google Scholar] [CrossRef]
- Ge, H.; Si, Y.; Roeder, R.G.; Bjorklund, S.; Kim, Y.; Burke, T.; Kadonaga, J.; Burke, T.; Kadonaga, J.; Burley, S.; et al. Isolation of CDNAs Encoding Novel Transcription Coactivators P52 and P75 Reveals an Alternate Regulatory Mechanism of Transcriptional Activation. EMBO J. 1998, 17, 6723–6729. [Google Scholar] [CrossRef]
- Cherepanov, P.; Devroe, E.; Silver, P.A.; Engelman, A. Identification of an Evolutionarily Conserved Domain in Human Lens Epithelium-Derived Growth Factor/Transcriptional Co-Activator P75 (LEDGF/P75) That Binds HIV-1 Integrase. J. Biol. Chem. 2004, 279, 48883–48892. [Google Scholar] [CrossRef]
- Cermakova, K.; Weydert, C.; Christ, F.; De Rijck, J.; Debyser, Z. Lessons Learned: HIV Points the Way Towards Precision Treatment of Mixed-Lineage Leukemia. Trends Pharmacol. Sci. 2016, 37, 660–671. [Google Scholar] [CrossRef] [PubMed]
- Cherepanov, P.; Ambrosio, A.L.B.; Rahman, S.; Ellenberger, T.; Engelman, A. Structural Basis for the Recognition between HIV-1 Integrase and Transcriptional Coactivator P75. Proc. Natl. Acad. Sci. USA 2005, 102, 17308–17313. [Google Scholar] [CrossRef] [PubMed]
- Hare, S.; Shun, M.-C.; Gupta, S.S.; Valkov, E.; Engelman, A.; Cherepanov, P. A Novel Co-Crystal Structure Affords the Design of Gain-of-Function Lentiviral Integrase Mutants in the Presence of Modified PSIP1/LEDGF/P75. PLoS Pathog. 2009, 5, e1000259. [Google Scholar] [CrossRef] [PubMed]
- Ciuffi, A.; Llano, M.; Poeschla, E.; Hoffmann, C.; Leipzig, J.; Shinn, P.; Ecker, J.R.; Bushman, F. A Role for LEDGF/P75 in Targeting HIV DNA Integration. Nat. Med. 2005, 11, 1287. [Google Scholar] [CrossRef]
- Shun, M.-C.; Raghavendra, N.K.; Vandegraaff, N.; Daigle, J.E.; Hughes, S.; Kellam, P.; Cherepanov, P.; Engelman, A. LEDGF/P75 Functions Downstream from Preintegration Complex Formation to Effect Gene-Specific HIV-1 Integration. Genes Dev. 2007, 21, 1767–1778. [Google Scholar] [CrossRef] [PubMed]
- Llano, M.; Saenz, D.T.; Meehan, A.; Wongthida, P.; Peretz, M.; Walker, W.H.; Teo, W.; Poeschla, E.M. An Essential Role for LEDGF/P75 in HIV Integration. Science 2006, 314, 461–464. [Google Scholar] [CrossRef] [PubMed]
- Vandekerckhove, L.; Christ, F.; Van Maele, B.; De Rijck, J.; Gijsbers, R.; Van den Haute, C.; Witvrouw, M.; Debyser, Z. Transient and Stable Knockdown of the Integrase Cofactor LEDGF/P75 Reveals Its Role in the Replication Cycle of Human Immunodeficiency Virus. J. Virol. 2006, 80, 1886–1896. [Google Scholar] [CrossRef] [PubMed]
- Schrijvers, R.; Vets, S.; De Rijck, J.; Malani, N.; Bushman, F.D.; Debyser, Z.; Gijsbers, R. HRP-2 Determines HIV-1 Integration Site Selection in LEDGF/P75 Depleted Cells. Retrovirology 2012, 9, 84. [Google Scholar] [CrossRef]
- Busschots, K.; Vercammen, J.; Emiliani, S.; Benarous, R.; Engelborghs, Y.; Christ, F.; Debyser, Z. The Interaction of LEDGF/P75 with Integrase Is Lentivirus-Specific and Promotes DNA Binding. J. Biol. Chem. 2005, 280, 17841–17847. [Google Scholar] [CrossRef]
- Llano, M.; Delgado, S.; Vanegas, M.; Poeschla, E.M. Lens Epithelium-Derived Growth Factor/P75 Prevents Proteasomal Degradation of HIV-1 Integrase. J. Biol. Chem. 2004, 279, 55570–55577. [Google Scholar] [CrossRef]
- Schrijvers, R.; De Rijck, J.; Demeulemeester, J.; Adachi, N.; Vets, S.; Ronen, K.; Christ, F.; Bushman, F.D.; Debyser, Z.; Gijsbers, R. LEDGF/P75-Independent HIV-1 Replication Demonstrates a Role for HRP-2 and Remains Sensitive to Inhibition by LEDGINs. PLoS Pathog. 2012, 8, e1002558. [Google Scholar] [CrossRef] [PubMed]
