The Odd “RB” Phage—Identification of Arabinosylation as a New Epigenetic Modification of DNA in T4-Like Phage RB69
Abstract
1. Introduction
2. Materials and Methods
2.1. Purification of RB69 DNA
2.2. Biochemical Analyses of RB69 DNA
2.3. Bioinformatic Analyses of Glucosyltransferases in T4-Related Phages
2.4. Cloning of the RB69 ORF053c–ORF052c Gene Region
3. Results
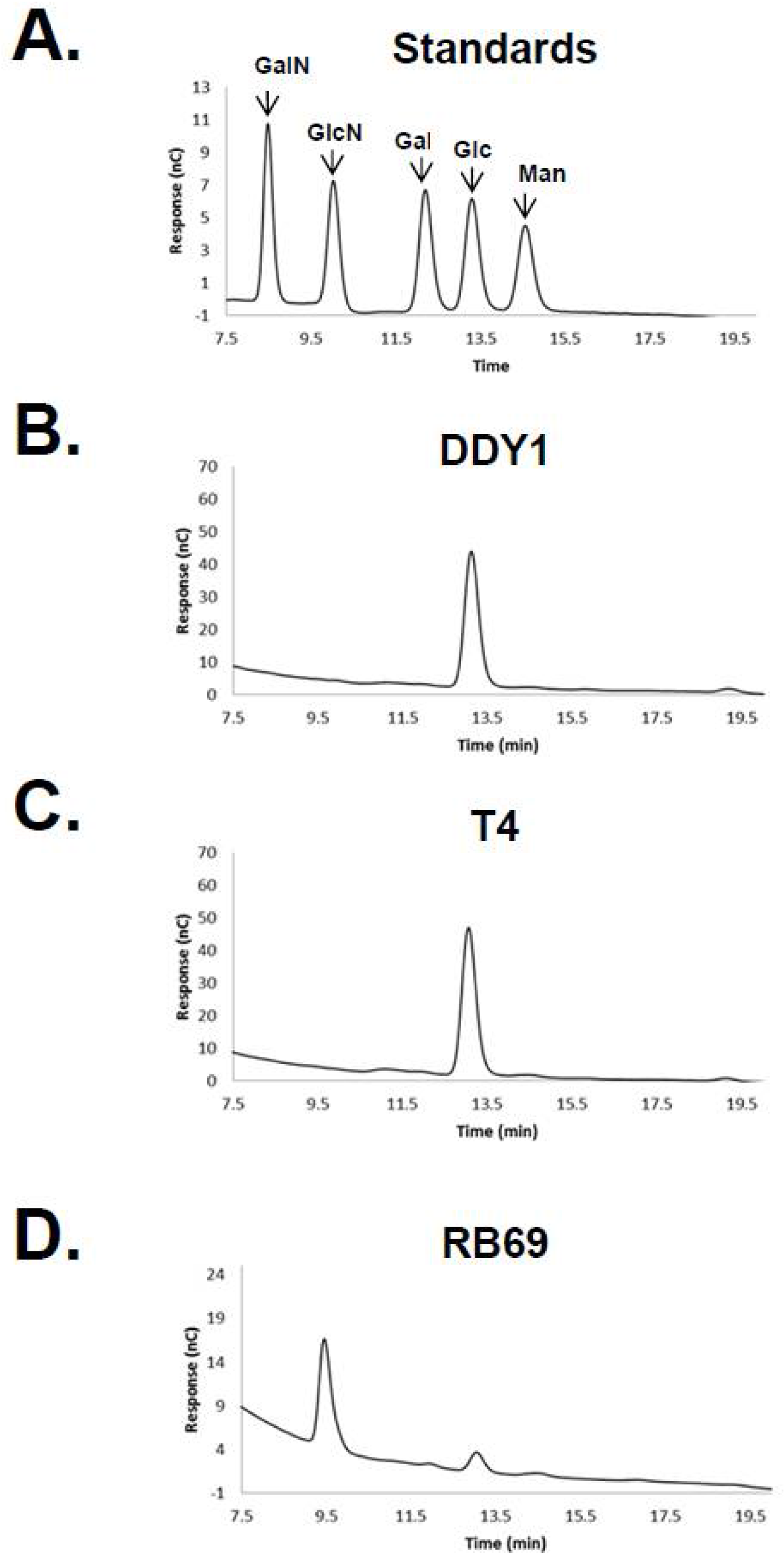
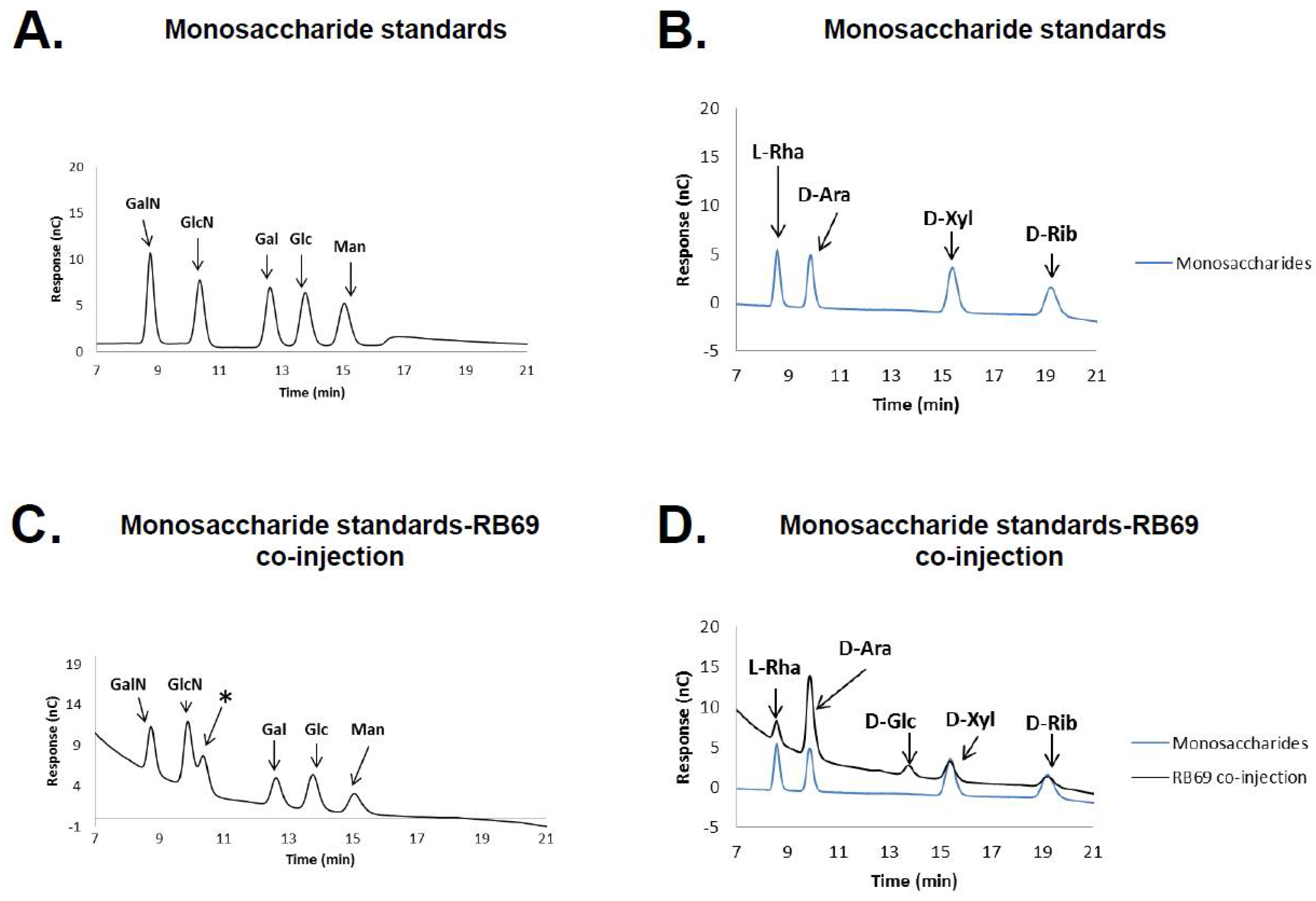
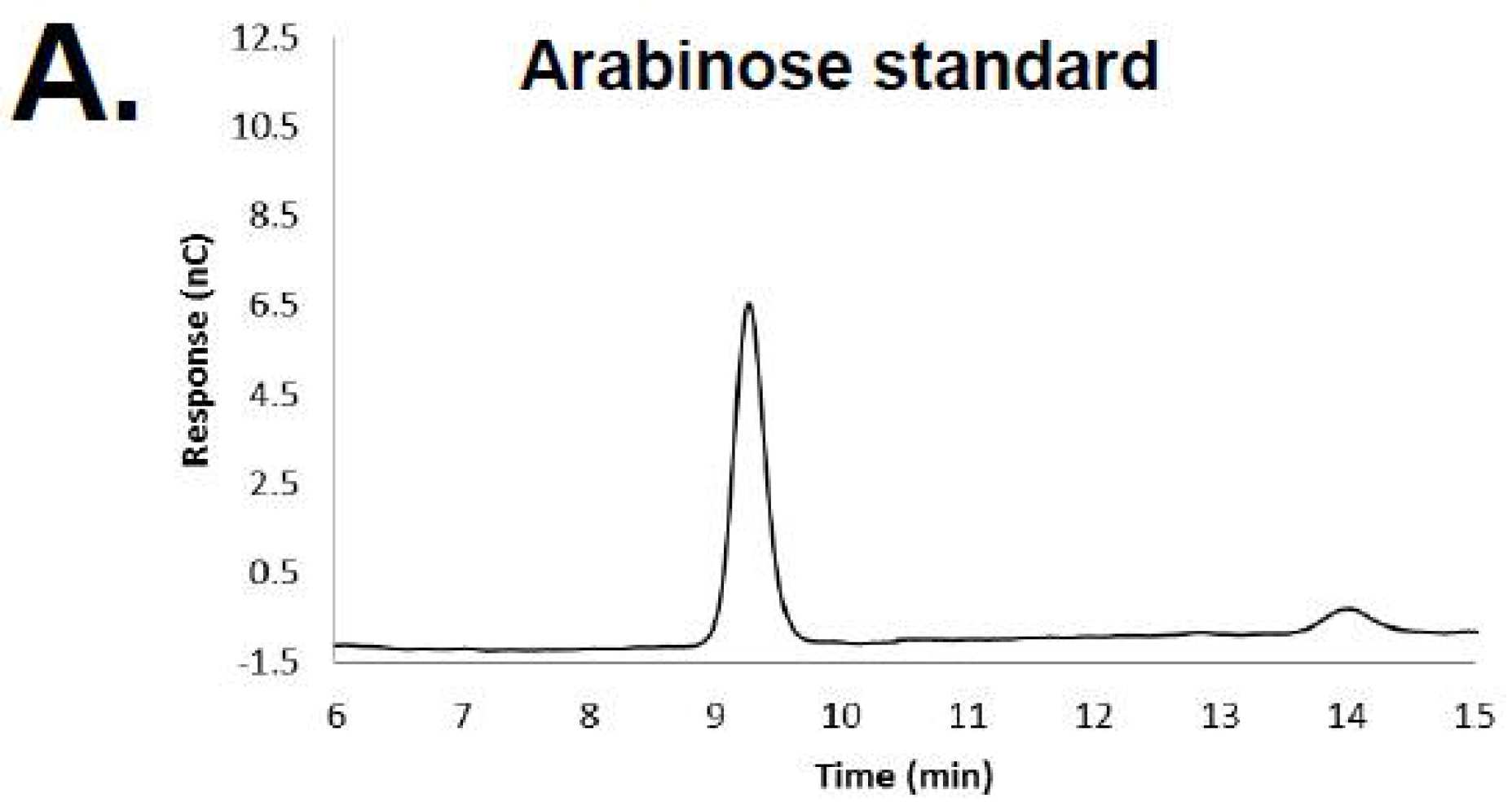
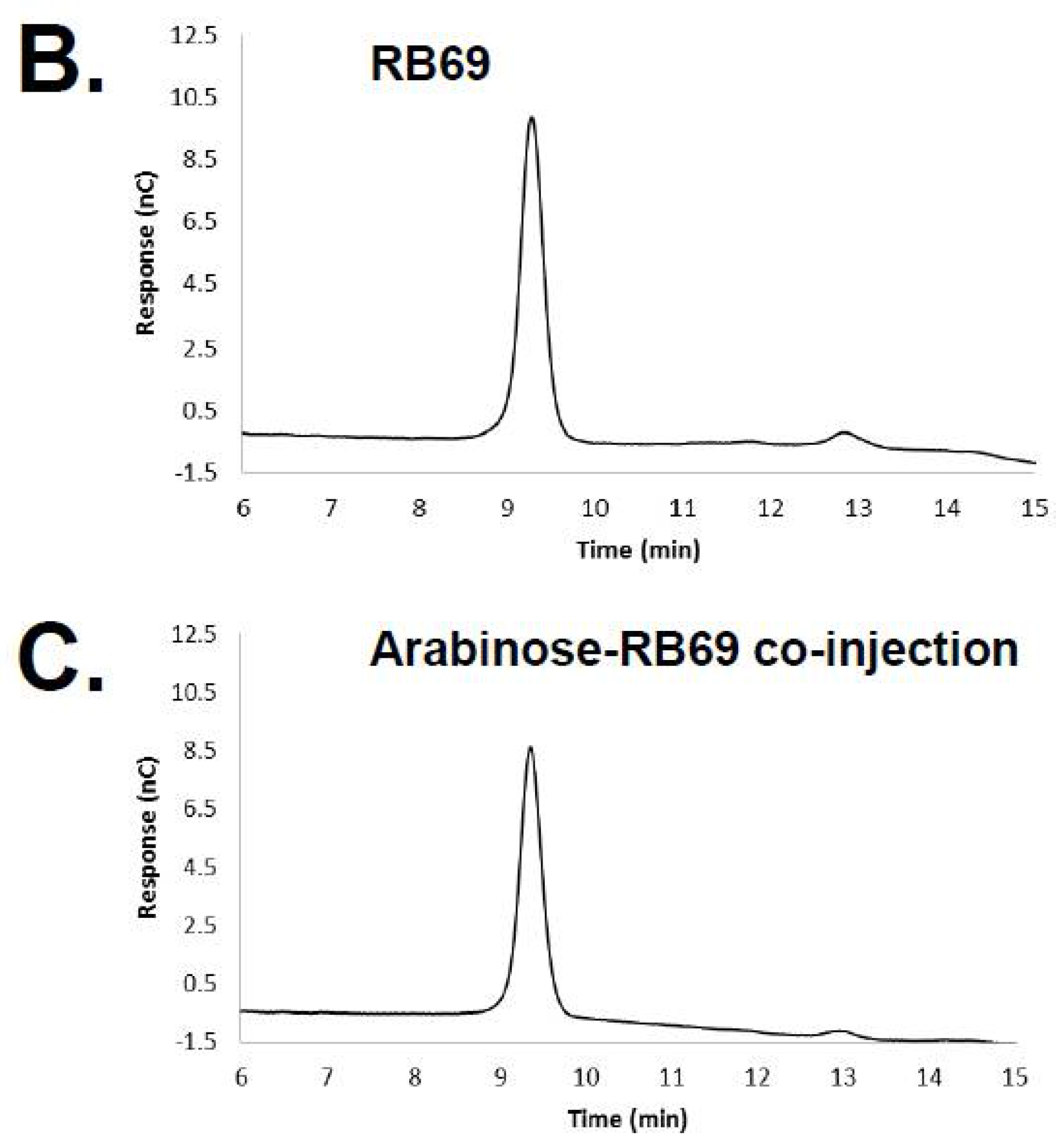
3.1. HPAEC-PAD and Mass Spectrometry of RB69 DNA Indicates It Contains Arabinose
3.2. Homologs of T-Even Glucosyltranferases in T4-Related and the “RB” Phages
3.3. Identification of RB69 ORF003c as a Putative Arabinosyltransferase
3.4. Identification of Candidate RB69 Genes for UDP-Arabinose Generation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Luria, S.E.; Human, M.L. A nonhereditary, host-induced variation of bacterial viruses. J. Bacteriol. 1952, 64, 557–569. [Google Scholar] [PubMed]
- Wyatt, G.R.; Cohen, S.S. The bases of the nucleic acids of some bacterial and animal viruses: The occurrence of 5-hydroxymethylcytosine. Biochem. J. 1953, 55, 774–782. [Google Scholar] [CrossRef] [PubMed]
- Miller, E.S.; Kutter, E.; Mosig, G.; Arisaka, F.; Kunisawa, T.; Ruger, W. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 2003, 67, 86–156. [Google Scholar] [CrossRef] [PubMed]
- JKutter, E.; Stidham, T.; Guttman, B.; Kutter, E.; Batts, D.; Peterson, S.; Djavakhishvili, T.; Arisaka, F.; Mesyanzhinov, V.; Ruger, W.; et al. Genomic map of bacteriophage T4. In Molecular Biology of Bacteriophage T4; Karam, J.D., Ed.; ASM Press: Washington, DC, USA, 1994; pp. 491–519. [Google Scholar]
- Loenen, W.A.; Dryden, D.T.; Raleigh, E.A.; Wilson, G.G.; Murray, N.E. Highlights of the DNA cutters: A short history of the restriction enzymes. Nucleic Acids Res. 2014, 42, 3–19. [Google Scholar] [CrossRef] [PubMed]
