Exploring Fungal Communities in the Needles of Marginal Conifer Tree Populations
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Sites and Sampling
2.2. Laboratory Work
2.3. Bioinformatics
2.4. Statistical Analyses
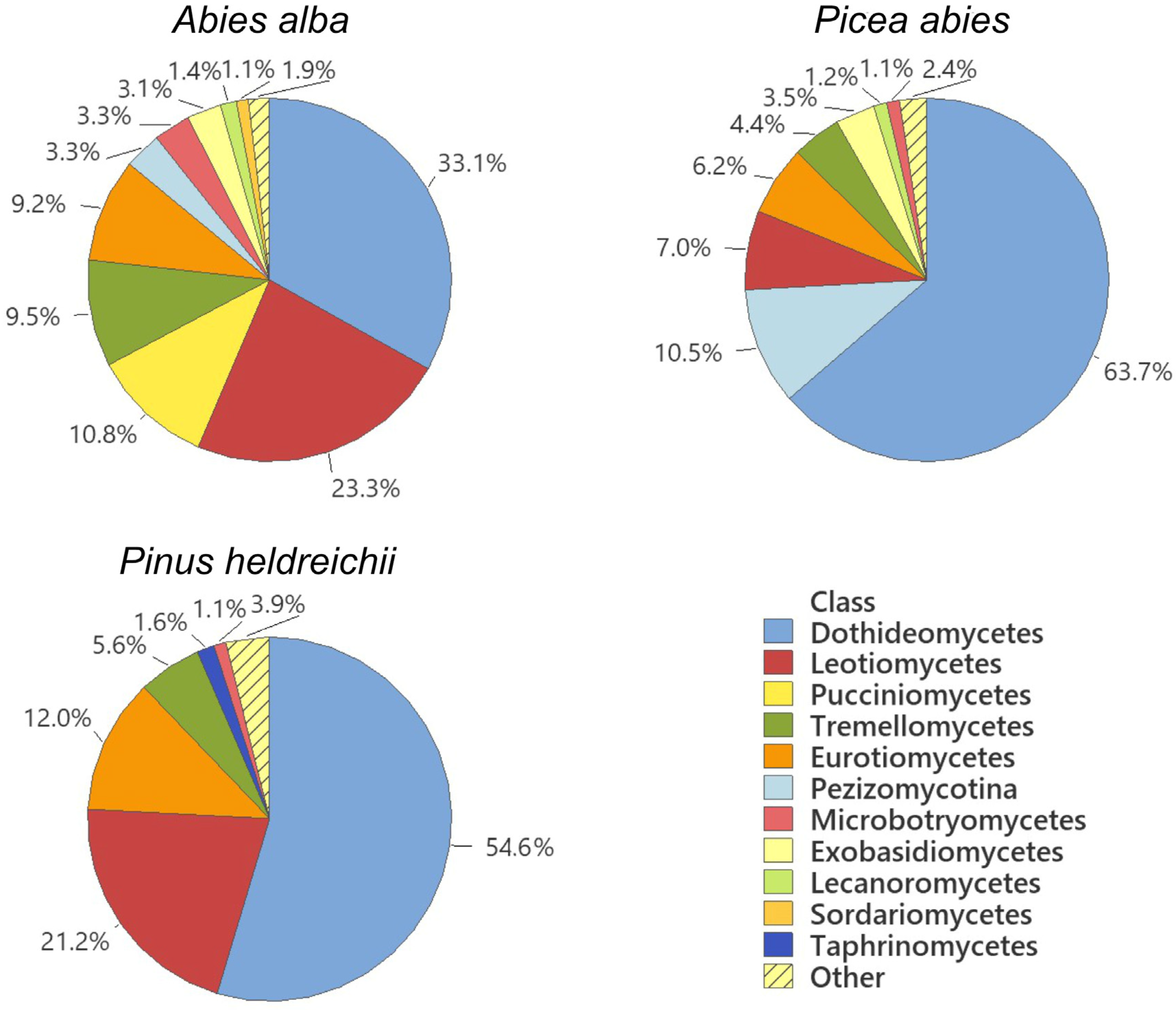
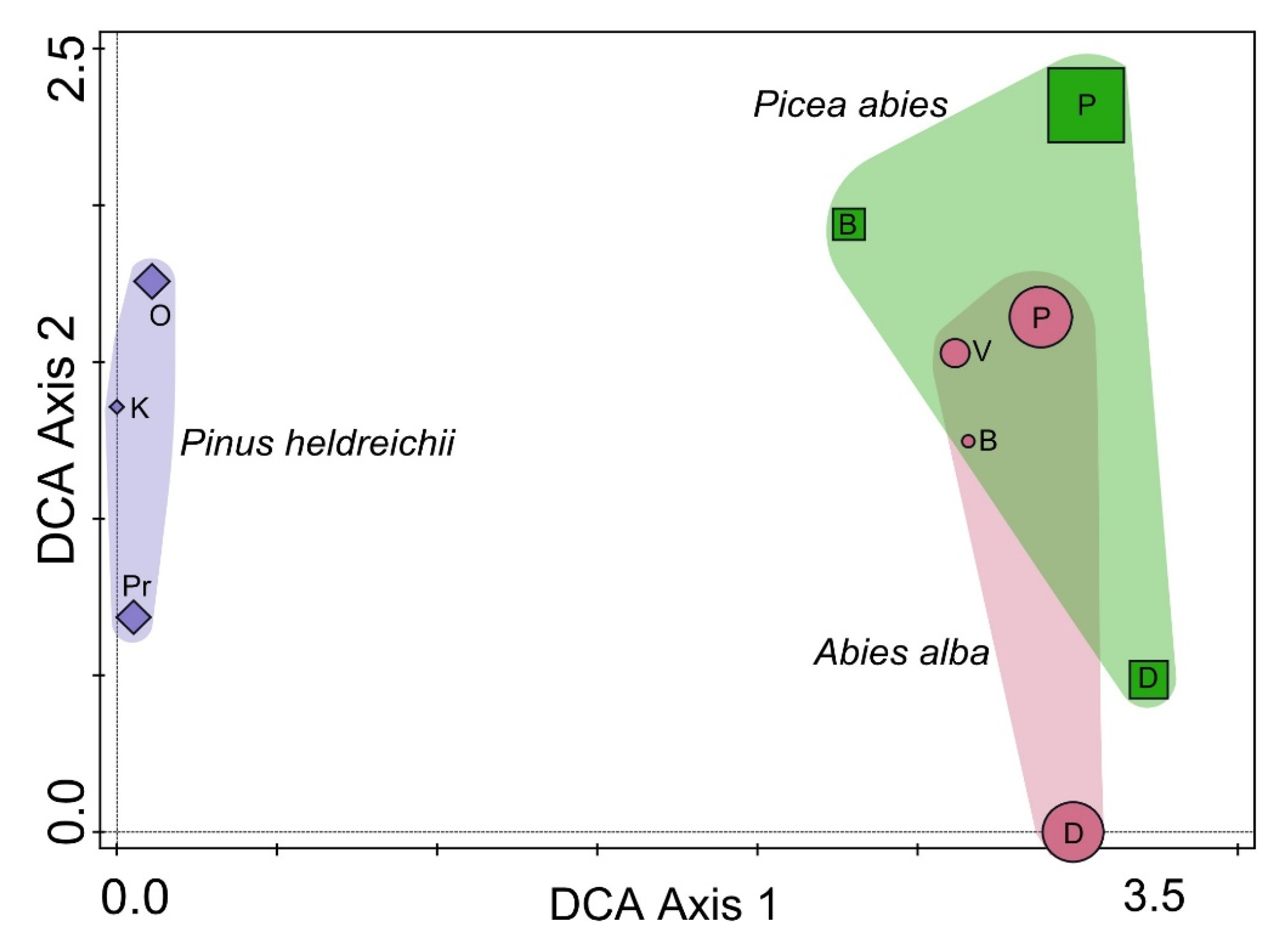
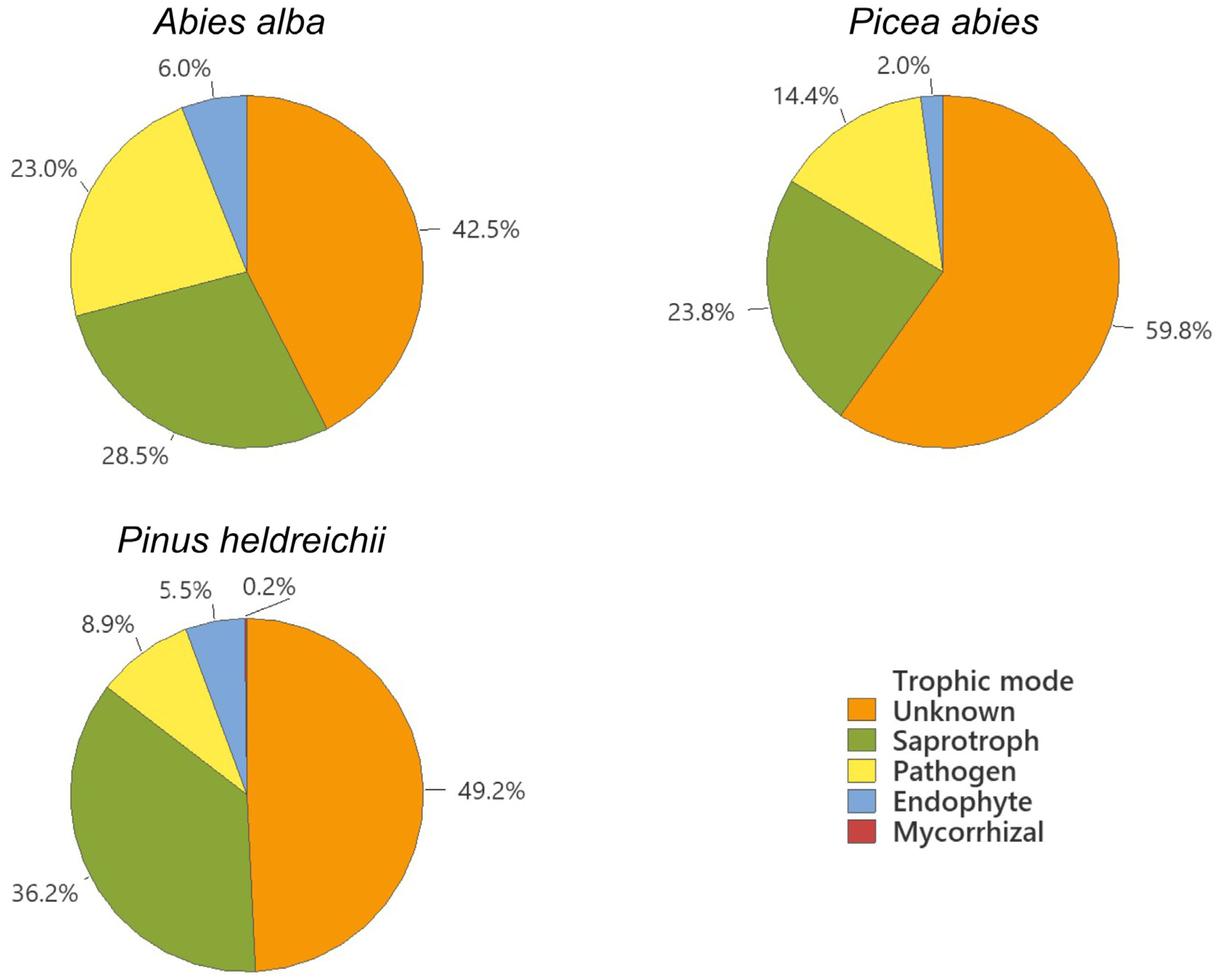
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Čurović, M.; Jovančević, M.; Balijagić, J. Wild fruit tree species of Montenegrin forests. In Forests of Southeast Europe Under a Changing Climate; Šijačić-Nikolić, M., Milovanović, J., Nonić, M., Eds.; Springer: Cham, Switzerland, 2018; pp. 21–28. [Google Scholar] [CrossRef]
- Skrøppa, T. EUFORGEN Technical Guidelines for Genetic Conservation and Use for Norway Spruce (Picea abies); International Plant Genetic Resources Institute: Rome, Italy, 2003; p. 6. [Google Scholar]