- De Rijck, J.; Vandekerckhove, L.; Gijsbers, R.; Hombrouck, A.; Hendrix, J.; Vercammen, J.; Engelborghs, Y.; Christ, F.; Debyser, Z. Overexpression of the Lens Epithelium-Derived Growth Factor/P75 Integrase Binding Domain Inhibits Human Immunodeficiency Virus Replication. J. Virol. 2006, 80, 11498–11509. [Google Scholar] [CrossRef] [PubMed]
- Christ, F.; Voet, A.; Marchand, A.; Nicolet, S.; Desimmie, B.A.; Marchand, D.; Bardiot, D.; Van der Veken, N.J.; Van Remoortel, B.; Strelkov, S.V.; et al. Rational Design of Small-Molecule Inhibitors of the LEDGF/P75-Integrase Interaction and HIV Replication. Nat. Chem. Biol. 2010, 6, 442–448. [Google Scholar] [CrossRef] [PubMed]
- Demeulemeester, J.; Chaltin, P.; Marchand, A.; De Maeyer, M.; Debyser, Z.; Christ, F. LEDGINs, Non-Catalytic Site Inhibitors of HIV-1 Integrase: A Patent Review (2006–2014). Expert Opin. Ther. Pat. 2014, 24, 609–632. [Google Scholar] [CrossRef] [PubMed]
- Christ, F.; Shaw, S.; Demeulemeester, J.; Desimmie, B.A.; Marchand, A.; Butler, S.; Smets, W.; Chaltin, P.; Westby, M.; Debyser, Z.; et al. Small-Molecule Inhibitors of the LEDGF/P75 Binding Site of Integrase Block HIV Replication and Modulate Integrase Multimerization. Antimicrob. Agents Chemother. 2012, 56, 4365–4374. [Google Scholar] [CrossRef] [PubMed]
- Kessl, J.J.; Jena, N.; Koh, Y.; Taskent-Sezgin, H.; Slaughter, A.; Feng, L.; de Silva, S.; Wu, L.; Le Grice, S.F.J.; Engelman, A.; et al. Multimode, Cooperative Mechanism of Action of Allosteric HIV-1 Integrase Inhibitors. J. Biol. Chem. 2012, 287, 16801–16811. [Google Scholar] [CrossRef] [PubMed]
- Desimmie, B.A.; Schrijvers, R.; Demeulemeester, J.; Borrenberghs, D.; Weydert, C.; Thys, W.; Vets, S.; Van Remoortel, B.; Hofkens, J.; De Rijck, J.; et al. LEDGINs Inhibit Late Stage HIV-1 Replication by Modulating Integrase Multimerization in the Virions. Retrovirology 2013, 10, 57. [Google Scholar] [CrossRef]
- Jurado, K.A.; Wang, H.; Slaughter, A.; Feng, L.; Kessl, J.J.; Koh, Y.; Wang, W.; Ballandras-Colas, A.; Patel, P.A.; Fuchs, J.R.; et al. Allosteric Integrase Inhibitor Potency Is Determined through the Inhibition of HIV-1 Particle Maturation. Proc. Natl. Acad. Sci. USA 2013, 110, 8690–8695. [Google Scholar] [CrossRef]
- Balakrishnan, M.; Yant, S.R.; Tsai, L.; O’Sullivan, C.; Bam, R.A.; Tsai, A.; Niedziela-Majka, A.; Stray, K.M.; Sakowicz, R.; Cihlar, T. Non-Catalytic Site HIV-1 Integrase Inhibitors Disrupt Core Maturation and Induce a Reverse Transcription Block in Target Cells. PLoS ONE 2013, 8, e74163. [Google Scholar] [CrossRef]
- Le Rouzic, E.; Bonnard, D.; Chasset, S.; Bruneau, J.-M.; Chevreuil, F.; Le Strat, F.; Nguyen, J.; Beauvoir, R.; Amadori, C.; Brias, J.; et al. Dual Inhibition of HIV-1 Replication by Integrase-LEDGF Allosteric Inhibitors Is Predominant at the Post-Integration Stage. Retrovirology 2013, 10, 144. [Google Scholar] [CrossRef]
- Vranckx, L.S.; Demeulemeester, J.; Saleh, S.; Boll, A.; Vansant, G.; Schrijvers, R.; Weydert, C.; Battivelli, E.; Verdin, E.; Cereseto, A.; et al. LEDGIN-Mediated Inhibition of Integrase–LEDGF/P75 Interaction Reduces Reactivation of Residual Latent HIV. EBioMedicine 2016, 8, 248–264. [Google Scholar] [CrossRef] [PubMed]
- Battivelli, E.; Dahabieh, M.S.; Abdel-Mohsen, M.; Svensson, J.P.; Tojal Da Silva, I.; Cohn, L.B.; Gramatica, A.; Deeks, S.; Greene, W.C.; Pillai, S.K.; et al. Distinct Chromatin Functional States Correlate with HIV Latency Reversal in Infected Primary CD4+ T Cells. eLife 2018, 7, e34655. [Google Scholar] [CrossRef]
