Comparison of Porcine Airway and Intestinal Epithelial Cell Lines for the Susceptibility and Expression of Pattern Recognition Receptors upon Influenza Virus Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Culture Conditions
2.2. Phenotyping of MK1-OSU Cells by Immunohistochemistry
2.3. Lectin Binding Assay
2.4. Culture of Swine and Human Influenza Virus and Infection of Cells
2.5. Indirect Immunoflourescence Assay for Virus Detection
2.6. Determination of Percentage of MK1-OSU Cells Infected Using Flow Cytometry
2.7. Growth Kinetics of Influenza Viruses in MK1-OSU, SD-PJEC and MDCK Cells
2.8. Estimation of Pattern Recognition Receptors (PRRs) Using Flow Cytometry
2.9. Real-Time RT-PCR Assays for Pattern-Recognition Receptors (PRRs)
2.10. Statistical Analysis
3. Results
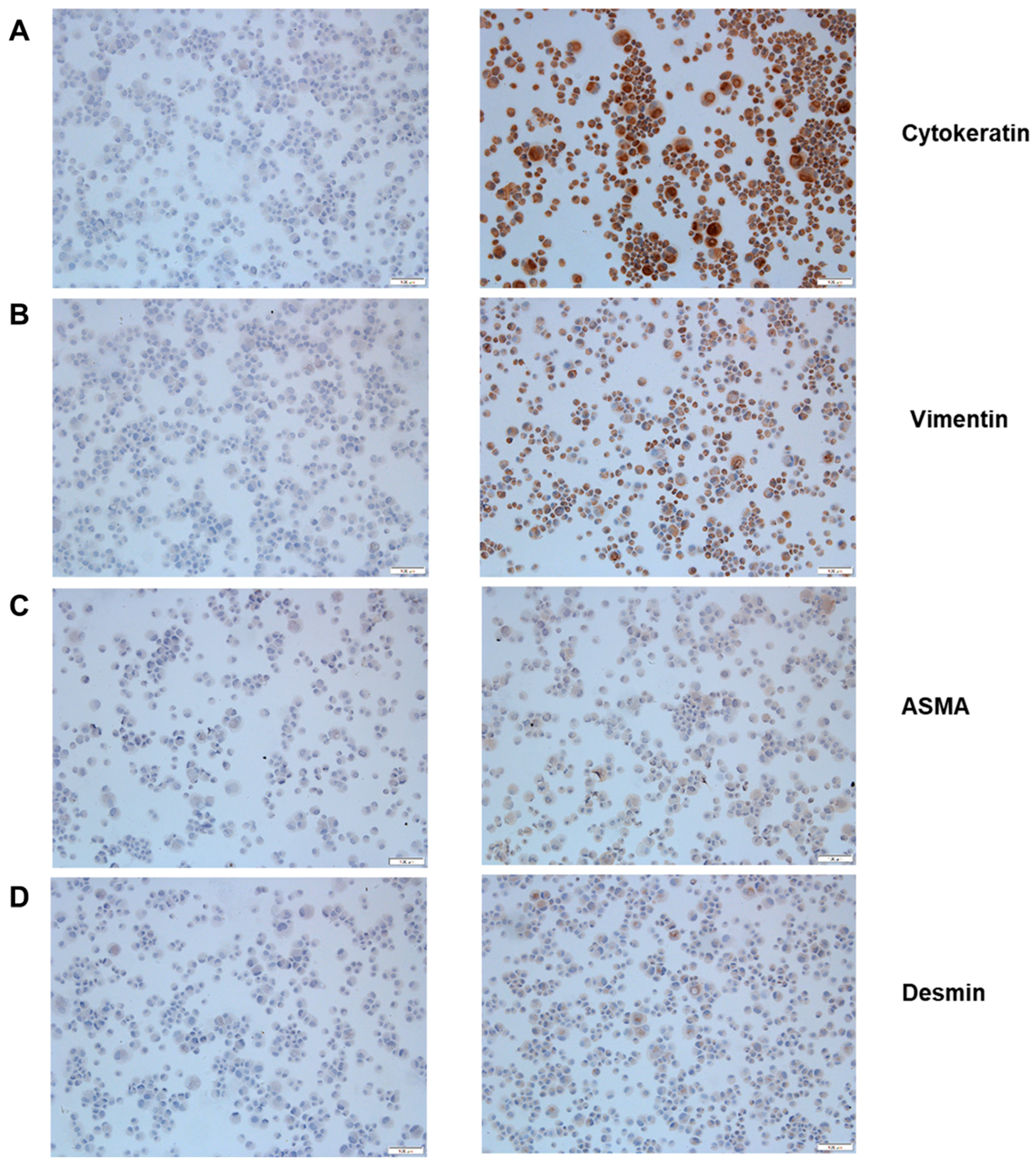
3.1. MK1-OSU Cells are Phenotypically Epithelial Cells
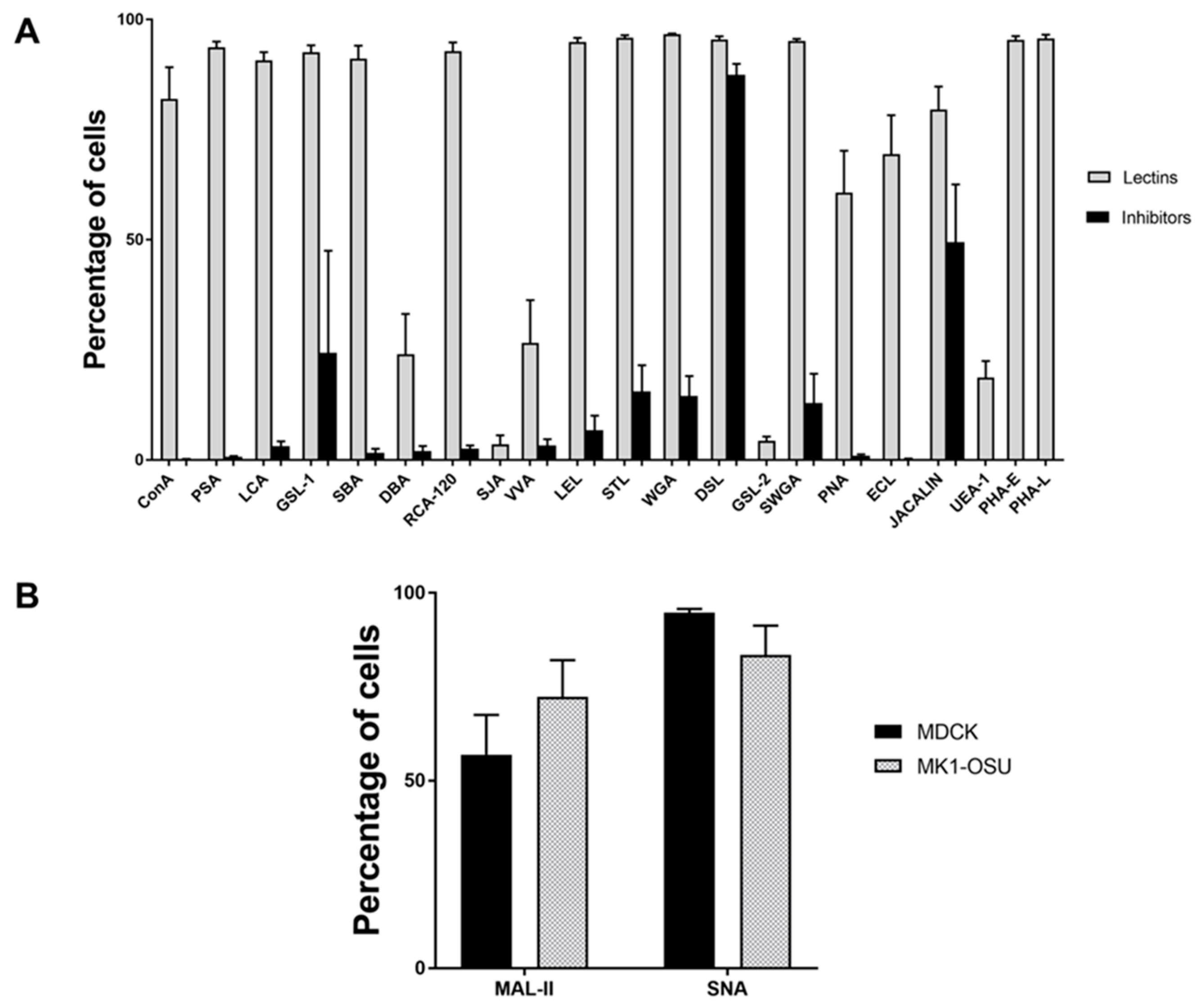
3.2. Lectin Binding Profile of MK1-OSU Cells Indicated the Presence of Heterogeneous Cell Surface Sugar Moieties
3.3. MK1-OSU Cells Expressed Sial2-3Gal and Sial2-6Gal Receptors
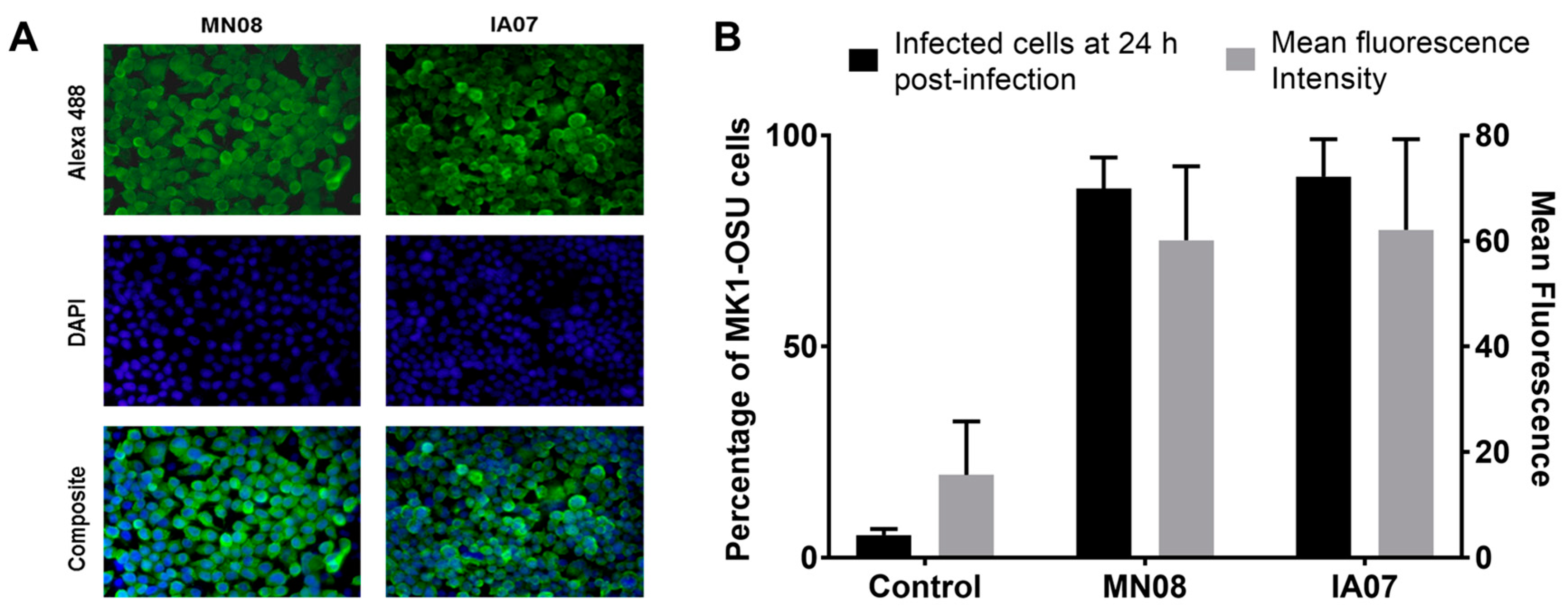
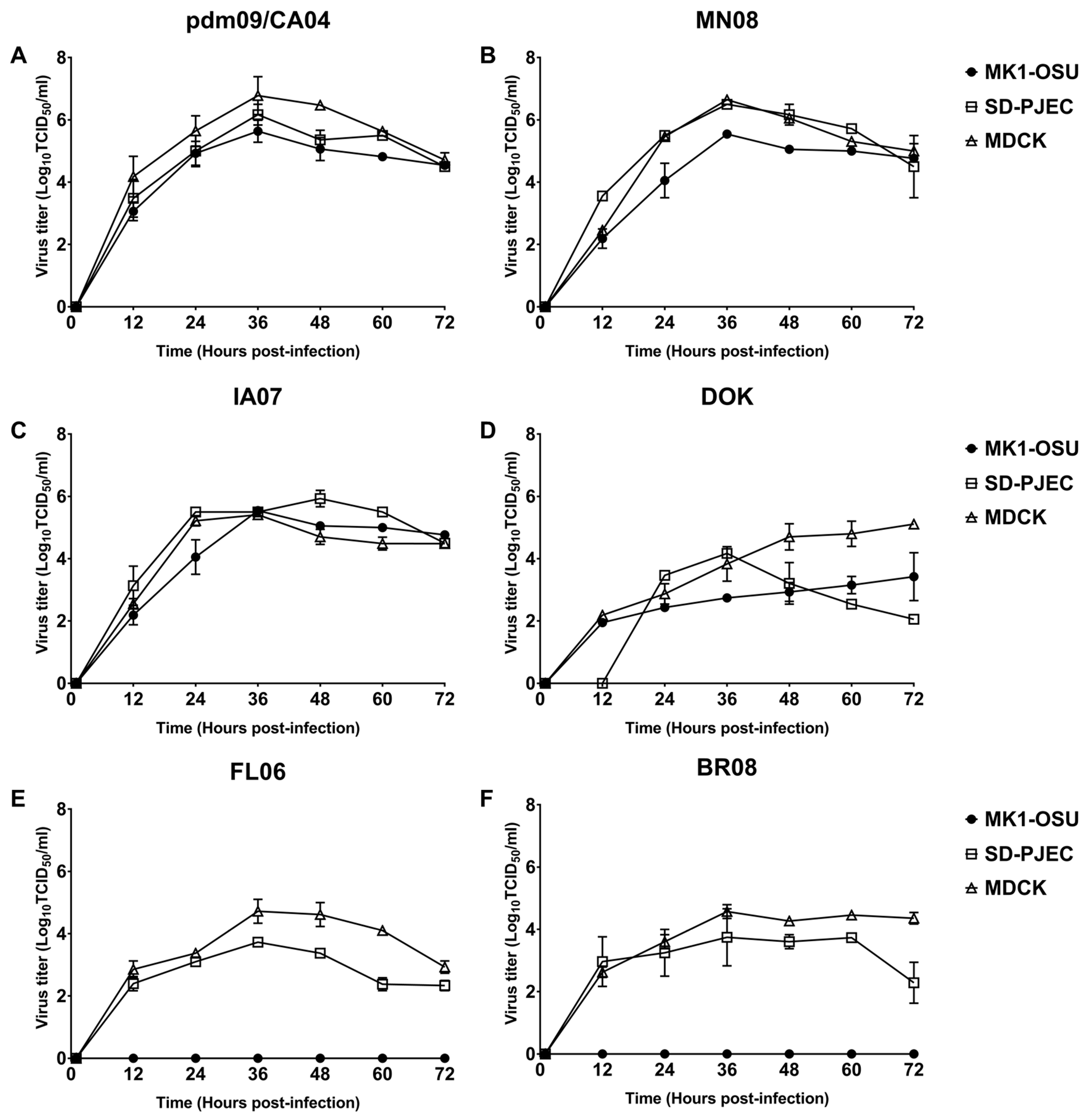
3.4. MK1-OSU Cells Were Susceptible to SIV Infection
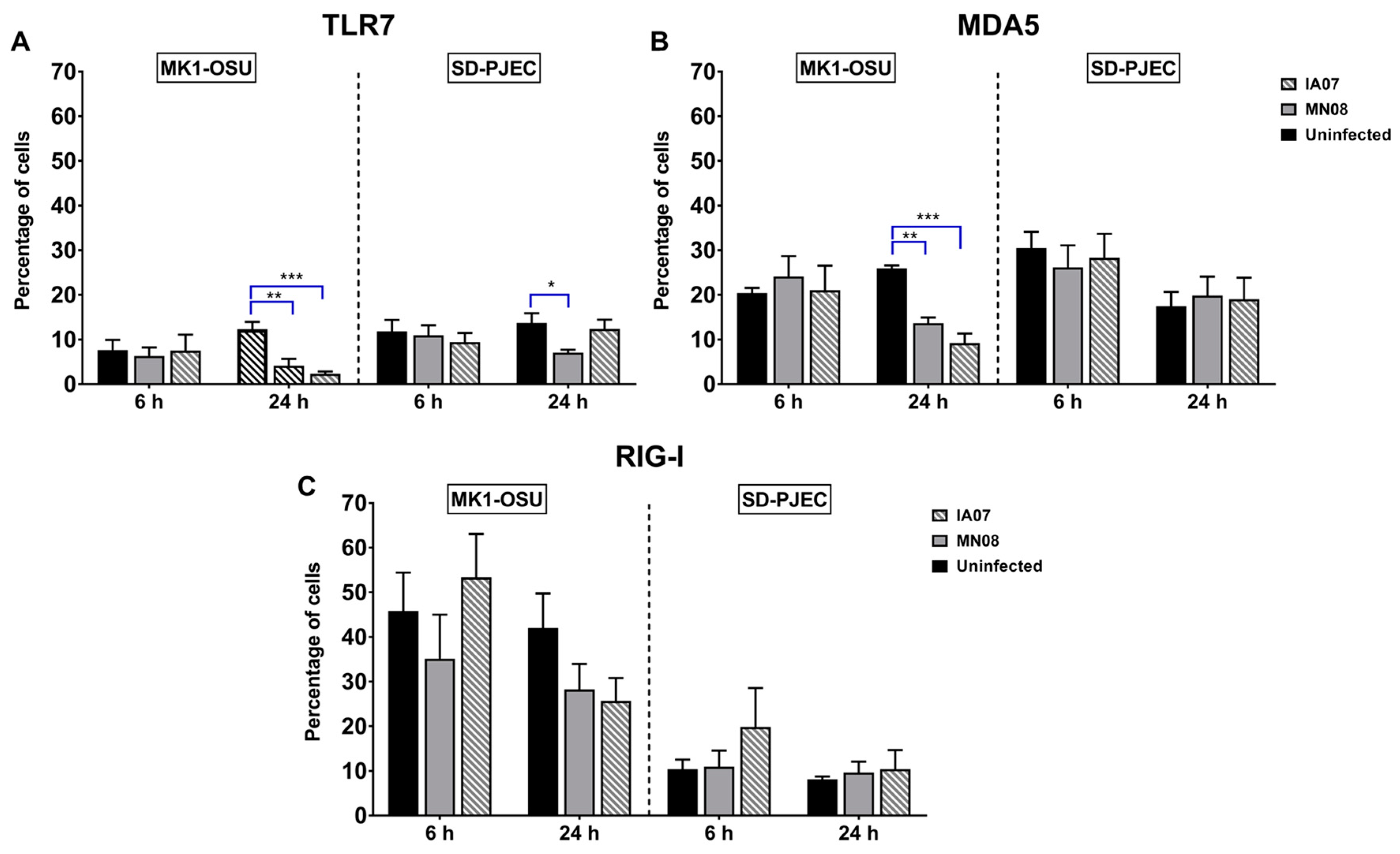
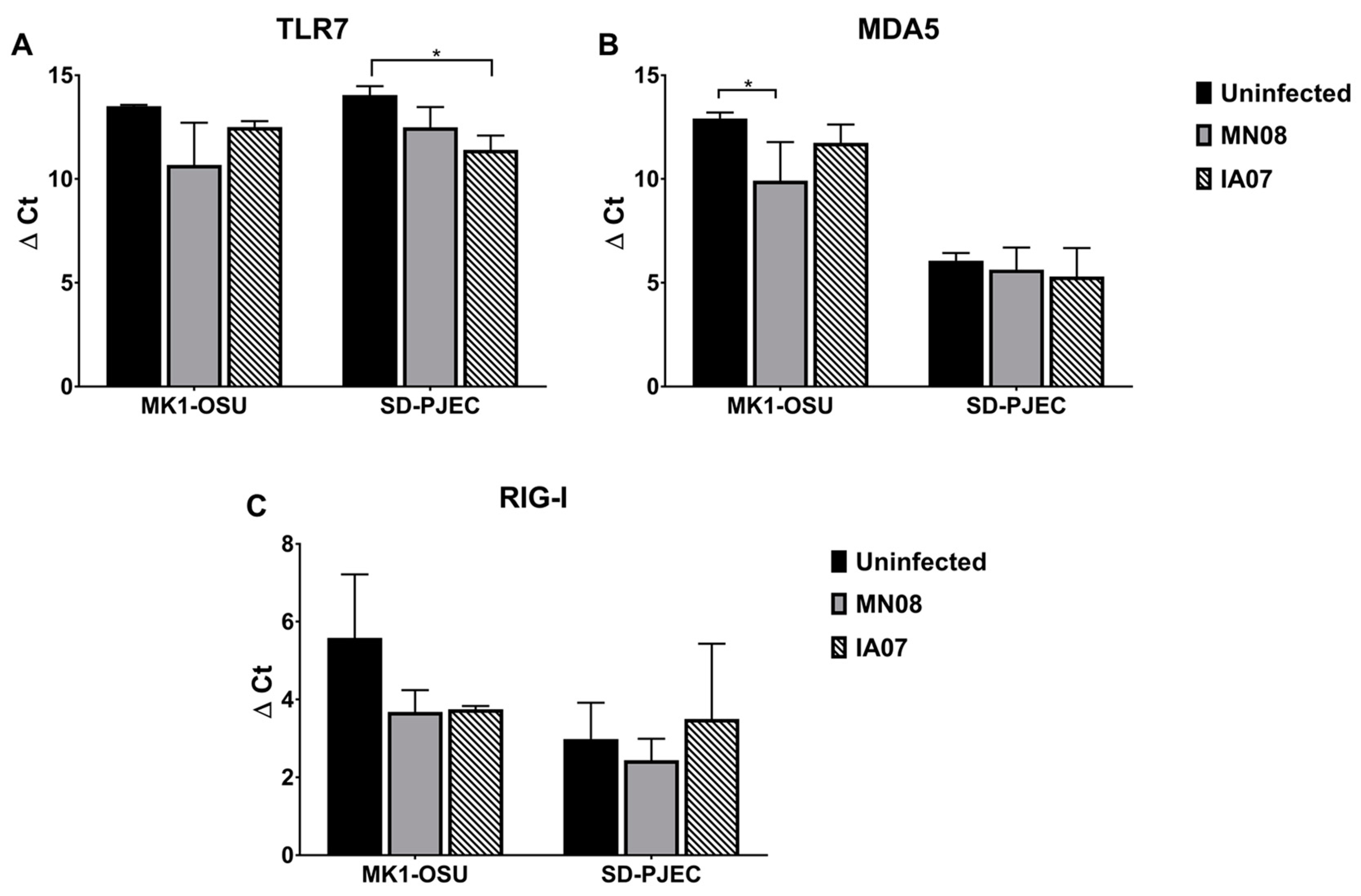
3.5. Influenza Infection Differentially Affected the Expression of TLRs and RLRs in MK1-OSU and SD-PJEC Cells at 24 h
4. Discussion
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Vincent, A.L.; Ma, W.; Lager, K.M.; Janke, B.H.; Richt, J.A. Swine influenza viruses A North American perspective. Adv. Virus Res. 2008, 72, 127–154. [Google Scholar] [PubMed]
- Brown, I.H. History and epidemiology of swine influenza in Europe. Curr. Top. Microbiol. Immunol. 2013, 370, 133–146. [Google Scholar] [PubMed]
- Nelli, R.K.; Kuchipudi, S.V.; White, G.A.; Perez, B.B.; Dunham, S.P.; Chang, K.C. Comparative distribution of human and avian type sialic acid influenza receptors in the pig. BMC Vet. Res. 2010, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Takatsy, G.; Farkas, E.; Romvary, J. Susceptibility of the domestic pig to influenza B virus. Nature 1969, 222, 184–185. [Google Scholar] [CrossRef] [PubMed]
- Ran, Z.; Shen, H.; Lang, Y.; Kolb, E.A.; Turan, N.; Zhu, L.; Ma, J.; Bawa, B.; Liu, Q.; Liu, H.; et al. Domestic pigs are susceptible to infection with influenza B viruses. J. Virol. 2015, 89, 4818–4826. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Abiko, C.; Peng, G.; Muraki, Y.; Sugawara, K.; Hongo, S.; Kitame, F.; Mizuta, K.; Numazaki, Y.; Suzuki, H.; et al. Interspecies transmission of influenza C virus between humans and pigs. Virus Res. 1997, 48, 71–79. [Google Scholar] [CrossRef]
- Guo, Y.J.; Jin, F.G.; Wang, P.; Wang, M.; Zhu, J.M. Isolation of influenza C virus from pigs and experimental infection of pigs with influenza C virus. J. Gen. Virol. 1983, 64 Pt 1, 177–182. [Google Scholar] [PubMed]