- Bair, C.L.; Black, L.W. A type IV modification dependent restriction nuclease that targets glucosylated hydroxymethyl cytosine modified dnas. J. Mol. Biol. 2007, 366, 768–778. [Google Scholar] [CrossRef] [PubMed]
- Bair, C.L.; Rifat, D.; Black, L.W. Exclusion of glucosyl-hydroxymethylcytosine DNA containing bacteriophages is overcome by the injected protein inhibitor IPI*. J. Mol. Biol. 2007, 366, 779–789. [Google Scholar] [CrossRef] [PubMed]
- Bryson, A.L.; Hwang, Y.; Sherrill-Mix, S.; Wu, G.D.; Lewis, J.D.; Black, L.; Clark, T.A.; Bushman, F.D. Covalent modification of bacteriophage T4 DNA inhibits CRISPR-Cas9. MBio 2015, 6, e00648. [Google Scholar] [CrossRef] [PubMed]
- Vlot, M.; Houkes, J.; Lochs, S.J.A.; Swarts, D.C.; Zheng, P.; Kunne, T.; Mohanraju, P.; Anders, C.; Jinek, M.; van der Oost, J.; et al. Bacteriophage DNA glucosylation impairs target DNA binding by type I and II but not by type V CRISPR-Cas effector complexes. Nucleic Acids Res. 2018, 46, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Weigele, P.; Raleigh, E.A. Biosynthesis and function of modified bases in bacteria and their viruses. Chem. Rev. 2016, 116, 12655–12687. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, G.R.; He, P.; Hilfinger, J.; Tseng, M.-J. Deoxyribonucleoside triphosphate synthesis and phage T4 DNA replication. In Molecular Biology of Bacteriophage T4; Karam, J.D., Ed.; ASM Press: Washington, DC, USA, 1994; pp. 14–27. [Google Scholar]
- Lamm, N.; Wang, Y.; Mathews, C.K.; Ruger, W. Deoxycytidylate hydroxymethylase gene of bacteriophage T4. Nucleotide sequence determination and over-expression of the gene. Eur. J. Biochem. 1988, 172, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Larivière, L.; Moréra, S. A base-flipping mechanism for the T4 phage β-glucosyltransferase and identification of a transition-state analog. J. Mol. Biol. 2002, 324, 483–490. [Google Scholar] [CrossRef]
