Molecular Basis of Bacterial Host Interactions by Gram-Positive Targeting Bacteriophages
Abstract
1. Introduction
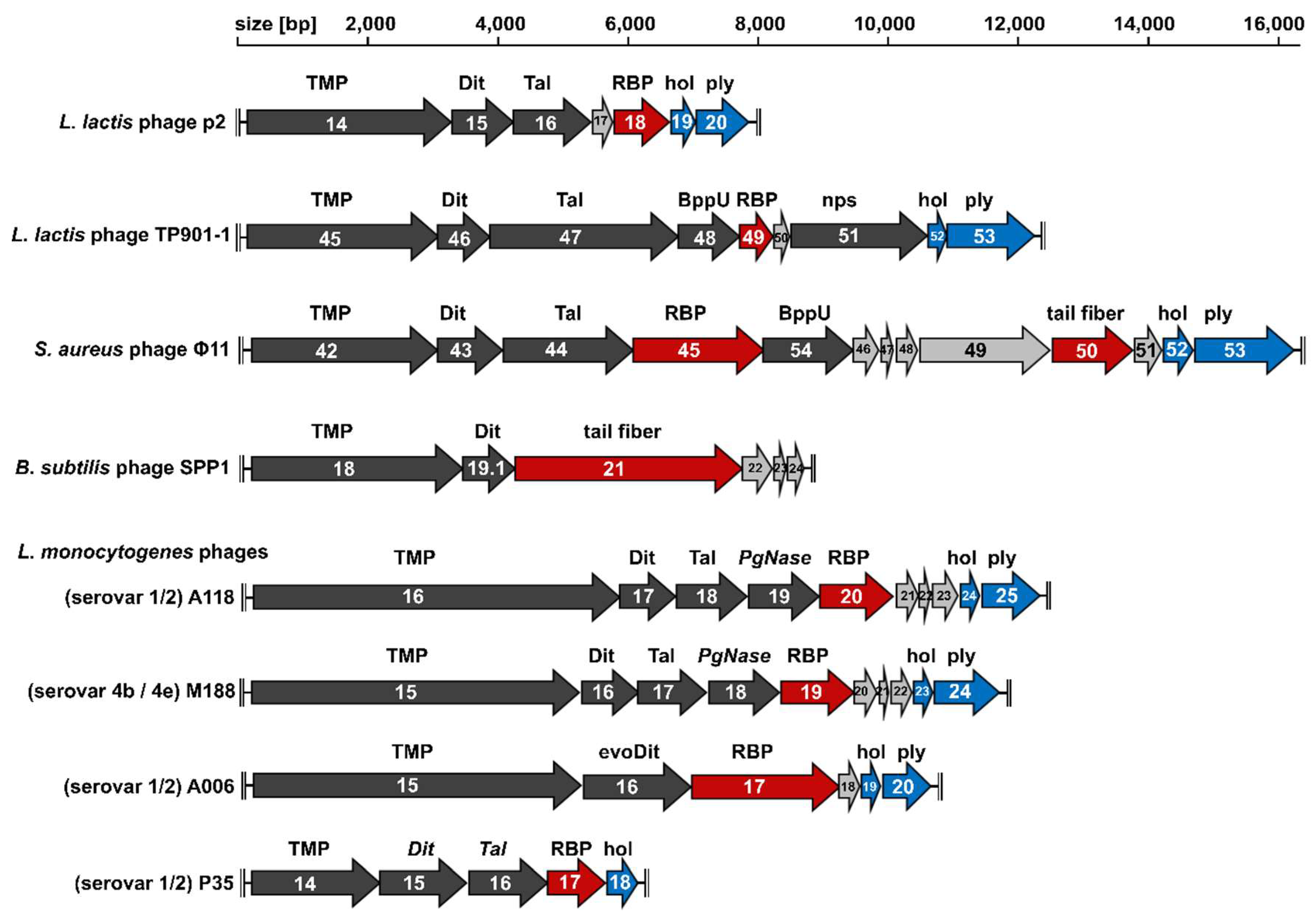
2. The Conserved Tail Morphogenesis Sub-Module of Gram-Positive-Targeting Siphoviridae
3. Distal Tail Protein (Dit)
4. Tail-Associated Lysin (Tal)
5. Upper Baseplate Protein (BppU)
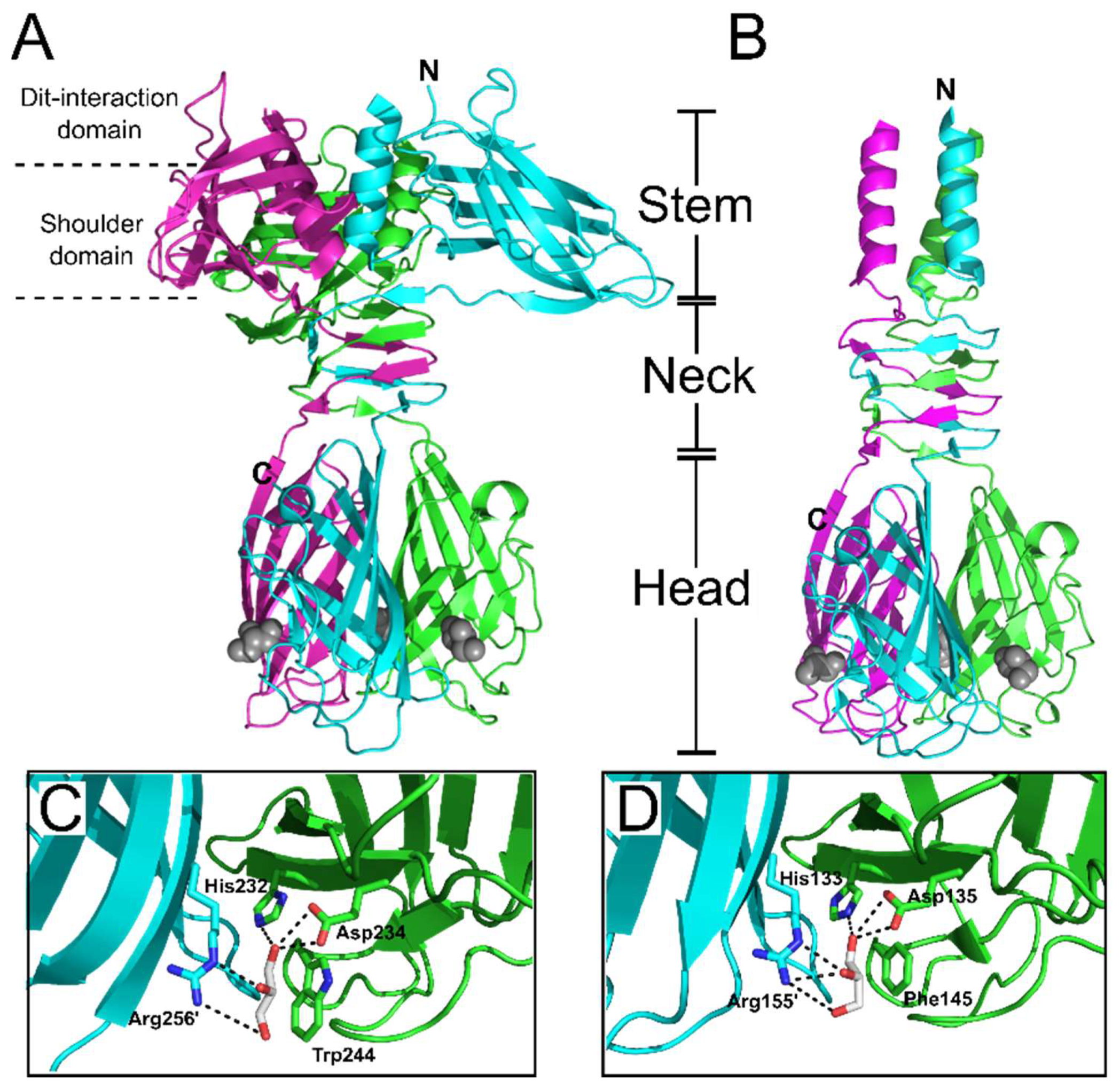
6. Receptor Binding Protein (RBP)
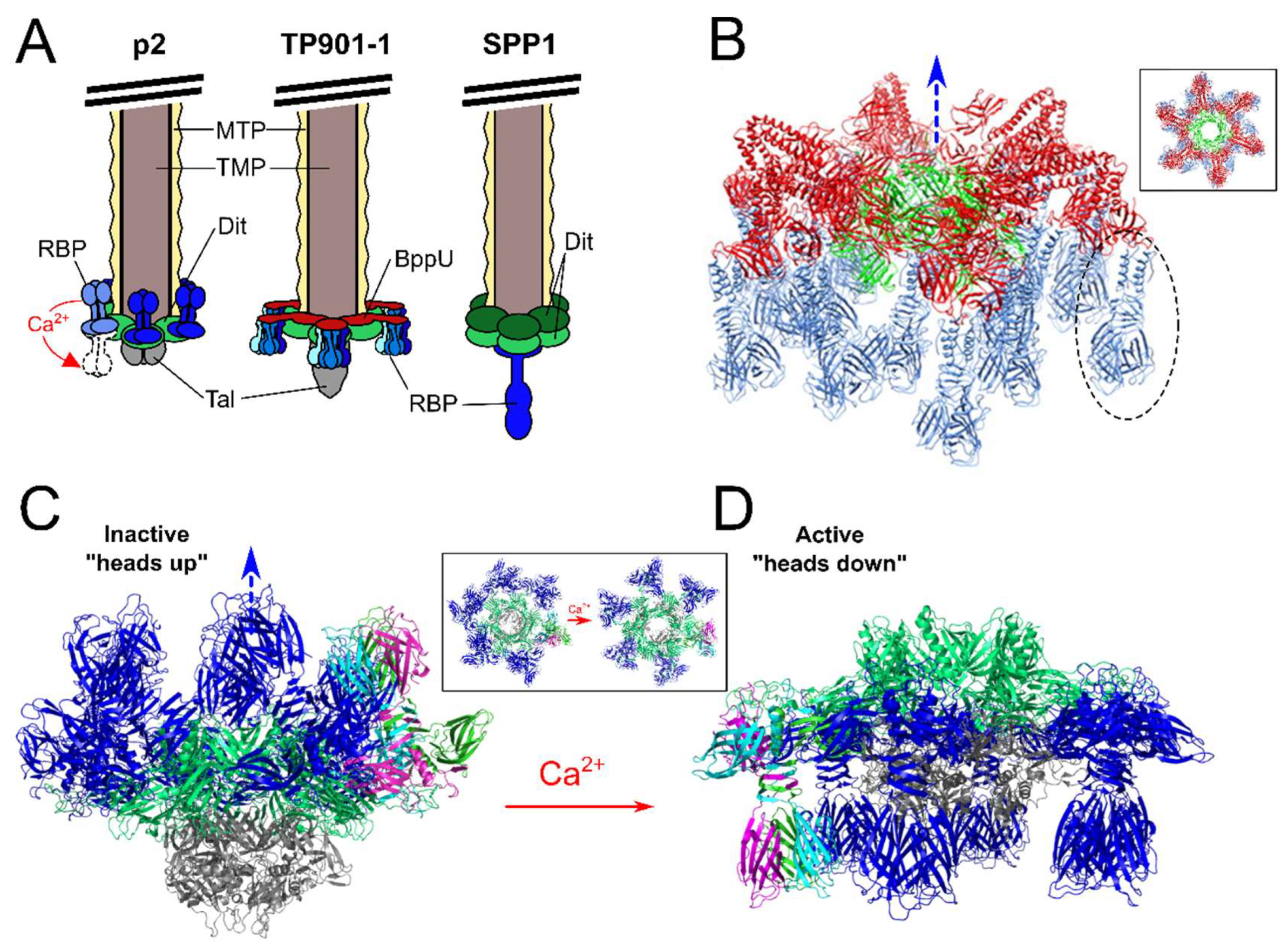
7. Different Baseplates Architectures of the Model Lactococcal Phages p2 and TP901-1
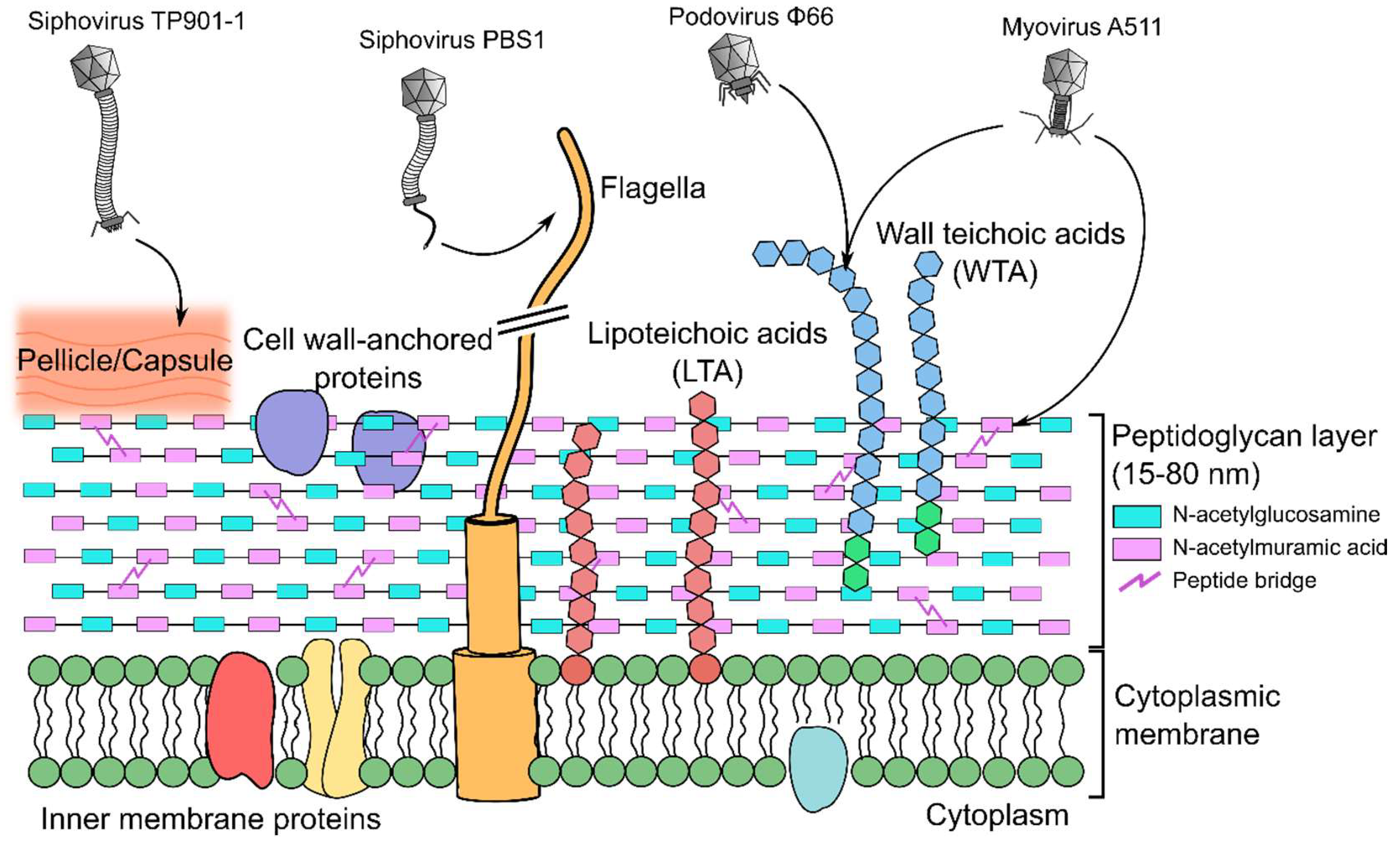
8. Peptidoglycan-Binding Phages
9. Teichoic Acid-Binding Phages
10. Lipoteichoic Acid-Binding Lactobacillus Phages
11. Wall Teichoic Acid-Binding Listeria Phages
12. Wall Teichoic Acid-Binding Staphylococcus Phages
13. Pellicle-Binding Lactococcal Phages