- Hause, B.M.; Ducatez, M.; Collin, E.A.; Ran, Z.; Liu, R.; Sheng, Z.; Armien, A.; Kaplan, B.; Chakravarty, S.; Hoppe, A.D.; et al. Isolation of a novel swine influenza virus from oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog. 2013, 9, e1003176. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Fu, X.; Li, G.; Kerlin, F.; Veit, M. Novel influenza D virus: Epidemiology, pathology, evolution and biological characteristics. Virulence 2017, 8, 1580–1591. [Google Scholar] [CrossRef] [PubMed]
- Collin, E.A.; Sheng, Z.; Lang, Y.; Ma, W.; Hause, B.M.; Li, F. Cocirculation of two distinct genetic and antigenic lineages of proposed influenza D virus in cattle. J. Virol. 2015, 89, 1036–1042. [Google Scholar] [CrossRef] [PubMed]
- Youil, R.; Su, Q.; Toner, T.J.; Szymkowiak, C.; Kwan, W.S.; Rubin, B.; Petrukhin, L.; Kiseleva, I.; Shaw, A.R.; DiStefano, D. Comparative study of influenza virus replication in Vero and MDCK cell lines. J. Virol. Methods 2004, 120, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Matlin, K.S.; Reggio, H.; Helenius, A.; Simons, K. Infectious entry pathway of influenza virus in a canine kidney cell line. J. Cell Biol. 1981, 91, 601–613. [Google Scholar] [CrossRef] [PubMed]
- Pleschka, S.; Wolff, T.; Ehrhardt, C.; Hobom, G.; Planz, O.; Rapp, U.R.; Ludwig, S. Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade. Nat. Cell Biol. 2001, 3, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Swindle, M.M.; Makin, A.; Herron, A.J.; Clubb, F.J., Jr.; Frazier, K.S. Swine as models in biomedical research and toxicology testing. Vet. Pathol. 2012, 49, 344–356. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, N.A.; Serao, N.V.; Elsbernd, A.J.; Hansen, S.L.; Walk, C.L.; Bedford, M.R.; Patience, J.F. Quantitative relationships between standardized total tract digestible phosphorus and total calcium intakes and their retention and excretion in growing pigs fed corn-soybean meal diets. J. Anim. Sci. 2015, 93, 2174–2182. [Google Scholar] [CrossRef] [PubMed]
- Dang, Y.; Lachance, C.; Wang, Y.; Gagnon, C.A.; Savard, C.; Segura, M.; Grenier, D.; Gottschalk, M. Transcriptional approach to study porcine tracheal epithelial cells individually or dually infected with swine influenza virus and streptococcus suis. BMC Vet. Res. 2014, 10, 86. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Ortega, M.; Melo, S.; Punyadarsaniya, D.; Rame, C.; Olivier, M.; Soubieux, D.; Marc, D.; Simon, G.; Herrler, G.; Berri, M.; et al. Innate immune response to a H3N2 subtype swine influenza virus in newborn porcine trachea cells, alveolar macrophages, and precision-cut lung slices. Vet. Res. 2014, 45, 42. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, M.; Scalvini, A.; Losio, M.N.; Corradi, A.; Soncini, M.; Bignotti, E.; Milanesi, E.; Ajmone-Marsan, P.; Barlati, S.; Bellotti, D.; et al. Establishment and characterization of two new pig cell lines for use in virological diagnostic laboratories. J. Virol. Methods 2003, 107, 205–212. [Google Scholar] [CrossRef]