- Gold, L.M.; Schweiger, M. Synthesis of phage-specific alpha- and beta-glucosyl transferases directed by T-even DNA in vitro. Proc. Natl. Acad. Sci. USA 1969, 62, 892–898. [Google Scholar] [CrossRef] [PubMed]
- Lariviere, L.; Sommer, N.; Morera, S. Structural evidence of a passive base-flipping mechanism for agt, an unusual GT-B glycosyltransferase. J. Mol. Biol. 2005, 352, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Lehman, I.R.; Pratt, E.A. On the structure of the glucosylated hydroxymethylcytosine nucleotides of coliphages T2, T4, and T6. J. Biol. Chem. 1960, 235, 3254–3259. [Google Scholar] [PubMed]
- Sommer, N.; Depping, R.; Piotrowski, M.; Ruger, W. Bacteriophage T4 alpha-glucosyltransferase: A novel interaction with gp45 and aspects of the catalytic mechanism. Biochem. Biophys. Res. Commun. 2004, 323, 809–815. [Google Scholar] [CrossRef] [PubMed]
- Carlson, K.; Raleigh, E.A.; Hattman, S. Restriction and modification. In Molecular Biology of Bacteriophage T4; Karam, J.D., Ed.; ASM Press: Washington, DC, USA, 1994; pp. 369–381. [Google Scholar]
- Russell, R.L. Speciation among the T-Even Bacteriophages; California Institute of Techonology: Pasadena, CA, USA, 1967. [Google Scholar]
- Petrov, V.; Ratnayaka, S.; Nolan, J.; Miller, E.; Karam, J. Genomes of the T4-related bacteriophages as windows on microbial genome evolution. Virol. J. 2010, 7, 292. [Google Scholar] [CrossRef] [PubMed]
- Rifat, D.; Wright, N.T.; Varney, K.M.; Weber, D.J.; Black, L.W. Restriction endonuclease inhibitor IPI* of bacteriophage T4: A novel structure for a dedicated target. J. Mol. Biol. 2008, 375, 720–734. [Google Scholar] [CrossRef] [PubMed]