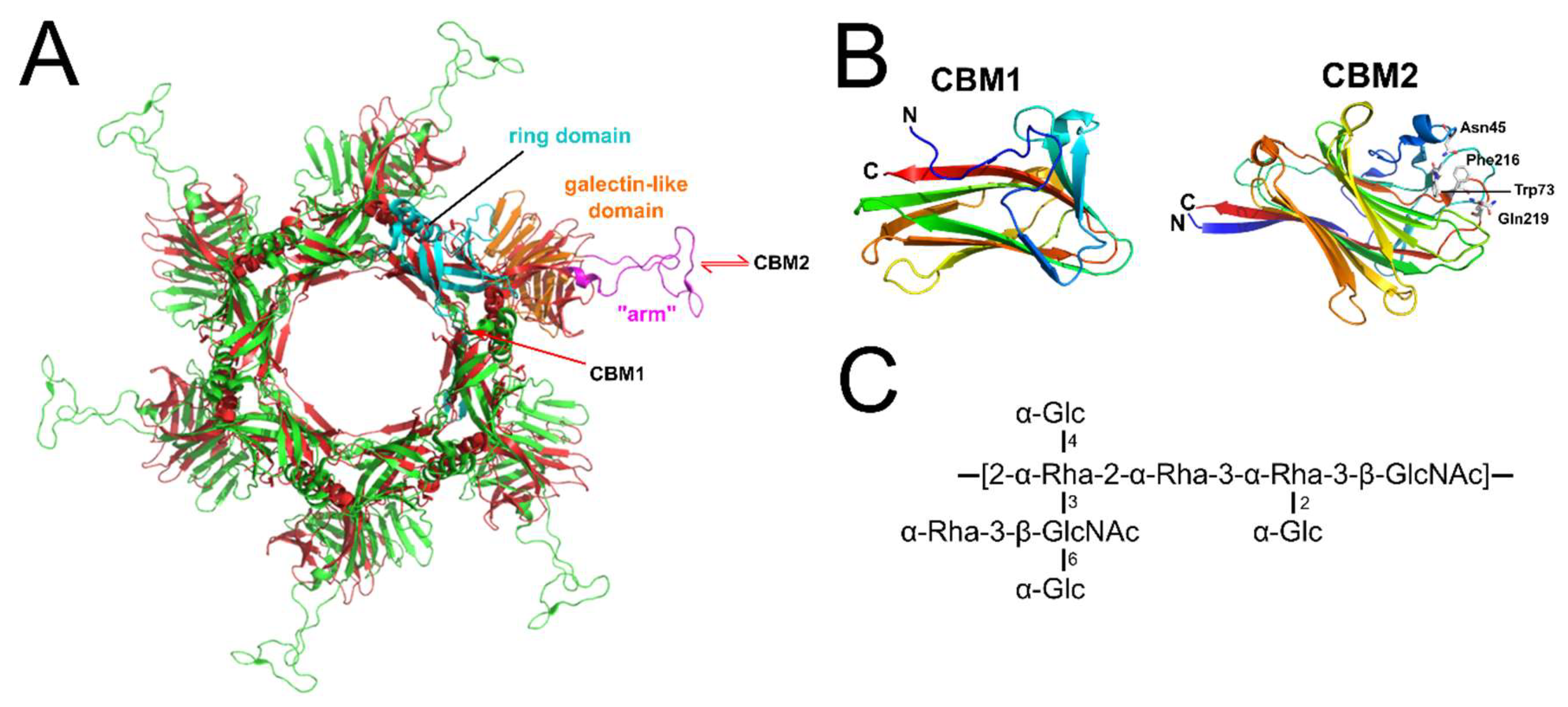
14. Receptor Binding Sites of Lactococcal Phage RBPs
15. Flagellotropic Phages
16. Future Perspective
Author Contributions
Funding
Conflicts of Interest
References
- Hatfull, G.F. Dark Matter of the Biosphere: The Amazing World of Bacteriophage Diversity. J. Virol. 2015, 89, 8107–8110. [Google Scholar] [CrossRef] [PubMed]
- Chibani-Chennoufi, S.; Bruttin, A.; Dillmann, M.-L.; Brüssow, H. Phage-host interaction: An ecological perspective. J. Bacteriol. 2004, 186, 3677–3686. [Google Scholar] [CrossRef] [PubMed]
- Riley, P.A. Phages: Their role in bacterial pathogenesis and biotechnology. J. Clin. Pathol. 2006, 59, 1003–1004. [Google Scholar] [CrossRef]
- Duckworth, D. History and Basic Properties of Bacterial Viruses. Available online: https://ci.nii.ac.jp/naid/10020227033/#cit (accessed on 1 July 2018).