- Chen, H.-C.; Martinez, J.P.; Zorita, E.; Meyerhans, A.; Filion, G.J. Position Effects Influence HIV Latency Reversal. Nat. Struct. Mol. Biol. 2017, 24, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-C.; Zorita, E.; Filion, G.J. Using Barcoded HIV Ensembles (B-HIVE) for Single Provirus Transcriptomics. Curr. Protoc. Mol. Biol. 2018, 122, e56. [Google Scholar] [CrossRef] [PubMed]
- Puray-Chavez, M.; Tedbury, P.R.; Huber, A.D.; Ukah, O.B.; Yapo, V.; Liu, D.; Ji, J.; Wolf, J.J.; Engelman, A.N.; Sarafianos, S.G. Multiplex Single-Cell Visualization of Nucleic Acids and Protein during HIV Infection. Nat. Commun. 2017, 8, 1882. [Google Scholar] [CrossRef] [PubMed]
- Rafati, H.; Parra, M.; Hakre, S.; Moshkin, Y.; Verdin, E.; Mahmoudi, T. Repressive LTR Nucleosome Positioning by the BAF Complex Is Required for HIV Latency. PLoS Biol. 2011, 9, e1001206. [Google Scholar] [CrossRef]
- Achuthan, V.; Perreira, J.M.; Sowd, G.A.; Puray-Chavez, M.; McDougall, W.M.; Paulucci-Holthauzen, A.; Wu, X.; Fadel, H.J.; Poeschla, E.M.; Multani, A.S.; et al. Capsid-CPSF6 Interaction Licenses Nuclear HIV-1 Trafficking to Sites of Viral DNA Integration. Cell Host Microbe 2018, 24, 392–404.e8. [Google Scholar] [CrossRef]
- Gérard, A.; Ségéral, E.; Naughtin, M.; Abdouni, A.; Charmeteau, B.; Cheynier, R.; Rain, J.-C.; Emiliani, S. The Integrase Cofactor LEDGF/P75 Associates with Iws1 and Spt6 for Postintegration Silencing of HIV-1 Gene Expression in Latently Infected Cells. Cell Host Microbe 2015, 17, 107–117. [Google Scholar] [CrossRef]
- Kessing, C.F.; Nixon, C.C.; Li, C.; Tsai, P.M.; Takata, H.; Mousseau, G.; Ho, P.T.; Honeycutt, J.B.; Fallahi, M.; Trautmann, L.; et al. In Vivo Suppression of HIV Rebound by Didehydro-Cortistatin A, a “Block-and-Lock” Strategy for HIV-1 Cure. Cell Rep. 2017, 21, 600–611. [Google Scholar] [CrossRef]
- Mousseau, G.; Kessing, C.F.; Fromentin, R.; Trautmann, L.; Chomont, N.; Valente, S.T. The Tat Inhibitor Didehydro-Cortistatin A Prevents HIV-1 Reactivation from Latency. mBio 2015, 6, e00465-15. [Google Scholar] [CrossRef]
- Mousseau, G.; Mediouni, S.; Valente, S.T. Targeting HIV Transcription: The Quest for a Functional Cure. Curr. Top. Microbiol. Immunol. 2015, 389, 121–145. [Google Scholar] [CrossRef] [PubMed]
- Hocqueloux, L.; Avettand-Fènoël, V.; Jacquot, S.; Prazuck, T.; Legac, E.; Mélard, A.; Niang, M.; Mille, C.; Le Moal, G.; Viard, J.-P.; et al. Long-Term Antiretroviral Therapy Initiated during Primary HIV-1 Infection Is Key to Achieving Both Low HIV Reservoirs and Normal T Cell Counts. J. Antimicrob. Chemother. 2013, 68, 1169–1178. [Google Scholar] [CrossRef] [PubMed]
- Malatinkova, E.; De Spiegelaere, W.; Bonczkowski, P.; Kiselinova, M.; Vervisch, K.; Trypsteen, W.; Johnson, M.; Verhofstede, C.; de Looze, D.; Murray, C.; et al. Impact of a Decade of Successful Antiretroviral Therapy Initiated at HIV-1 Seroconversion on Blood and Rectal Reservoirs. eLife 2015, 4, e09115. [Google Scholar] [CrossRef] [PubMed]
- Buzon, M.J.; Martin-Gayo, E.; Pereyra, F.; Ouyang, Z.; Sun, H.; Li, J.Z.; Piovoso, M.; Shaw, A.; Dalmau, J.; Zangger, N.; et al. Long-Term Antiretroviral Treatment Initiated at Primary HIV-1 Infection Affects the Size, Composition, and Decay Kinetics of the Reservoir of HIV-1-Infected CD4 T Cells. J. Virol. 2014, 88, 10056–10065. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).