- Revel, H.R.; Luria, S.E. DNA-glucosylation in T-even phage: Genetic determination and role in phagehost interaction. Annu. Rev. Genet. 1970, 4, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped blast and psi-blast: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Hughey, R.; Krogh, A. Hidden markov models for sequence analysis: Extension and analysis of the basic method. Comput. Appl. Biosci. 1996, 12, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Karplus, K.; Barrett, C.; Hughey, R. Hidden markov models for detecting remote protein homologies. Bioinformatics 1998, 14, 846–856. [Google Scholar] [CrossRef] [PubMed]
- Söding, J. Protein homology detection by HMM-HMM comparison. Bioinformatics 2005, 21, 951–960. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845. [Google Scholar] [CrossRef] [PubMed]
- Qiu, D.; Damron, F.H.; Mima, T.; Schweizer, H.P.; Yu, H.D. Pbad-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl. Environ. Microbiol. 2008, 74, 7422–7426. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.C. High-performance anion-exchange chromatography for carbohydrate analysis. Anal. Biochem. 1990, 189, 151–162. [Google Scholar] [CrossRef]
- Yaung, S.J.; Esvelt, K.M.; Church, G.M. Complete genome sequences of T4-like bacteriophages RB3, RB5, RB6, RB7, RB9, RB10, RB27, RB33, RB55, RB59, and RB68. Genome Announc. 2015, 3, e01122. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.A.; Weintraub, S.T.; Wu, W.; Winkler, D.C.; Cheng, N.; Steven, A.C.; Black, L.W. Extensive proteolysis of head and inner body proteins by a morphogenetic protease in the giant Pseudomonas aeruginosa phage φkz. Mol. Microbiol. 2012, 84, 324–339. [Google Scholar] [CrossRef] [PubMed]
- Hardies, S.C.; Thomas, J.A.; Serwer, P. Comparative genomics of Bacillus thuringiensis phage 0305phi8-36: Patterns of descent in a novel ancient phage lineage. J. Mol. Biol. 2007, 4, 97. [Google Scholar]