- Dowah, A.S.A.; Clokie, M.R.J. Review of the nature, diversity and structure of bacteriophage receptor binding proteins that target Gram-positive bacteria. Biophys. Rev. 2018, 10, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Bertozzi Silva, J.; Storms, Z.; Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef] [PubMed]
- Molineux, I.J.; Panja, D. Popping the cork: Mechanisms of phage genome ejection. Nat. Rev. Microbiol. 2013, 11, 194–204. [Google Scholar] [CrossRef] [PubMed]
- Taylor, N.M.I.; Prokhorov, N.S.; Guerrero-Ferreira, R.C.; Shneider, M.M.; Browning, C.; Goldie, K.N.; Stahlberg, H.; Leiman, P.G. Structure of the T4 baseplate and its function in triggering sheath contraction. Nature 2016, 533, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Ge, P.; Scholl, D.; Leiman, P.G.; Yu, X.; Miller, J.F.; Zhou, Z.H. Atomic structures of a bactericidal contractile nanotube in its pre- and postcontraction states. Nat. Struct. Mol. Biol. 2015, 22, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Leiman, P.G.; Basler, M.; Ramagopal, U.A.; Bonanno, J.B.; Sauder, J.M.; Pukatzki, S.; Burley, S.K.; Almo, S.C.; Mekalanos, J.J. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. USA 2009, 106, 4154–4159. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Margolin, W.; Molineux, I.J.; Liu, J. Structural remodeling of bacteriophage T4 and host membranes during infection initiation. Proc. Natl. Acad. Sci. USA 2015, 112, E4919–E4928. [Google Scholar] [CrossRef] [PubMed]
- Veesler, D.; Robin, G.; Lichière, J.; Auzat, I.; Tavares, P.; Bron, P.; Campanacci, V.; Cambillau, C. Crystal Structure of Bacteriophage SPP1 Distal Tail Protein (gp19.1) A BASEPLATE HUB PARADIGM IN GRAM-POSITIVE INFECTING PHAGES. J. Biol. Chem. 2010, 285, 36666–36673. [Google Scholar] [CrossRef] [PubMed]
- Plisson, C.; White, H.E.; Auzat, I.; Zafarani, A.; São-José, C.; Lhuillier, S.; Tavares, P.; Orlova, E.V. Structure of bacteriophage SPP1 tail reveals trigger for DNA ejection. EMBO J. 2007, 26, 3720–3728. [Google Scholar] [CrossRef] [PubMed]
- Baptista, C.; Santos, M.A.; São-José, C. Phage SPP1 reversible adsorption to Bacillus subtilis cell wall teichoic acids accelerates virus recognition of membrane receptor YueB. J. Bacteriol. 2008, 190, 4989–4996. [Google Scholar] [CrossRef] [PubMed]
- Vinga, I.; Baptista, C.; Auzat, I.; Petipas, I.; Lurz, R.; Tavares, P.; Santos, M.A.; São-José, C. Role of bacteriophage SPP1 tail spike protein gp21 on host cell receptor binding and trigger of phage DNA ejection. Mol. Microbiol. 2012, 83, 289–303. [Google Scholar] [CrossRef] [PubMed]
- Sciara, G.; Bebeacua, C.; Bron, P.; Tremblay, D.; Ortiz-Lombardia, M.; Lichière, J.; van Heel, M.; Campanacci, V.; Moineau, S.; Cambillau, C. Structure of lactococcal phage p2 baseplate and its mechanism of activation. Proc. Natl. Acad. Sci. USA 2010, 107, 6852–6857. [Google Scholar] [CrossRef] [PubMed]
- Bebeacua, C.; Tremblay, D.; Farenc, C.; Chapot-Chartier, M.-P.; Sadovskaya, I.; van Heel, M.; Veesler, D.; Moineau, S.; Cambillau, C. Structure, Adsorption to Host, and Infection Mechanism of Virulent Lactococcal Phage p2. J. Virol. 2013, 87, 12302–12312. [Google Scholar] [CrossRef] [PubMed]
- Veesler, D.; Spinelli, S.; Mahony, J.; Lichière, J.; Blangy, S.; Bricogne, G.; Legrand, P.; Ortiz-Lombardia, M.; Campanacci, V.; van Sinderen, D.; et al. Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism. Proc. Natl. Acad. Sci. USA 2012, 109, 8954–8958. [Google Scholar] [CrossRef] [PubMed]
- Bebeacua, C.; Bron, P.; Lai, L.; Vegge, C.S.; Brøndsted, L.; Spinelli, S.; Campanacci, V.; Veesler, D.; van Heel, M.; Cambillau, C. Structure and molecular assignment of lactococcal phage TP901-1 baseplate. J. Biol. Chem. 2010, 285, 39079–39086. [Google Scholar] [CrossRef] [PubMed]
- Habann, M.; Leiman, P.G.; Vandersteegen, K.; Van den Bossche, A.; Lavigne, R.; Shneider, M.M.; Bielmann, R.; Eugster, M.R.; Loessner, M.J.; Klumpp, J. Listeria phage A511, a model for the contractile tail machineries of SPO1-related bacteriophages. Mol. Microbiol. 2014, 92, 84–99. [Google Scholar] [CrossRef] [PubMed]
- Koç, C.; Xia, G.; Kühner, P.; Spinelli, S.; Roussel, A.; Cambillau, C.; Stehle, T. Structure of the host-recognition device of Staphylococcus aureus phage ϕ11. Sci. Rep. 2016, 6, 27581. [Google Scholar] [CrossRef] [PubMed]
- Chapot-Chartier, M.-P.; Vinogradov, E.; Sadovskaya, I.; Andre, G.; Mistou, M.-Y.; Trieu-Cuot, P.; Furlan, S.; Bidnenko, E.; Courtin, P.; Péchoux, C.; et al. Cell Surface of Lactococcus lactis Is Covered by a Protective Polysaccharide Pellicle. J. Biol. Chem. 2010, 285, 10464–10471. [Google Scholar] [CrossRef] [PubMed]
- Raimondo, L.M.; Lundh, N.P.; Martinez, R.J. Primary Adsorption Site of Phage PBS1: The Flagellum of Bacillus subtilis. J. Virol. 1968, 2, 256–264. [Google Scholar] [PubMed]
- Li, X.; Gerlach, D.; Du, X.; Larsen, J.; Stegger, M.; Kühner, P.; Peschel, A.; Xia, G.; Winstel, V. An accessory wall teichoic acid glycosyltransferase protects Staphylococcus aureus from the lytic activity of Podoviridae. Sci. Rep. 2015, 5, 17219. [Google Scholar] [CrossRef] [PubMed]
- Schooley, R.T.; Biswas, B.; Gill, J.J.; Hernandez-Morales, A.; Lancaster, J.; Lessor, L.; Barr, J.J.; Reed, S.L.; Rohwer, F.; Benler, S.; et al. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob. Agents Chemother. 2017. [Google Scholar] [CrossRef] [PubMed]
- Golkar, Z.; Bagasra, O.; Pace, D.G. Bacteriophage therapy: A potential solution for the antibiotic resistance crisis. J. Infect. Dev. Ctries. 2014, 8, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Borie, C.; Albala, I.; Sánchez, P.; Sánchez, M.L.; Ramírez, S.; Navarro, C.; Morales, M.A.; Retamales, A.J.; Robeson, J. Bacteriophage treatment reduces Salmonella colonization of infected chickens. Avian Dis. 2008, 52, 64–67. [Google Scholar] [CrossRef] [PubMed]