- Jones, J.C.; Baranovich, T.; Zaraket, H.; Guan, Y.; Shu, Y.; Webby, R.J.; Webster, R.G. Human H7N9 influenza A viruses replicate in swine respiratory tissue explants. J. Virol. 2013, 87, 12496–12498. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Wu, N.H.; Nerlich, A.; Herrler, G.; Valentin-Weigand, P.; Seitz, M. Dynamic virus-bacterium interactions in a porcine precision-cut lung slice coinfection model: Swine influenza virus paves the way for streptococcus suis infection in a two-step process. Infect. Immun. 2015, 83, 2806–2815. [Google Scholar] [CrossRef] [PubMed]
- Punyadarsaniya, D.; Liang, C.H.; Winter, C.; Petersen, H.; Rautenschlein, S.; Hennig-Pauka, I.; Schwegmann-Wessels, C.; Wu, C.Y.; Wong, C.H.; Herrler, G. Infection of differentiated porcine airway epithelial cells by influenza virus: Differential susceptibility to infection by porcine and avian viruses. PLoS ONE 2011, 6, e28429. [Google Scholar] [CrossRef] [PubMed]
- Khatri, M.; Goyal, S.M.; Saif, Y.M. Oct4+ stem/progenitor swine lung epithelial cells are targets for influenza virus replication. J. Virol. 2012, 86, 6427–6433. [Google Scholar] [CrossRef] [PubMed]
- Khatri, M.; Saif, Y.M. Epithelial cells derived from swine bone marrow express stem cell markers and support influenza virus replication in vitro. PLoS ONE 2011, 6, e29567. [Google Scholar] [CrossRef] [PubMed]
- Mogensen, T.H. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 2009, 22, 240–273. [Google Scholar] [CrossRef] [PubMed]
- Diebold, S.S.; Kaisho, T.; Hemmi, H.; Akira, S.; Reis e Sousa, C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 2004, 303, 1529–1531. [Google Scholar] [CrossRef] [PubMed]
- Guillot, L.; Le Goffic, R.; Bloch, S.; Escriou, N.; Akira, S.; Chignard, M.; Si-Tahar, M. Involvement of toll-like receptor 3 in the immune response of lung epithelial cells to double-stranded RNA and influenza A virus. J. Biol. Chem. 2005, 280, 5571–5580. [Google Scholar] [CrossRef] [PubMed]
- Loo, Y.M.; Fornek, J.; Crochet, N.; Bajwa, G.; Perwitasari, O.; Martinez-Sobrido, L.; Akira, S.; Gill, M.A.; Garcia-Sastre, A.; Katze, M.G.; et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 2008, 82, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Pichlmair, A.; Schulz, O.; Tan, C.P.; Naslund, T.I.; Liljestrom, P.; Weber, F.; Reis e Sousa, C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 2006, 314, 997–1001. [Google Scholar] [CrossRef] [PubMed]
- Mordstein, M.; Neugebauer, E.; Ditt, V.; Jessen, B.; Rieger, T.; Falcone, V.; Sorgeloos, F.; Ehl, S.; Mayer, D.; Kochs, G.; et al. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J. Virol. 2010, 84, 5670–5677. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Li, X.W.; Mao, Y.; Wang, J.; Lu, H.Z.; Chen, Y.S.; Liang, Z.A.; Liang, L.; Zhang, S.J.; Zhang, B.; et al. Clinical features of the initial cases of 2009 pandemic influenza A (H1N1) virus infection in China. N. Engl. J. Med. 2009, 361, 2507–2517. [Google Scholar] [CrossRef] [PubMed]
- Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team; Dawood, F.S.; Jain, S.; Finelli, L.; Shaw, M.W.; Lindstrom, S.; Garten, R.J.; Gubareva, L.V.; Xu, X.; Bridges, C.B.; et al. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 2009, 360, 2605–2615. [Google Scholar]