- Thomas, J.A.; Hardies, S.C.; Rolando, M.; Hayes, S.J.; Lieman, K.; Carroll, C.A.; Weintraub, S.T.; Serwer, P. Complete genomic sequence and mass spectrometric analysis of highly diverse, atypical Bacillus thuringiensis phage 0305φ8-36. Virology 2007, 368, 405–421. [Google Scholar] [CrossRef] [PubMed]
- Söding, J.; Biegert, A.; Lupas, A.N. The hhpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005, 33, W244–W248. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Rudd, K.E. Ecogene 3.0. Nucleic Acids Res. 2013, 41, D613–D624. [Google Scholar] [CrossRef] [PubMed]
- Meredith, T.C.; Woodard, R.W. Identification of gutq from Escherichia coli as a d-arabinose 5-phosphate isomerase. J. Bacteriol. 2005, 187, 6936–6942. [Google Scholar] [CrossRef] [PubMed]
- Iyer, L.M.; Zhang, D.; Burroughs, A.M.; Aravind, L. Computational identification of novel biochemical systems involved in oxidation, glycosylation and other complex modifications of bases in DNA. Nucleic Acids Res. 2013, 41, 7635–7655. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Hull, V.; Thomas, J.A.; Fu, X.; Gidwani, S.; Gupta, Y.K.; Black, L.W.; Xu, S.Y. Expression and purification of a single-chain type IV restriction enzyme eco94gmrsd and determination of its substrate preference. Sci. Rep. 2015, 5, 9747. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Lee, S.G.; Bar-Peled, M. Biosynthesis of UDP-xylose and UDP-arabinose in sinorhizobium meliloti 1021: First characterization of a bacterial UDP-xylose synthase, and UDP-xylose 4-epimerase. Microbiology 2011, 157, 260–269. [Google Scholar] [CrossRef] [PubMed]
- Trent, M.S.; Ribeiro, A.A.; Lin, S.; Cotter, R.J.; Raetz, C.R.H. An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-l-arabinose to lipid a: Induction in polymyxin-resistant mutants and role of a novel lipid-linked donor. J. Biol. Chem. 2001, 276, 43122–43131. [Google Scholar] [CrossRef] [PubMed]