- Chajęcka-Wierzchowska, W.; Zadernowska, A.; Kłębukowska, L.; Łaniewska-Trokenheim, Ł. Salmonella detection in poultry meat–validation of VIDAS Xpress Automatic Enzyme-Linked Fluorescent Immunoassay-Based Method. J. Food Saf. 2012, 32, 407–414. [Google Scholar] [CrossRef]
- Bao, H.; Zhang, P.; Zhang, H.; Zhou, Y.; Zhang, L.; Wang, R. Bio-Control of Salmonella Enteritidis in Foods Using Bacteriophages. Viruses 2015, 7, 4836–4853. [Google Scholar] [CrossRef] [PubMed]
- Denyes, J.M.; Dunne, M.; Steiner, S.; Mittelviefhaus, M.; Weiss, A.; Schmidt, H.; Klumpp, J.; Loessner, M.J. Modified bacteriophage S16 long tail fiber proteins for rapid and specific immobilization and detection of Salmonella cells. Appl. Environ. Microbiol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Torres, N.; Dunne, M.; Garde, S.; Meijers, R.; Narbad, A.; Ávila, M.; Mayer, M.J. Development of a specific fluorescent phage endolysin for in situ detection of Clostridium species associated with cheese spoilage. Microb. Biotechnol. 2018, 11, 332–345. [Google Scholar] [CrossRef] [PubMed]
- Kilcher, S.; Studer, P.; Muessner, C.; Klumpp, J.; Loessner, M.J. Cross-genus rebooting of custom-made, synthetic bacteriophage genomes in L-form bacteria. Proc. Natl. Acad. Sci. USA 2018, 115, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Adley, C.C. Past, Present and Future of Sensors in Food Production. Foods 2014, 3, 491–510. [Google Scholar] [CrossRef] [PubMed]
- Brzozowska, E.; Śmietana, M.; Koba, M.; Górska, S.; Pawlik, K.; Gamian, A.; Bock, W.J. Recognition of bacterial lipopolysaccharide using bacteriophage-adhesin-coated long-period gratings. Biosens. Bioelectron. 2015, 67, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Kittler, S.; Fischer, S.; Abdulmawjood, A.; Glünder, G.; Klein, G. Effect of bacteriophage application on Campylobacter jejuni loads in commercial broiler flocks. Appl. Environ. Microbiol. 2013, 79, 7525–7533. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Hosseindoust, A.; Lee, S.H.; Choi, Y.H.; Kim, M.J.; Lee, J.H.; Kwon, I.K.; Chae, B.J. Bacteriophage cocktail and multi-strain probiotics in the feed for weanling pigs: Effects on intestine morphology and targeted intestinal coliforms and Clostridium. Animal 2017, 11, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Guenther, S.; Herzig, O.; Fieseler, L.; Klumpp, J.; Loessner, M.J. Biocontrol of Salmonella Typhimurium in RTE foods with the virulent bacteriophage FO1-E2. Int. J. Food Microbiol. 2012, 154, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Guenther, S.; Huwyler, D.; Richard, S.; Loessner, M.J. Virulent bacteriophage for efficient biocontrol of Listeria monocytogenes in ready-to-eat foods. Appl. Environ. Microbiol. 2009, 75, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Kazi, M.; Annapure, U.S. Bacteriophage biocontrol of foodborne pathogens. J. Food Sci. Technol. 2016, 53, 1355–1362. [Google Scholar] [CrossRef] [PubMed]
- Klumpp, J.; Loessner, M.J. Listeria phages. Bacteriophage 2013, 3. [Google Scholar] [CrossRef] [PubMed]
- Zinno, P.; Devirgiliis, C.; Ercolini, D.; Ongeng, D.; Mauriello, G. Bacteriophage P22 to challenge Salmonella in foods. Int. J. Food Microbiol. 2014, 191, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, N.; Bhardwaj, S.K.; Mehta, J.; Kim, K.-H.; Deep, A. MOF-Bacteriophage Biosensor for Highly Sensitive and Specific Detection of Staphylococcus aureus. ACS Appl. Mater. Interfaces 2017, 9, 33589–33598. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, S.; Sorokulova, I.B.; Vodyanoy, V.J.; Simonian, A.L. Lytic phage as a specific and selective probe for detection of Staphylococcus aureus—A surface plasmon resonance spectroscopic study. Biosens. Bioelectron. 2007, 22, 948–955. [Google Scholar] [CrossRef] [PubMed]
- Lakshmanan, R.S.; Guntupalli, R.; Hu, J.; Kim, D.-J.; Petrenko, V.A.; Barbaree, J.M.; Chin, B.A. Phage immobilized magnetoelastic sensor for the detection of Salmonella typhimurium. J. Microbiol. Methods 2007, 71, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Lakshmanan, R.S.; Guntupalli, R.; Hu, J.; Petrenko, V.A.; Barbaree, J.M.; Chin, B.A. Detection of Salmonella typhimurium in fat free milk using a phage immobilized magnetoelastic sensor. Sens. Actuators B Chem. 2007, 126, 544–550. [Google Scholar] [CrossRef]
- Singh, A.; Poshtiban, S.; Evoy, S. Recent Advances in Bacteriophage Based Biosensors for Food-Borne Pathogen Detection. Sensors 2013, 13, 1763–1786. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-J.; Lin, H.; Yang, X. Industrial production of recombinant therapeutics in Escherichia coli and its recent advancements. J. Ind. Microbiol. Biotechnol. 2012, 39, 383–399. [Google Scholar] [CrossRef] [PubMed]
- Garneau, J.E.; Moineau, S. Bacteriophages of lactic acid bacteria and their impact on milk fermentations. Microb. Cell Factories 2011, 10 (Suppl. 1), S20. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, H.-W. Phage classification and characterization. Methods Mol. Biol. 2009, 501, 127–140. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, R.W.; Smith, M.C.M.; Burns, R.N.; Ford, M.E.; Hatfull, G.F. Evolutionary relationships among diverse bacteriophages and prophages: All the world’s a phage. Proc. Natl. Acad. Sci. USA 1999, 96, 2192–2197. [Google Scholar] [CrossRef] [PubMed]
- Flayhan, A.; Vellieux, F.M.D.; Lurz, R.; Maury, O.; Contreras-Martel, C.; Girard, E.; Boulanger, P.; Breyton, C. Crystal Structure of pb9, the Distal Tail Protein of Bacteriophage T5: A Conserved Structural Motif among All Siphophages. J. Virol. 2014, 88, 820–828. [Google Scholar] [CrossRef] [PubMed]