- Peiris, J.S.; Poon, L.L.; Guan, Y. Emergence of a novel swine-origin influenza A virus (S-OIV) H1N1 virus in humans. J. Clin. Virol. 2009, 45, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Pinsky, B.A.; Mix, S.; Rowe, J.; Ikemoto, S.; Baron, E.J. Long-term shedding of influenza A virus in stool of immunocompromised child. Emerg. Infect. Dis. 2010, 16, 1165–1167. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.J.; Moon, S.J.; Kuak, E.Y.; Yoo, H.M.; Kim, C.K.; Chey, M.J.; Shin, B.M. Frequent detection of pandemic (H1N1) 2009 virus in stools of hospitalized patients. J. Clin. Microbiol. 2010, 48, 2314–2315. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.; Wang, Z.; Sreenivasan, C.C.; Hause, B.M.; Gourapura, J.R.; Li, F.; Francis, D.H.; Kaushik, R.S.; Khatri, M. Poly I:C adjuvanted inactivated swine influenza vaccine induces heterologous protective immunity in pigs. Vaccine 2015, 33, 542–548. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Huber, V.C.; McCormick, K.; Kaushik, R.S.; Boon, A.C.; Zhu, L.; Hause, B.; Webby, R.J.; Fang, Y. Characterization of a porcine intestinal epithelial cell line for influenza virus production. J. Gen. Virol. 2012, 93, 2008–2016. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, R.S.; Begg, A.A.; Wilson, H.L.; Aich, P.; Abrahamsen, M.S.; Potter, A.; Babiuk, L.A.; Griebel, P. Establishment of fetal bovine intestinal epithelial cell cultures susceptible to bovine rotavirus infection. J. Virol. Methods 2008, 148, 182–196. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, J.M.; Chen, W.; Chu, P.; Berschneider, H.M.; Argenzio, R.A.; Paradiso, A.M. l-glutamine and l-asparagine stimulate Na+-H+ exchange in porcine jejunal enterocytes. Am. J. Physiol. 1994, 266, G828–G838. [Google Scholar] [CrossRef] [PubMed]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Chan, L.Y.; Yim, E.K.; Choo, A.B. Normalized median fluorescence: An alternative flow cytometry analysis method for tracking human embryonic stem cell states during differentiation. Tissue Eng. Part C Methods 2013, 19, 156–165. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Manual on Animal Influenza Diagnosis and Surveillance; WHO: Geneva, Switzerland, 2002. [Google Scholar]
- Ito, T.; Couceiro, J.N.; Kelm, S.; Baum, L.G.; Krauss, S.; Castrucci, M.R.; Donatelli, I.; Kida, H.; Paulson, J.C.; Webster, R.G.; et al. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J. Virol. 1998, 72, 7367–7373. [Google Scholar] [PubMed]
- Pearce, M.B.; Jayaraman, A.; Pappas, C.; Belser, J.A.; Zeng, H.; Gustin, K.M.; Maines, T.R.; Sun, X.; Raman, R.; Cox, N.J.; et al. Pathogenesis and transmission of swine origin A(H3N2)v influenza viruses in ferrets. Proc. Natl. Acad. Sci. USA 2012, 109, 3944–3949. [Google Scholar] [CrossRef] [PubMed]
- McFawn, P.K.; Mitchell, H.W. Bronchial compliance and wall structure during development of the immature human and pig lung. Eur. Respir. J. 1997, 10, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Kaeffer, B.; Bottreau, E.; Velge, P.; Pardon, P. Epithelioid and fibroblastic cell lines derived from the ileum of an adult histocompatible miniature boar (d/d haplotype) and immortalized by SV40 plasmid. Eur. J. Cell Biol. 1993, 62, 152–162. [Google Scholar] [PubMed]