- Iyer, L.M.; Zhang, D.; de Souza, R.F.; Pukkila, P.J.; Rao, A.; Aravind, L. Lineage-specific expansions of tet/jbp genes and a new class of DNA transposons shape fungal genomic and epigenetic landscapes. Proc. Natl. Acad. Sci. USA 2014, 111, 1676–1683. [Google Scholar] [CrossRef] [PubMed]
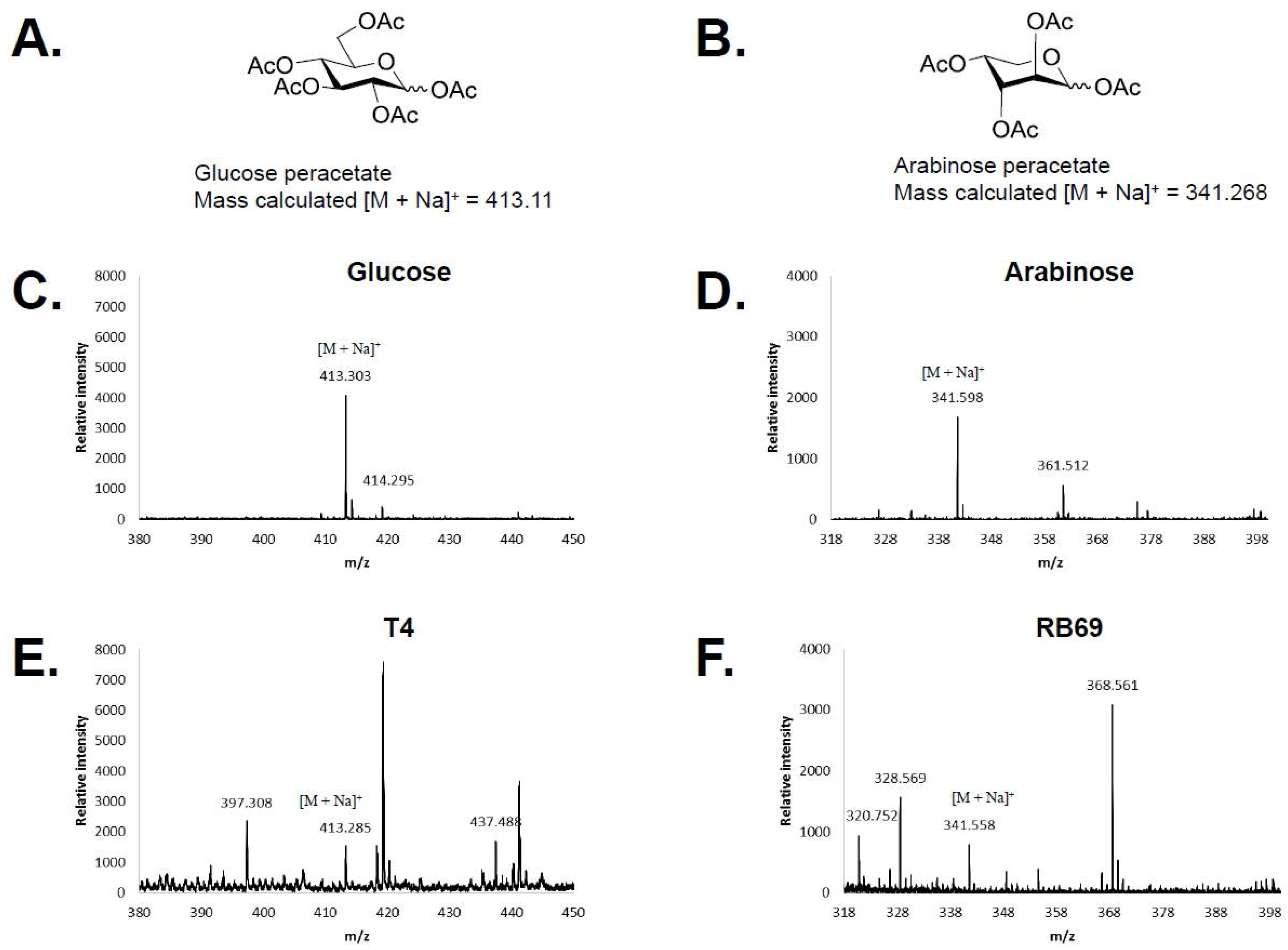
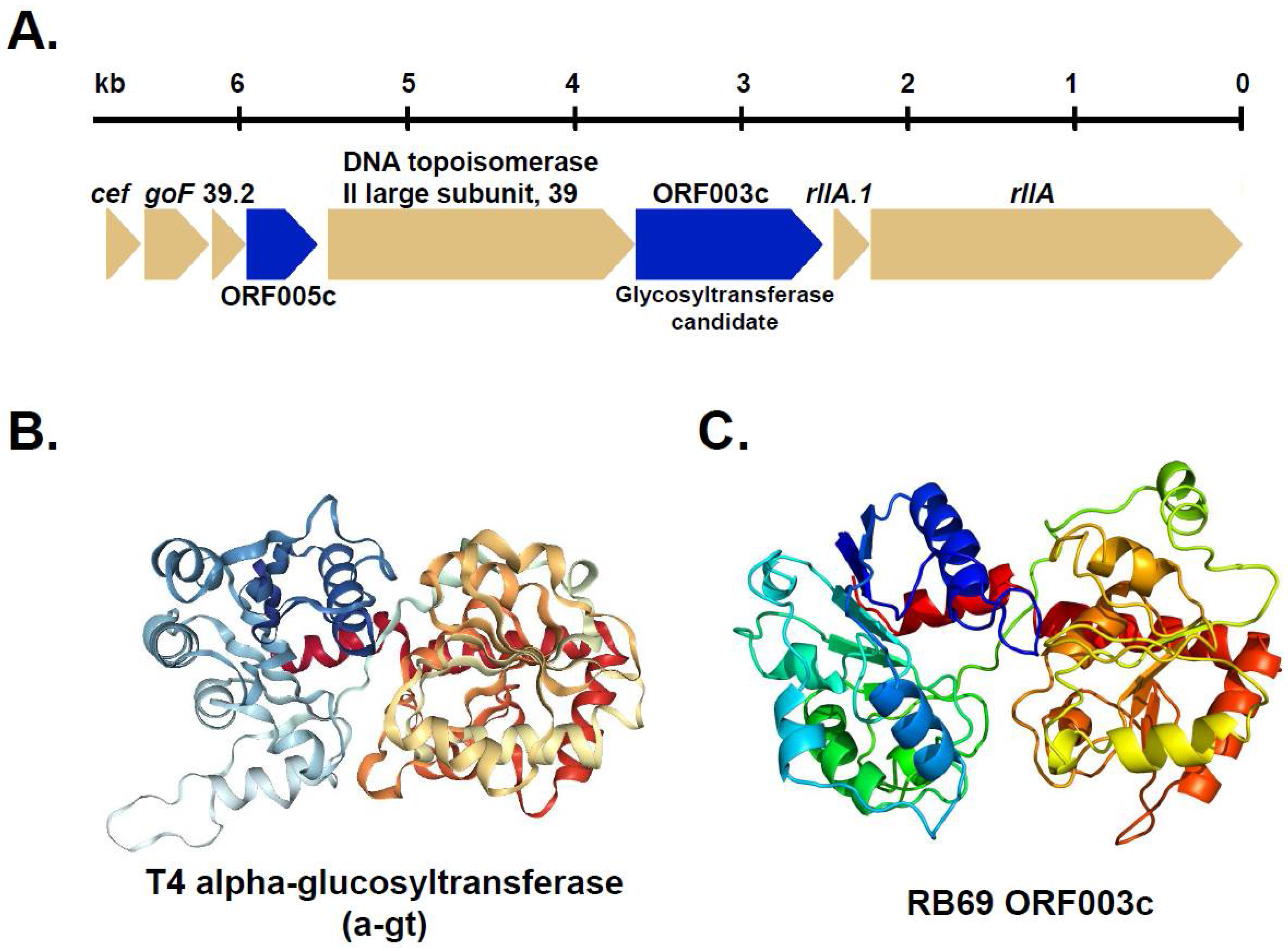

| Phage | T4 a-gt (400 aa) | T4 b-gt (351 aa) | T2 ba-gt (280 aa) | |||
|---|---|---|---|---|---|---|
| Protein Accession | Match Identity, % | Protein Accession | Match Identity, % | Protein Accession/Name | Match Identity, % | |
| T4 | NP_049673.1 | 100 | NP_049658.1 | 100 | n/a | n/a |
| T2 1 | a-gt_T2 | 91 | n/a | n/a | Q06717.1 | 100 |
| T6 1 | a-gt_T6 | 99 | n/a | n/a | Q06718.1 | 98 |
| RB3 | YP_009098445 | 91 | n/a | n/a | YP_009098428.1 | 99 |
| RB5 | AIT73068.1 | 91 | n/a | n/a | AIT73051.1 | 99 |
| RB6 | AIT73339.1 | 91 | n/a | n/a | AIT73322.1 | 99 |
| RB7 | AIT73610.1 | 91 | n/a | n/a | AIT73593.1 | 99 |
| RB9 | AIT73882.1 | 91 | n/a | n/a | AIT73865.1 | 99 |
| RB10 | AIT74154.1 | 91 | n/a | n/a | AIT74137.1 | 99 |
| RB14 | YP_002854395.1 | 92 | n/a | n/a | YP_002854377.1 | 99 |
| RB27 | YP_009102266.1 | 91 | n/a | n/a | YP_009102248.1 | 99 |
| RB32 | YP_803002.1 | 91 | n/a | n/a | YP_802983.1 | 98 |
| RB33 | AIT74699.1 | 91 | n/a | n/a | AIT74680.1 | 98 |
| RB51 | YP_002854018.1 | 91 | n/a | n/a | YP_002853998.1 | 99 |
| RB55 | AIT74973.1 | 100 | AIT74956.1 | 100 | n/a | n/a |
| RB59 | AIT75245.1 | 100 | AIT75228.1 | 100 | n/a | n/a |
| RB68 | YP_009167432.1 | 91 | n/a | n/a | YP_009167412.1 | 99 |
| RB69 | n/a | n/a | n/a | n/a | n/a | n/a |
| Examples of homologs in other T4-related phages | ||||||
| Yersinia phage PST | YP_009153660.1 | 99 | n/a | n/a | YP_009153643.1 | 97 |
| Shigella phage Shfl2 | YP_004414960.1 | 91 | n/a | n/a | YP_004414944.1 | 99 |