- Pell, L.G.; Liu, A.; Edmonds, L.; Donaldson, L.W.; Howell, P.L.; Davidson, A.R. The X-Ray Crystal Structure of the Phage λ Tail Terminator Protein Reveals the Biologically Relevant Hexameric Ring Structure and Demonstrates a Conserved Mechanism of Tail Termination among Diverse Long-Tailed Phages. J. Mol. Biol. 2009, 389, 938–951. [Google Scholar] [CrossRef] [PubMed]
- Dieterle, M.-E.; Spinelli, S.; Sadovskaya, I.; Piuri, M.; Cambillau, C. Evolved distal tail carbohydrate binding modules of Lactobacillus phage J-1: A novel type of anti-receptor widespread among lactic acid bacteria phages. Mol. Microbiol. 2017, 104, 608–620. [Google Scholar] [CrossRef] [PubMed]
- Dieterle, M.E.; Fina Martin, J.; Durán, R.; Nemirovsky, S.I.; Sanchez Rivas, C.; Bowman, C.; Russell, D.; Hatfull, G.F.; Cambillau, C.; Piuri, M. Characterization of prophages containing “evolved” Dit/Tal modules in the genome of Lactobacillus casei BL23. Appl. Microbiol. Biotechnol. 2016, 100, 9201–9215. [Google Scholar] [CrossRef] [PubMed]
- Cid, M.; Pedersen, H.L.; Kaneko, S.; Coutinho, P.M.; Henrissat, B.; Willats, W.G.T.; Boraston, A.B. Recognition of the helical structure of β-1,4-galactan by a new family of carbohydrate-binding modules. J. Biol. Chem. 2010, 285, 35999–36009. [Google Scholar] [CrossRef] [PubMed]
- Hudson, K.L.; Bartlett, G.J.; Diehl, R.C.; Agirre, J.; Gallagher, T.; Kiessling, L.L.; Woolfson, D.N. Carbohydrate-Aromatic Interactions in Proteins. J. Am. Chem. Soc. 2015, 137, 15152–15160. [Google Scholar] [CrossRef] [PubMed]
- Bourne, Y.; Roussel, A.; Frey, M.; Rougé, P.; Fontecilla-Camps, J.C.; Cambillau, C. Three-dimensional structures of complexes of Lathyrus ochrus isolectin I with glucose and mannose: Fine specificity of the monosaccharide-binding site. Proteins 1990, 8, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Vinogradov, E.; Sadovskaya, I.; Grard, T.; Chapot-Chartier, M.-P. Structural studies of the rhamnose-rich cell wall polysaccharide of Lactobacillus casei BL23. Carbohydr. Res. 2016, 435, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Dorscht, J.; Klumpp, J.; Bielmann, R.; Schmelcher, M.; Born, Y.; Zimmer, M.; Calendar, R.; Loessner, M.J. Comparative Genome Analysis of Listeria Bacteriophages Reveals Extensive Mosaicism, Programmed Translational Frameshifting, and a Novel Prophage Insertion Site. J. Bacteriol. 2009, 191, 7206–7215. [Google Scholar] [CrossRef] [PubMed]
- Veesler, D.; Cambillau, C. A Common Evolutionary Origin for Tailed-Bacteriophage Functional Modules and Bacterial Machineries. Microbiol. Mol. Biol. Rev. 2011, 75, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Kenny, J.G.; McGrath, S.; Fitzgerald, G.F.; van Sinderen, D. Bacteriophage Tuc2009 Encodes a Tail-Associated Cell Wall-Degrading Activity. J. Bacteriol. 2004, 186, 3480–3491. [Google Scholar] [CrossRef] [PubMed]
- Stockdale, S.R.; Mahony, J.; Courtin, P.; Chapot-Chartier, M.-P.; Pijkeren, J.-P.; van Britton, R.A.; Neve, H.; Heller, K.J.; Aideh, B.; Vogensen, F.K.; et al. The Lactococcal Phages Tuc2009 and TP901-1 Incorporate Two Alternate Forms of Their Tail Fiber into Their Virions for Infection Specialization. J. Biol. Chem. 2013, 288, 5581–5590. [Google Scholar] [CrossRef] [PubMed]
- Mahony, J.; Martel, B.; Tremblay, D.M.; Neve, H.; Heller, K.J.; Moineau, S.; van Sinderen, D. Identification of a new P335 subgroup through molecular analysis of lactococcal phages Q33 and BM13. Appl. Environ. Microbiol. 2013, 79, 4401–4409. [Google Scholar] [CrossRef] [PubMed]
- Kanamaru, S.; Leiman, P.G.; Kostyuchenko, V.A.; Chipman, P.R.; Mesyanzhinov, V.V.; Arisaka, F.; Rossmann, M.G. Structure of the cell-puncturing device of bacteriophage T4. Nature 2002, 415, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Kondou, Y.; Kitazawa, D.; Takeda, S.; Tsuchiya, Y.; Yamashita, E.; Mizuguchi, M.; Kawano, K.; Tsukihara, T. Structure of the central hub of bacteriophage Mu baseplate determined by X-ray crystallography of gp44. J. Mol. Biol. 2005, 352, 976–985. [Google Scholar] [CrossRef] [PubMed]
- Legrand, P.; Collins, B.; Blangy, S.; Murphy, J.; Spinelli, S.; Gutierrez, C.; Richet, N.; Kellenberger, C.; Desmyter, A.; Mahony, J.; et al. The Atomic Structure of the Phage Tuc2009 Baseplate Tripod Suggests that Host Recognition Involves Two Different Carbohydrate Binding Modules. MBio 2016, 7, e01781-15. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Koç, C.; Kühner, P.; Stierhof, Y.-D.; Krismer, B.; Enright, M.C.; Penadés, J.R.; Wolz, C.; Stehle, T.; Cambillau, C.; et al. An essential role for the baseplate protein Gp45 in phage adsorption to Staphylococcus aureus. Sci. Rep. 2016, 6, 26455. [Google Scholar] [CrossRef] [PubMed]
- Cambillau, C. Bacteriophage module reshuffling results in adaptive host range as exemplified by the baseplate model of listerial phage A118. Virology 2015, 484, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Mahony, J.; van Sinderen, D. Structural Aspects of the Interaction of Dairy Phages with Their Host Bacteria. Viruses 2012, 4, 1410–1424. [Google Scholar] [CrossRef] [PubMed]
- Flayhan, A.; Wien, F.; Paternostre, M.; Boulanger, P.; Breyton, C. New insights into pb5, the receptor binding protein of bacteriophage T5, and its interaction with its Escherichia coli receptor FhuA. Biochimie 2012, 94, 1982–1989. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, D.M.; Tegoni, M.; Spinelli, S.; Campanacci, V.; Blangy, S.; Huyghe, C.; Desmyter, A.; Labrie, S.; Moineau, S.; Cambillau, C. Receptor-binding protein of Lactococcus lactis phages: Identification and characterization of the saccharide receptor-binding site. J. Bacteriol. 2006, 188, 2400–2410. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, S.; Campanacci, V.; Blangy, S.; Moineau, S.; Tegoni, M.; Cambillau, C. Modular Structure of the Receptor Binding Proteins of Lactococcus lactis Phages the RBP Structure of the temperate Phage TP901-1. J. Biol. Chem. 2006, 281, 14256–14262. [Google Scholar] [CrossRef] [PubMed]