- Kedinger, M.; Haffen, K.; Simon-Assmann, P. Intestinal tissue and cell cultures. Differentiation 1987, 36, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Rusu, D.; Loret, S.; Peulen, O.; Mainil, J.; Dandrifosse, G. Immunochemical, biomolecular and biochemical characterization of bovine epithelial intestinal primocultures. BMC Cell Biol. 2005, 6, 42. [Google Scholar] [CrossRef] [PubMed]
- Kasper, M.; Stosiek, P. The expression of vimentin in epithelial cells from human nasal mucosa. Eur. Arch. Otorhinolaryngol. 1990, 248, 53–56. [Google Scholar] [CrossRef] [PubMed]
- Rogel, M.R.; Soni, P.N.; Troken, J.R.; Sitikov, A.; Trejo, H.E.; Ridge, K.M. Vimentin is sufficient and required for wound repair and remodeling in alveolar epithelial cells. FASEB J. 2011, 25, 3873–3883. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.W.; Wang, H.W.; Chang, C.W.; Chu, H.W.; Chen, C.Y.; Yu, J.C.; Chao, J.I.; Liu, H.F.; Ding, S.L.; Shen, C.Y. MicroRNA-30a inhibits cell migration and invasion by downregulating vimentin expression and is a potential prognostic marker in breast cancer. Breast Cancer Res. Treat. 2012, 134, 1081–1093. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Fang, W.; Li, S.; Yao, K. Aberrant expression of nuclear vimentin and related epithelial-mesenchymal transition markers in nasopharyngeal carcinoma. Int. J. Cancer 2012, 131, 1863–1873. [Google Scholar] [CrossRef] [PubMed]
- Carrillo, M.P.; Martinez, N.M.; Patino Mdel, P.; Iregui, C.A. Inhibition of Pasteurella multocida adhesion to rabbit respiratory epithelium using lectins. Vet. Med. Int. 2015, 2015, 365428. [Google Scholar] [CrossRef] [PubMed]
- Perfumo, C.J.; Mores, N.; Armocida, A.D.; Piffer, I.A.; Massone, A.R.; Itagaki, S. Histochemical and lectinhistochemical studies on nasal mucosa of pigs with or without respiratory diseases. J. Vet. Med. Sci. 1998, 60, 1021–1023. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, K.; Woodiga, S.A.; Dwivedi, V.; Buckwalter, C.M.; Singh, A.K.; Binjawadagi, B.; Hiremath, J.; Manickam, C.; Schleappi, R.; Khatri, M.; et al. Pretreatment of epithelial cells with live streptococcus pneumoniae has no detectable effect on influenza A virus replication in vitro. PLoS ONE 2014, 9, e90066. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Zhang, W.; Duggan, E.S.; Booth, J.L.; Zou, M.-H.; Metcalf, J.P. RIG-I and TLR3 are both required for maximum interferon induction by influenza virus in human lung alveolar epithelial cells. Virology 2015, 482, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Liniger, M.; Summerfield, A.; Zimmer, G.; McCullough, K.C.; Ruggli, N. Chicken cells sense influenza A virus infection through MDA5 and CARDIF signaling involving LGP2. J. Virol. 2012, 86, 705–717. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Cui, J.; Song, Y.; Zhang, S.; Han, F.; Yuan, R.; Gong, L.; Jiao, P.; Liao, M. Duck MDA5 functions in innate immunity against H5N1 highly pathogenic avian influenza virus infections. Vet. Res. 2014, 45, 66. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Wong, C.K.; Hui, D.S.; Lee, S.K.; Wong, R.Y.; Ngai, K.L.; Chan, M.C.; Chu, Y.J.; Ho, A.W.; Lui, G.C.; et al. Role of human toll-like receptors in naturally occurring influenza A infections. Influenza Other Respir. Viruses 2013, 7, 666–675. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Kim, C.H.; Kim, M.J.; Ryu, J.H.; Seong, S.Y.; Kim, S.; Lim, S.J.; Holtzman, M.J.; Yoon, J.H. The induction of pattern-recognition receptor expression against influenza A virus through Duox2-derived reactive oxygen species in nasal mucosa. Am. J. Respir. Cell Mol. Biol. 2015, 53, 525–535. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Miao, J.; Hou, J.; Lu, C. The effects of H3N2 swine influenza virus infection on TLRS and RLRS signaling pathways in porcine alveolar macrophages. Virol. J. 2015, 12, 61. [Google Scholar] [CrossRef] [PubMed]
- Gack, M.U.; Albrecht, R.A.; Urano, T.; Inn, K.S.; Huang, I.C.; Carnero, E.; Farzan, M.; Inoue, S.; Jung, J.U.; Garcia-Sastre, A. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 2009, 5, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Billharz, R.; Zeng, H.; Proll, S.C.; Korth, M.J.; Lederer, S.; Albrecht, R.; Goodman, A.G.; Rosenzweig, E.; Tumpey, T.M.; Garcia-Sastre, A.; et al. The NS1 protein of the 1918 pandemic influenza virus blocks host interferon and lipid metabolism pathways. J. Virol. 2009, 83, 10557–10570. [Google Scholar] [CrossRef] [PubMed]
- Fortes, P.; Beloso, A.; Ortin, J. Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks mRNA nucleocytoplasmic transport. EMBO J. 1994, 13, 704–712. [Google Scholar] [PubMed]
- Ruckle, A.; Haasbach, E.; Julkunen, I.; Planz, O.; Ehrhardt, C.; Ludwig, S. The NS1 protein of influenza A virus blocks RIG-I-mediated activation of the noncanonical NF-κB pathway and p52/ReLB-dependent gene expression in lung epithelial cells. J. Virol. 2012, 86, 10211–10217. [Google Scholar] [CrossRef] [PubMed]
- Nemeroff, M.E.; Barabino, S.M.; Li, Y.; Keller, W.; Krug, R.M. Influenza virus NS1 protein interacts with the cellular 30 KDA subunit of CPSF and inhibits 3’end formation of cellular pre-mRNAs. Mol. Cell 1998, 1, 991–1000. [Google Scholar] [CrossRef]
- Geiss, G.K.; Salvatore, M.; Tumpey, T.M.; Carter, V.S.; Wang, X.; Basler, C.F.; Taubenberger, J.K.; Bumgarner, R.E.; Palese, P.; Katze, M.G.; et al. Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: The role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc. Natl. Acad. Sci. USA 2002, 99, 10736–10741. [Google Scholar] [CrossRef] [PubMed]
- Papon, L.; Oteiza, A.; Imaizumi, T.; Kato, H.; Brocchi, E.; Lawson, T.G.; Akira, S.; Mechti, N. The viral RNA recognition sensor RIG-I is degraded during encephalomyocarditis virus (EMCV) infection. Virology 2009, 393, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Solis, M.; Nakhaei, P.; Jalalirad, M.; Lacoste, J.; Douville, R.; Arguello, M.; Zhao, T.; Laughrea, M.; Wainberg, M.A.; Hiscott, J. RIG-I-mediated antiviral signaling is inhibited in HIV-1 infection by a protease-mediated sequestration of RIG-I. J. Virol. 2011, 85, 1224–1236. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Han, C.; Xie, B.; Hu, X.; Yu, Q.; Shi, L.; Wang, Q.; Li, D.; Wang, J.; Zheng, P.; et al. Induction of Siglec-G by RNA viruses inhibits the innate immune response by promoting RIG-I degradation. Cell 2013, 152, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Benitez, A.A.; Panis, M.; Xue, J.; Varble, A.; Shim, J.V.; Frick, A.L.; López, C.B.; Sachs, D.; tenOever, B.R. In vivo RNAi screening identifies MDA5 as a significant contributor to the cellular defense against influenza A virus. Cell Rep. 2015, 11, 1714–1726. [Google Scholar] [CrossRef] [PubMed]
| Lectins | Concentration (µg/mL) | Inhibitor | Concentration (mM) |
|---|---|---|---|
| 1. Glucose/mannose group | |||
| Concanavalin-A (ConA) | 10 | α-methyl mannoside, α-methyl glucoside | 200 each |
| Pisum Sativum agglutinin (PSA) | 20 | α-methyl mannoside, α-methyl glucoside | 200 each |
| Lens culinaris agglutinin (LCA) | 20 | α-methyl mannoside, α-methyl glucoside | 200 each |
| 2. N-acetylgalactosamine group | |||
| Griffonia simplicifolia lectin I (GSL-1) | 10 | Galactose | 400 |
| Soybean agglutinin (SBA) | 10 | N-acetyl galactosamine | 200 |
| Dolichohs Biflorus agglutinin (DBA) | 10 | N-acetyl galactosamine | 200 |
| Ricinus communis agglutinin (RCA-120) | 10 | Galactose | 200 |
| Sophora japonica agglutinin (SJA) | 20 | No inhibitor used | |
| Vicia villosa agglutinin (VVA) | 20 | N-acetyl galactosamine | 200 |
| 3. N-acetylglucosamine group | |||
| Lycopersicon esculentum (tomato) lectin (LEL) | 20 | Chitin hydrosylate | 200 |
| Solanum tuberosum(potato) lectin (STL) | 20 | Chitin hydrosylate | 200 |
| Wheat germ agglutinin (WGA) | 10 | Chitin hydrosylate | 200 |
| Datura stramonium lectin (DSL) | 20 | Chitin hydrosylate | 200 |
| Griffornia simplicifolia lectin II (GSL-2) | 20 | No inhibitor used | |
| Succinylated WGA (SWGA) | 20 | Chitin hydrosylate | 200 |
| 4. Galactose group | |||
| Peanut agglutinin (PNA) | 20 | Galactose | 200 |
| Erythrina cristagalli lectin (ECL) | 20 | Lactose | 200 |
| Jacalin (JACALIN) | 20 | Galactose | 400 |
| 5. Fucose group | |||
| Ulex europaeus agglutinin I (UEA-1) | 10 | No inhibitor used | |
| 6. Oligosaccharide group | |||
| Phaseolus vulgaris erythroagglutinin (PHA-E) | 20 | No inhibitor used | |
| Phaseolus vulgaris Leucoagglutinin (PHA-L) | 20 | No inhibitor used | |
| 7. Sialic acid group | |||
| Sambucus nigra lectin (SNA) | 10 | No inhibitor used | |
| Maackia amurensis lectin II (MAL-II) | 10 | No inhibitor used |
| PRR | Primary Antibody | Isotype Control | Secondary Antibody |
|---|---|---|---|
| TLR2 | Mouse anti-human/mouse (ebiosciences) | Mouse IgG1 (ebiosciences) | Goat anti-mouse IgG Alexa Fluor 488 (Invitrogen) |
| TLR3 | Mouse anti-human (ebiosciences) | ||
| TLR4 | Mouse anti-human (ebiosciences | Mouse IgG2a (ebiosciences) | |
| TLR5 | Goat anti-human (Santa Cruz biotechnology) | Goat IgG (Santa Cruz biotechnology) | Chicken anti-goat IgG Alexa Fluor 488 (Invitrogen) |
| TLR6 | Rat anti-human (ebiosciences) | Rat IgG2a, kappa (ebiosciences) | Mouse anti-rat IgG FITC (ebiosciences) |
| TLR7 | Goat anti-human (Santa Cruz biotechnology) | Goat IgG (Santa Cruz biotechnology) | Chicken anti-goat IgG Alexa Fluor 488 (Invitrogen) |