| Escherichia phage wV7 | YP_007004802.1 | 91 | n/a | n/a | YP_007004785.1 | 99 |
| Escherichia phage AR1 | YP_009167872.1 | 91 | n/a | n/a | YP_009167855.1 | 99 |
| Escherichia phage HY01 | YP_009148507.1 | 91 | n/a | n/a | YP_009148490.1 | 98 |
| E. coli phage e11/2 (slur02) | YP_009210250.1 | 91 | n/a | n/a | YP_009210234.1 | 99 |
| E. coli ACG-C40 | YP_006986613.1 | 94 | n/a | n/a | YP_006986593.1 | 98 |
| Examples of more highly diverged homologs in other phages | ||||||
| Enterobacteria phage CC31 | n/a | n/a | YP_004009897.1 | 49 | YP_004009898.1 | 65 |
| Enterobacter phage PG7 | n/a | n/a | YP_009005302.1 | 49 | YP_009005303.1 | 65 |
| Salmonella phage S16 | n/a | n/a | YP_007501076.1 | 50 | YP_007501077.1 | 64 |
| Salmonella phage STML-198 | n/a | n/a | YP_009148028.1 | 50 | YP_009148029.1 | 64 |
| Citrobacter phage Moon | n/a | n/a | YP_009146477.1 | 50 | YP_009146478.1 | 64 |
| Serratia phage PS2 | n/a | n/a | YP_009030087.1 | 53 | YP_009030088.1 | 48 |
| Salmonella phage STP4-a | n/a | n/a | YP_009126243.1 | 50 | YP_009126244.1 | 64 |
| Citrobacter phage Merlin | n/a | n/a | YP_009203756.1 | 48 | YP_009203757.1 | 64 |
| Bacillus phage G | YP_009015609.1 (gp306) | 24 | n/a | n/a | n/a | n/a |
| Bacillus phage G | YP_009015609.1 (gp313) | 24 | n/a | n/a | n/a | n/a |
| Phage, Protein | Protein Accession | E-Value for Protein When Scored Against 1 | |
|---|---|---|---|
| T4 a-gt HMM | Phage G gp306 HMM | ||
| T4 a-gt | NP_049673.1 | 3.06e-168 * | 3.62e-38 * |
| T4 b-gt | NP_049658.1 | 1.07e-06 | 1.46e-13 |
| Phage G, gp306 | YP_009015609.1 | 9.92e-133 * | 6.19e-46 * |
| Phage G, gp313 | YP_009015609.1 | 1.16e-132 * | 5.30e-45 * |
| RB69, RB69ORF003c | NP_861693.1 | 3.24e-05 | 4.61e-19 |
| RB69, ORF052_53c | Supplementary Figure S3 | 1.82e-02 9.12e+01 2 | 9.85e-01 2.07e+02 2 |
| JS09, JS09_0177 | YP_009037500.1 | 5.45e-05 | 2.32e-18 |