- Farenc, C.; Spinelli, S.; Vinogradov, E.; Tremblay, D.; Blangy, S.; Sadovskaya, I.; Moineau, S.; Cambillau, C. Molecular Insights on the Recognition of a Lactococcus lactis Cell Wall Pellicle by the Phage 1358 Receptor Binding Protein. J. Virol. 2014, 88, 7005–7015. [Google Scholar] [CrossRef] [PubMed]
- Ricagno, S.; Campanacci, V.; Blangy, S.; Spinelli, S.; Tremblay, D.; Moineau, S.; Tegoni, M.; Cambillau, C. Crystal Structure of the Receptor-Binding Protein Head Domain from Lactococcus lactis Phage bIL170. J. Virol. 2006, 80, 9331–9335. [Google Scholar] [CrossRef] [PubMed]
- Siponen, M.; Spinelli, S.; Blangy, S.; Moineau, S.; Cambillau, C.; Campanacci, V. Crystal Structure of a Chimeric Receptor Binding Protein Constructed from Two Lactococcal Phages. J. Bacteriol. 2009, 191, 3220–3225. [Google Scholar] [CrossRef] [PubMed]
- Van Raaij, M.J.; Louis, N.; Chroboczek, J.; Cusack, S. Structure of the human adenovirus serotype 2 fiber head domain at 1.5 A resolution. Virology 1999, 262, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Chappell, J.D.; Prota, A.E.; Dermody, T.S.; Stehle, T. Crystal structure of reovirus attachment protein σ1 reveals evolutionary relationship to adenovirus fiber. EMBO J. 2002, 21, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, S.; Desmyter, A.; Verrips, C.T.; de Haard, H.J.W.; Moineau, S.; Cambillau, C. Lactococcal bacteriophage p2 receptor-binding protein structure suggests a common ancestor gene with bacterial and mammalian viruses. Nat. Struct. Mol. Biol. 2006, 13, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, S.; Veesler, D.; Bebeacua, C.; Cambillau, C. Structures and host-adhesion mechanisms of lactococcal siphophages. Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Mahony, J.; Tremblay, D.M.; Labrie, S.J.; Moineau, S.; van Sinderen, D. Investigating the requirement for calcium during lactococcal phage infection. Int. J. Food Microbiol. 2015, 201, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Chhibber, S.; Kaur, T.; Kaur, S. Essential role of calcium in the infection process of broad-spectrum methicillin-resistant Staphylococcus aureus bacteriophage. J. Basic Microbiol. 2014, 54, 775–780. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, S.; Bebeacua, C.; Orlov, I.; Tremblay, D.; Klaholz, B.P.; Moineau, S.; Cambillau, C. Cryo-Electron Microscopy Structure of Lactococcal Siphophage 1358 Virion. J. Virol. 2014, 88, 8900–8910. [Google Scholar] [CrossRef] [PubMed]
- Dupuis, M.-È.; Moineau, S. Genome Organization and Characterization of the Virulent Lactococcal Phage 1358 and Its Similarities to Listeria Phages. Appl. Environ. Microbiol. 2010, 76, 1623–1632. [Google Scholar] [CrossRef] [PubMed]
- Archibald, A.R. Phage Receptors in Gram-positive Bacteria. In Virus Receptors; Receptors and Recognition; Springer: Dordrecht, The Netherlands, 1980; pp. 5–26. ISBN 978-94-011-6920-2. [Google Scholar]
- Vollmer, W.; Blanot, D.; de Pedro, M.A. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 2008, 32, 149–167. [Google Scholar] [CrossRef] [PubMed]
- Takumi, K.; Takeoka, A.; Kinouchi, T.; Kawata, T. Solubilization and Partial Properties of Receptor Substance for Bacteriophage α2 Induced from Clostridium botulinum Type A 190L. Microbiol. Immunol. 1985, 29, 1185–1195. [Google Scholar] [CrossRef] [PubMed]
- Gaidelyte, A.; Cvirkaite-Krupovic, V.; Daugelavicius, R.; Bamford, J.K.H.; Bamford, D.H. The entry mechanism of membrane-containing phage Bam35 infecting Bacillus thuringiensis. J. Bacteriol. 2006, 188, 5925–5934. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.; Santa Maria, J.P.; Walker, S. Wall teichoic acids of gram-positive bacteria. Annu. Rev. Microbiol. 2013, 67, 313–336. [Google Scholar] [CrossRef] [PubMed]
- Kojima, N.; Araki, Y.; Ito, E. Structure of the linkage units between ribitol teichoic acids and peptidoglycan. J. Bacteriol. 1985, 161, 299–306. [Google Scholar] [PubMed]
- Xia, G.; Kohler, T.; Peschel, A. The wall teichoic acid and lipoteichoic acid polymers of Staphylococcus aureus. Int. J. Med. Microbiol. 2010, 300, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Bera, A.; Biswas, R.; Herbert, S.; Kulauzovic, E.; Weidenmaier, C.; Peschel, A.; Götz, F. Influence of wall teichoic acid on lysozyme resistance in Staphylococcus aureus. J. Bacteriol. 2007, 189, 280–283. [Google Scholar] [CrossRef] [PubMed]
- Davison, S.; Couture-Tosi, E.; Candela, T.; Mock, M.; Fouet, A. Identification of the Bacillus anthracis (gamma) phage receptor. J. Bacteriol. 2005, 187, 6742–6749. [Google Scholar] [CrossRef] [PubMed]
- Munsch-Alatossava, P.; Alatossava, T. The extracellular phage-host interactions involved in the bacteriophage LL-H infection of Lactobacillus delbrueckii ssp. lactis ATCC 15808. Front. Microbiol. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Monteville, M.R.; Ardestani, B.; Geller, B.L. Lactococcal bacteriophages require a host cell wall carbohydrate and a plasma membrane protein for adsorption and ejection of DNA. Appl. Environ. Microbiol. 1994, 60, 3204–3211. [Google Scholar] [PubMed]
- Lubbers, M.W.; Waterfield, N.R.; Beresford, T.P.; Page, R.W.L.; Jarvis, A.W. Sequencing and analysis of the prolate-headed lactococcal bacteriophage c2 genome and identification of the structural genes. Appl. Environ. Microbiol. 1995, 61, 4348–4356. [Google Scholar] [PubMed]
- Bielmann, R.; Habann, M.; Eugster, M.R.; Lurz, R.; Calendar, R.; Klumpp, J.; Loessner, M.J. Receptor binding proteins of Listeria monocytogenes bacteriophages A118 and P35 recognize serovar-specific teichoic acids. Virology 2015, 477, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Räisänen, L.; Schubert, K.; Jaakonsaari, T.; Alatossava, T. Characterization of lipoteichoic acids as Lactobacillus delbrueckii phage receptor components. J. Bacteriol. 2004, 186, 5529–5532. [Google Scholar] [CrossRef] [PubMed]