| TLR8 | Rabbit anti-human (Santa Cruz biotechnology) | Rabbit IgG (Santa Cruz biotechnology) | Goat anti-rabbit IgG Alexa Fluor 488 (Invitrogen) |
| TLR9 | Rat anti-human (ebiosciences) | Rat IgG2a, kappa (ebiosciences) | Mouse anti-rat IgG FITC (ebiosciences) |
| MDA5, RIG-I | Goat anti-human (Santa Cruz biotechnology) | Goat IgG (Santa Cruz biotechnology) | Chicken anti-goat IgG Alexa Fluor 488 (Invitrogen) |
| Gene | Primer Sequences | Annealing Temp (°C) | Accession Number |
|---|---|---|---|
| TLR-7 | 5′ACA ATG ATA TCG CCA CCT CCA CCA3′ | 55 | NM_001097434 |
| 3′TGG CCA AGG AGA GAG TCT TCA GAT5′ | |||
| RIG-I | 5′TAT CCG AGC AGC AGG CTT TGA3′ | 58 | NM_213804 |
| 3′TGA AGT TTA GGG TTC TCG TTG CTG GGA5′ | |||
| MDA5 | 5′TGC CCT TTC CCA GTG GAT TAC TGA3′ | 58 | NM_001100194 |
| 3′TGT GTC CAG CTC CAA TCA GAT GGT5′ | |||
| RPL4 | 5′CAA GAG TAA CTA CAA CCT TC3′ | 57 | XM_003121741 |
| 3′GAA CTC TAC GAT GAA TCT TC5′ |
| TLRs | MK1-OSU | SD-PJEC | ||||
|---|---|---|---|---|---|---|
| Uninfected | MN08 | IA07 | Uninfected | MN08 | IA07 | |
| TLR-2 | 65.1 ± 3.5 | 64.5 ± 12.5 | 73.9 ± 13.7 | 56.5 ± 14.0 | 52.1 ± 11.1 | 57.2 ± 14.1 |
| TLR-4 | 7.3 ± 1.5 | 6.6 ± 0.7 | 3.9 ± 3.4 | 17.2 ± 6.0 | 22.3 ± 8.7 | 21.5 ± 12.5 |
| TLR-5 | 23.6 ± 10.2 | 28.7 ± 12.4 | 26.9 ± 12.5 | 25.8 ± 20.2 | 21.8 ± 11.3 | 27.5 ± 26.1 |
| TLR-6 | 26.7 ± 21.3 | 22.3 ± 12.4 | 26.4 ± 19.0 | 66.8 ± 7.2 | 66.8 ± 10.8 | 67.4 ± 10.9 |
| TLR-8 | 17.2 ± 2.8 | 10.1 ± 2.2 | 11.5 ± 4.9 | 4.9 ± 3.5 | 8.2 ± 2.3 | 16.5 ± 4.1 |
| TLR-9 | 32.7 ± 13.3 | 46.1 ± 16.6 | 35.5 ± 21.1 | 33.8 ± 22.4 | 31.8 ± 22.9 | 37.4 ± 16.1 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thomas, M.; Pierson, M.; Uprety, T.; Zhu, L.; Ran, Z.; Sreenivasan, C.C.; Wang, D.; Hause, B.; Francis, D.H.; Li, F.; et al. Comparison of Porcine Airway and Intestinal Epithelial Cell Lines for the Susceptibility and Expression of Pattern Recognition Receptors upon Influenza Virus Infection. Viruses 2018, 10, 312. https://doi.org/10.3390/v10060312
Thomas M, Pierson M, Uprety T, Zhu L, Ran Z, Sreenivasan CC, Wang D, Hause B, Francis DH, Li F, et al. Comparison of Porcine Airway and Intestinal Epithelial Cell Lines for the Susceptibility and Expression of Pattern Recognition Receptors upon Influenza Virus Infection. Viruses. 2018; 10(6):312. https://doi.org/10.3390/v10060312
Chicago/Turabian StyleThomas, Milton, Max Pierson, Tirth Uprety, Laihua Zhu, Zhiguang Ran, Chithra C. Sreenivasan, Dan Wang, Ben Hause, David H. Francis, Feng Li, and et al. 2018. "Comparison of Porcine Airway and Intestinal Epithelial Cell Lines for the Susceptibility and Expression of Pattern Recognition Receptors upon Influenza Virus Infection" Viruses 10, no. 6: 312. https://doi.org/10.3390/v10060312
APA StyleThomas, M., Pierson, M., Uprety, T., Zhu, L., Ran, Z., Sreenivasan, C. C., Wang, D., Hause, B., Francis, D. H., Li, F., & Kaushik, R. S. (2018). Comparison of Porcine Airway and Intestinal Epithelial Cell Lines for the Susceptibility and Expression of Pattern Recognition Receptors upon Influenza Virus Infection. Viruses, 10(6), 312. https://doi.org/10.3390/v10060312