| Shfl125875, BI097_gp055 | YP_009289016 | 4.17e+00 | 1.50e-16 |
| Acj61, Acj61p077 | YP_004009694.1 | 3.50e+00 | 3.37e-11 |
| Acj61, Acj61p076 | YP_004009693.1 | 11.82e-04 | 2.90e-15 |
| Organism | PDB ID | Glycosyltransferase | E-Value for Phage Protein | |||
|---|---|---|---|---|---|---|
| T4 a-gt | T4 b-gt | Phage G gp306 | RB69 ORF003c | |||
| T4 phage | 1XV5_A | DNA alpha-glucosyltransferase | 2.10e-39 | 3.10e-20 | 5.30e-33 | 1.90e-24 |
| Chlorella virus NY2A | 3OY2_A | Glycosyltransferase B736L; GDP-mannose, GT4 | 1.60e-35 | 3.60e-21 | 4.70e-32 | 4.00e-26 |
| Halothermothrix | 2R60_A | glycosyl transferase, group 1 | 3.30e-35 | 8.30e-22 | 8.90e-34 | 5.60e-26 |
| Campylobacter | 6EJI_A | WlaC protein; glycosyltransferase | 4.90e-34 | 2.70e-22 | 1.80e-31 | 2.90e-27 |
| Geobacillus | 2X0D_A | WSAF; GT4 family, transferase | 1.80e-33 | 3.20e-22 | 1.30e-31 | 4.90e-26 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thomas, J.A.; Orwenyo, J.; Wang, L.-X.; Black, L.W. The Odd “RB” Phage—Identification of Arabinosylation as a New Epigenetic Modification of DNA in T4-Like Phage RB69. Viruses 2018, 10, 313. https://doi.org/10.3390/v10060313
Thomas JA, Orwenyo J, Wang L-X, Black LW. The Odd “RB” Phage—Identification of Arabinosylation as a New Epigenetic Modification of DNA in T4-Like Phage RB69. Viruses. 2018; 10(6):313. https://doi.org/10.3390/v10060313
Chicago/Turabian StyleThomas, Julie A., Jared Orwenyo, Lai-Xi Wang, and Lindsay W. Black. 2018. "The Odd “RB” Phage—Identification of Arabinosylation as a New Epigenetic Modification of DNA in T4-Like Phage RB69" Viruses 10, no. 6: 313. https://doi.org/10.3390/v10060313
APA StyleThomas, J. A., Orwenyo, J., Wang, L.-X., & Black, L. W. (2018). The Odd “RB” Phage—Identification of Arabinosylation as a New Epigenetic Modification of DNA in T4-Like Phage RB69. Viruses, 10(6), 313. https://doi.org/10.3390/v10060313