- Räisänen, L.; Draing, C.; Pfitzenmaier, M.; Schubert, K.; Jaakonsaari, T.; Aulock, S. von Hartung, T.; Alatossava, T. Molecular Interaction between Lipoteichoic Acids and Lactobacillus delbrueckii Phages Depends on D-Alanyl and α-Glucose Substitution of Poly(Glycerophosphate) Backbones. J. Bacteriol. 2007, 189, 4135–4140. [Google Scholar] [CrossRef] [PubMed]
- Mahony, J.; Cambillau, C.; van Sinderen, D. Host recognition by lactic acid bacterial phages. FEMS Microbiol. Rev. 2017, 41, S16–S26. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Boulos, S.; Sumrall, E.; Gerber, B.; Julian-Rodero, A.; Eugster, M.R.; Fieseler, L.; Nyström, L.; Ebert, M.-O.; Loessner, M.J. Structural and functional diversity in Listeria cell wall teichoic acids. J. Biol. Chem. 2017. [Google Scholar] [CrossRef] [PubMed]
- Farber, J.M.; Peterkin, P.I. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 1991, 55, 476–511. [Google Scholar] [PubMed]
- Eugster, M.R.; Loessner, M.J. Wall teichoic acids restrict access of bacteriophage endolysin Ply118, Ply511, and PlyP40 cell wall binding domains to the Listeria monocytogenes peptidoglycan. J. Bacteriol. 2012, 194, 6498–6506. [Google Scholar] [CrossRef] [PubMed]
- Casey, A.; Jordan, K.; Neve, H.; Coffey, A.; McAuliffe, O. A tail of two phages: Genomic and functional analysis of Listeria monocytogenes phages vB_LmoS_188 and vB_LmoS_293 reveal the receptor-binding proteins involved in host specificity. Front. Microbiol. 2015, 6, 1107. [Google Scholar] [CrossRef] [PubMed]
- Deghorain, M.; Van Melderen, L. The Staphylococci Phages Family: An Overview. Viruses 2012, 4, 3316–3335. [Google Scholar] [CrossRef] [PubMed]
- Bartual, S.G.; Otero, J.M.; Garcia-Doval, C.; Llamas-Saiz, A.L.; Kahn, R.; Fox, G.C.; van Raaij, M.J. Structure of the bacteriophage T4 long tail fiber receptor-binding tip. Proc. Natl. Acad. Sci. USA 2010, 107, 20287–20292. [Google Scholar] [CrossRef] [PubMed]
- Trojet, S.N.; Caumont-Sarcos, A.; Perrody, E.; Comeau, A.M.; Krisch, H.M. The gp38 Adhesins of the T4 Superfamily: A Complex Modular Determinant of the Phage’s Host Specificity. Genome Biol. Evol. 2011, 3, 674–686. [Google Scholar] [CrossRef] [PubMed]
- Marti, R.; Zurfluh, K.; Hagens, S.; Pianezzi, J.; Klumpp, J.; Loessner, M.J. Long tail fibres of the novel broad-host-range T-even bacteriophage S16 specifically recognize Salmonella OmpC. Mol. Microbiol. 2013, 87, 818–834. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Doval, C.; van Raaij, M.J. Structure of the receptor-binding carboxy-terminal domain of bacteriophage T7 tail fibers. Proc. Natl. Acad. Sci. USA 2012, 109, 9390–9395. [Google Scholar] [CrossRef] [PubMed]
- Eugster, M.R.; Loessner, M.J. Rapid Analysis of Listeria monocytogenes Cell Wall Teichoic Acid Carbohydrates by ESI-MS/MS. PLoS ONE 2011, 6, e21500. [Google Scholar] [CrossRef] [PubMed]
- Wendlinger, G.; Loessner, M.J.; Scherer, S. Bacteriophage receptors on Listeria monocytogenes cells are the N-acetylglucosamine and rhamnose substituents of teichoic acids or the peptidoglycan itself. Microbiology 1996, 142, 985–992. [Google Scholar] [CrossRef] [PubMed]
- Xia, G.; Corrigan, R.M.; Winstel, V.; Goerke, C.; Gründling, A.; Peschel, A. Wall Teichoic Acid-Dependent Adsorption of Staphylococcal Siphovirus and Myovirus. J. Bacteriol. 2011, 193, 4006–4009. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, I.; Osada, K.; Azam, A.H.; Asakawa, H.; Miyanaga, K.; Tanji, Y. The Presence of Two Receptor-Binding Proteins Contributes to the Wide Host Range of Staphylococcal Twort-Like Phages. Appl. Environ. Microbiol. 2016, 82, 5763–5774. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.; Xia, G.; Luhachack, L.G.; Campbell, J.; Meredith, T.C.; Chen, C.; Winstel, V.; Gekeler, C.; Irazoqui, J.E.; Peschel, A.; Walker, S. Methicillin resistance in Staphylococcus aureus requires glycosylated wall teichoic acids. Proc. Natl. Acad. Sci. USA 2012, 109, 18909–18914. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Benson, S.D.; Butcher, S.J.; Bamford, D.H.; Burnett, R.M. The receptor binding protein P2 of PRD1, a virus targeting antibiotic-resistant bacteria, has a novel fold suggesting multiple functions. Structure 2003, 11, 309–322. [Google Scholar] [CrossRef]
- Sycheva, L.V.; Shneider, M.M.; Sykilinda, N.N.; Ivanova, M.A.; Miroshnikov, K.A.; Leiman, P.G. Crystal structure and location of gp131 in the bacteriophage phiKZ virion. Virology 2012, 434, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Deveau, H.; Labrie, S.J.; Chopin, M.-C.; Moineau, S. Biodiversity and classification of lactococcal phages. Appl. Environ. Microbiol. 2006, 72, 4338–4346. [Google Scholar] [CrossRef] [PubMed]
- Babu, K.S.; Spence, W.S.; Monteville, M.R.; Geller, B.L. Characterization of a cloned gene (pip) from lactococcus lactis required for phage infection. Dev. Biol. Stand. 1995, 85, 569–575. [Google Scholar] [PubMed]
- Mooney, D.T.; Jann, M.; Geller, B.L. Subcellular location of phage infection protein (Pip) in Lactococcus lactis. Can. J. Microbiol. 2006, 52, 664–672. [Google Scholar] [CrossRef] [PubMed]
- São-José, C.; Lhuillier, S.; Lurz, R.; Melki, R.; Lepault, J.; Santos, M.A.; Tavares, P. The ectodomain of the viral receptor YueB forms a fiber that triggers ejection of bacteriophage SPP1 DNA. J. Biol. Chem. 2006, 281, 11464–11470. [Google Scholar] [CrossRef] [PubMed]
- Shiraishi, T.; Yokota, S.; Fukiya, S.; Yokota, A. Structural diversity and biological significance of lipoteichoic acid in Gram-positive bacteria: Focusing on beneficial probiotic lactic acid bacteria. Biosci. Microbiota Food Health 2016, 35, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Ainsworth, S.; Sadovskaya, I.; Vinogradov, E.; Courtin, P.; Guerardel, Y.; Mahony, J.; Grard, T.; Cambillau, C.; Chapot-Chartier, M.-P.; van Sinderen, D. Differences in lactococcal cell wall polysaccharide structure are major determining factors in bacteriophage sensitivity. MBio 2014, 5, e00880-14. [Google Scholar] [CrossRef] [PubMed]
- McCabe, O.; Spinelli, S.; Farenc, C.; Labbé, M.; Tremblay, D.; Blangy, S.; Oscarson, S.; Moineau, S.; Cambillau, C. The targeted recognition of Lactococcus lactis phages to their polysaccharide receptors. Mol. Microbiol. 2015, 96, 875–886. [Google Scholar] [CrossRef] [PubMed]
- Grahn, A.M.; Caldentey, J.; Bamford, J.K.H.; Bamford, D.H. Stable Packaging of Phage PRD1 DNA Requires Adsorption Protein P2, Which Binds to the IncP Plasmid-Encoded Conjugative Transfer Complex. J. Bacteriol. 1999, 181, 6689–6696. [Google Scholar] [PubMed]
- Mahony, J.; van Sinderen, D. Gram-Positive Phages: From Isolation to Application; Frontiers Media SA: Lausanne, Switzerland, 2015; ISBN 978-2-88919-493-3. [Google Scholar]
- Sertic, V.; Boulgakov, N. Bactériophages spécifiques pour des variétés bactériennes flagellées. Compt. Rend. Soc. Biol. 1936, 123, 887–888. [Google Scholar]
- Choi, Y.; Shin, H.; Lee, J.-H.; Ryu, S. Identification and characterization of a novel flagellum-dependent Salmonella-infecting bacteriophage, iEPS5. Appl. Environ. Microbiol. 2013, 79, 4829–4837. [Google Scholar] [CrossRef] [PubMed]
- Joys, T.M. Correlation between Susceptibility to Bacteriophage PBS1 and Motility in Bacillus subtilis. J. Bacteriol. 1965, 90, 1575–1577. [Google Scholar] [PubMed]
- Guerrero-Ferreira, R.C.; Viollier, P.H.; Ely, B.; Poindexter, J.S.; Georgieva, M.; Jensen, G.J.; Wright, E.R. Alternative mechanism for bacteriophage adsorption to the motile bacterium Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 2011, 108, 9963–9968. [Google Scholar] [CrossRef] [PubMed]
- Meynell, E.W. A phage, phi chi, which attacks motile bacteria. J. Gen. Microbiol. 1961, 25, 253–290. [Google Scholar] [CrossRef] [PubMed]
- Lovett, P.S. PBP1: A flagella specific bacteriophage mediating transduction in Bacillus pumilus. Virology 1972, 47, 743–752. [Google Scholar] [CrossRef]
- Pires, D.P.; Cleto, S.; Sillankorva, S.; Azeredo, J.; Lu, T.K. Genetically Engineered Phages: A Review of Advances over the Last Decade. Microbiol. Mol. Biol. Rev. 2016, 80, 523–543. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.K.; Collins, J.J. Dispersing biofilms with engineered enzymatic bacteriophage. Proc. Natl. Acad. Sci. USA 2007, 104, 11197–11202. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.K.; Collins, J.J. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc. Natl. Acad. Sci. USA 2009, 106, 4629–4634. [Google Scholar] [CrossRef] [PubMed]
- Ando, H.; Lemire, S.; Pires, D.P.; Lu, T.K. Engineering Modular Viral Scaffolds for Targeted Bacterial Population Editing. Cell Syst. 2015, 1, 187–196. [Google Scholar] [CrossRef] [PubMed]
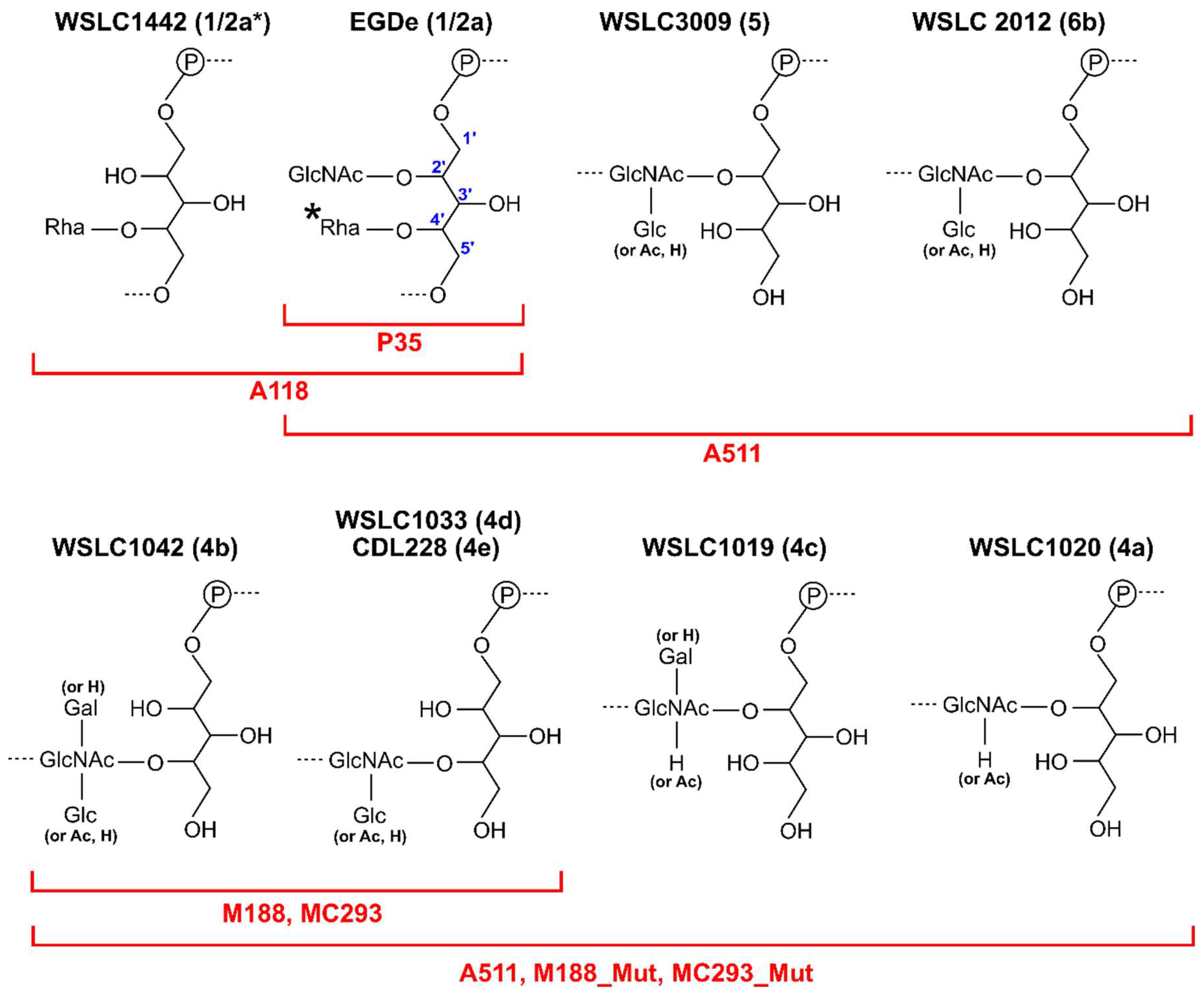
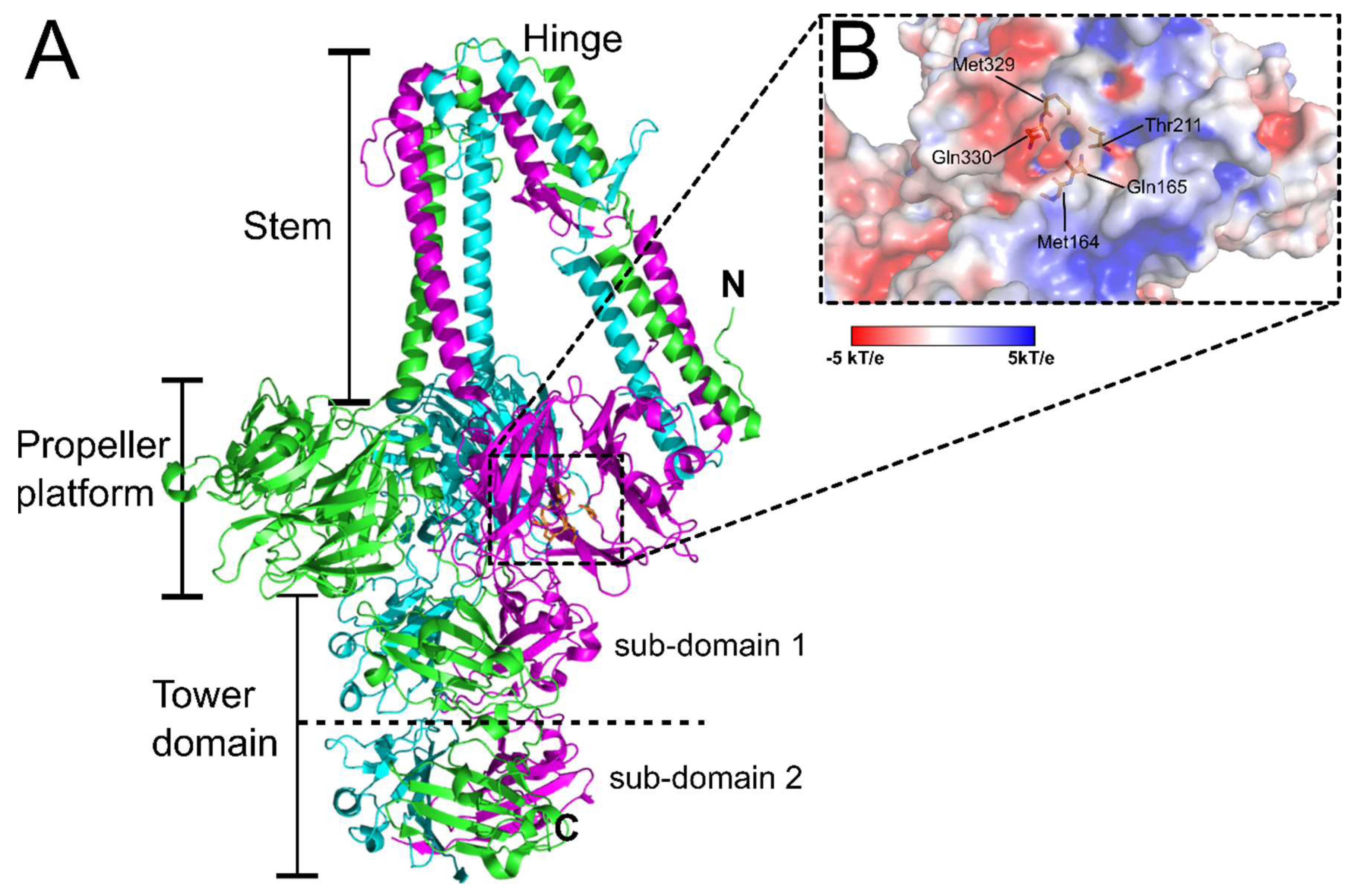
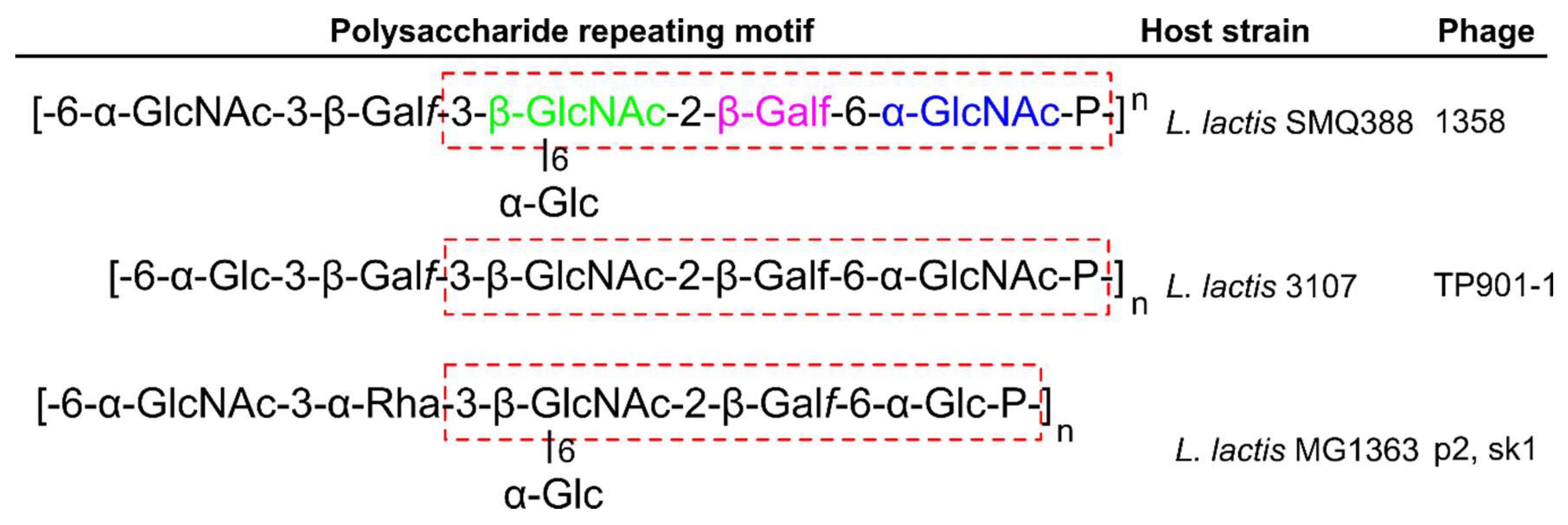
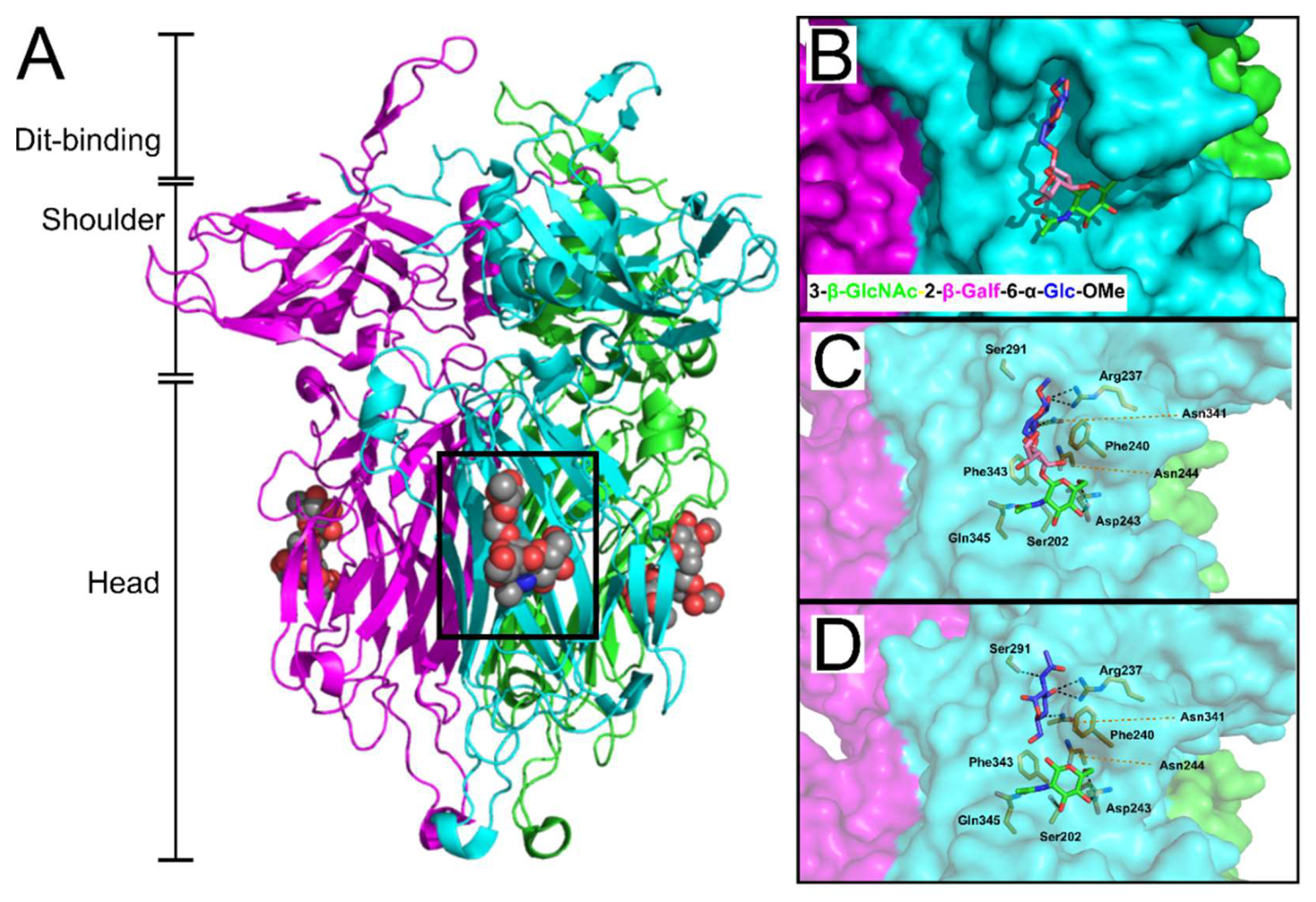
| Phage | Family | Main Host | Receptor(s) | Receptor Details | RBP Known (?) | References |
|---|---|---|---|---|---|---|
| γ | Siphoviridae | Bacillus anthracis | cell-wall-anchored protein | gamma phage receptor (GamR) | - | [92] |
| SPP1 | Siphoviridae | Bacillus subtilis | wall teichoic acid (WTA) and membrane protein | Reversible binding: GlcNAc substituted poly(glycerophosphate) of WTA. Irreversible binding: membrane protein YueB | ORF21 | [14] |
| PBS1 | Siphoviridae | Bacillus subtilis | Flagella | Reversible binding to flagella filament (only to motile cells) | - | [23] |
| LL-H | Siphoviridae | Lactoccus delbrueckii | LTA | Reversible binding: Glucose substituted LTA. Irreversible binding: negatively charged glycerol phosphate groups of LTA | SP58 (g71) | [93] |
| c2 | Lactoccus lactis | membrane protein and PG | Reversible binding: Rhamnose residues of PG. Irreversible binding: membrane “phage infection protein” (PIP) | ORFl10 | [94,95] | |
| p2 | Siphoviridae | Lactoccus lactis | Pellicle | phosphohexasaccharide pellicle | ORF18 | [17] |
| TP901-1 | Siphoviridae | Lactoccus lactis | Pellicle | phosphopentasaccharide pellicle | ORF49 | [18] |
| A118 | Siphoviridae | Listeria monocytogenes | WTA | GlcNAc substituted WTA | ORF20 and ORF19 | [96] |
| P35 | Siphoviridae | Listeria monocytogenes | WTA | GlcNAc and Rhamnose substituted WTA | ORF17 | [96] |
| A511 | Myoviridae | Listeria monocytogenes | WTA and PG | WTA and peptidoglycan | ORF108 | [20] |
| ϕ66 | Podoviridae | Staphylococcus aureus | WTA | β-O-GlcNAc substituted WTA | - | [24] |
| ϕ11 | Siphoviridae | Staphylococcus aureus | WTA and PG | α- or β-GlcNAc substituted WTA and 6-O-acetylated MurNAc of PG | ORF45 and ORF50 (tail fiber) | [21,67] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dunne, M.; Hupfeld, M.; Klumpp, J.; Loessner, M.J. Molecular Basis of Bacterial Host Interactions by Gram-Positive Targeting Bacteriophages. Viruses 2018, 10, 397. https://doi.org/10.3390/v10080397
Dunne M, Hupfeld M, Klumpp J, Loessner MJ. Molecular Basis of Bacterial Host Interactions by Gram-Positive Targeting Bacteriophages. Viruses. 2018; 10(8):397. https://doi.org/10.3390/v10080397
Chicago/Turabian StyleDunne, Matthew, Mario Hupfeld, Jochen Klumpp, and Martin J. Loessner. 2018. "Molecular Basis of Bacterial Host Interactions by Gram-Positive Targeting Bacteriophages" Viruses 10, no. 8: 397. https://doi.org/10.3390/v10080397
APA StyleDunne, M., Hupfeld, M., Klumpp, J., & Loessner, M. J. (2018). Molecular Basis of Bacterial Host Interactions by Gram-Positive Targeting Bacteriophages. Viruses, 10(8), 397. https://doi.org/10.3390/v10080397