- Mauri, A.; de Rigo, D.; Caudullo, G. Abies alba in Europe: Distribution, habitat, usage and threats. In European Atlas of Forest Tree Species; San-Miguel-Ayanz, J., de Rigo, D., Caudullo, G., Houston Durrant, T., Mauri, A., Eds.; Publ. Off. EU: Luxembourg, 2016; p. e01493b. [Google Scholar]
- Seppä, H.; Alenius, T.; Bradshaw, R.; Giesecke, T.; Heikkilä, M.; Muukkonen, P. Invasion of Norway spruce (Picea abies) and the rise of the boreal ecosystem in Fennoscandia. J. Ecol. 2009, 97, 629–640. [Google Scholar] [CrossRef]
- Vendramin, G.G.; Fineschi, S.; Fady, B. EUFORGEN Technical Guidelines for Genetic Conservation and Use for Bosnian Pine (Pinus heldreichii); Bioversity International: Rome, Italy, 2008; p. 6. [Google Scholar]
- Lazarević, J.; Menkis, A. Fungi inhabiting fine roots of Pinus heldreichii in the Montenegrin montane forests. Symbiosis 2018, 74, 189–197. [Google Scholar] [CrossRef]
- Rajala, T.; Velmala, S.M.; Vesala, R.; Smolander, A.; Pennanen, T. The community of needle endophytes reflects the current physiological state of Norway spruce. Fungal Biol. 2014, 118, 309–315. Available online: https://www.sciencedirect.com/science/article/pii/S1878614614000099 (accessed on 5 April 2025). [CrossRef]
- Terhonen, E.; Blumenstein, K.; Kovalchuk, A.; Asiegbu, F. Forest tree microbiomes and associated fungal endophytes: Functional roles and impact on forest health. Forests 2019, 10, 42. [Google Scholar] [CrossRef]
- Velagić-Hajrudinović, A. Regional approach to biodiversity information management and reporting in Southeast Europe. Biodivers. Inf. Sci. Stand. 2019, 3, e38190. [Google Scholar] [CrossRef]
- Lakušić, R. Forest communities of Jugoslavia- SR Crna Gora. In Encyclopedia of Forestry; Potočić, Z., Ed.; Jugoslovenski Leksikografski Zavod Miroslav Krleža: Zagreb, Croatia, 1987; pp. 388–395. [Google Scholar]
- Fuštić, B.; Đuretić, G. Soils of Montenegro; Biotechnical Institute, University of Montenegro: Podgorica, Montenegro, 2000. [Google Scholar]
- Burić, M.; Micev, B.; Mitrović, L. Atlas of Climate of Montenegro; Montenegrin Academy of Science and Arts: Podgorica, Montenegro, 2012. [Google Scholar]
- Lakušić, D.; Šinžar-Sekulić, J. Overview map of the potential vegetation of Montenegro. In Botanical Lexicon of Montenegro; Montenegrin Academy of Science and Arts: Podgorica, Montenegro, 2022; p. 679. [Google Scholar]
- Ihrmark, K.; Bodeker, I.T.M.; Cruz-Martinez, K.; Friberg, H.; Kubartova, A.; Schenck, J.; Strid, Y.; Stenlid, J.; Brandstrom-Durling, M.; Clemmensen, K.E.; et al. New primers to amplify the fungal ITS2 region—Evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 2012, 82, 666–677. [Google Scholar] [CrossRef]
- Mischerikova, V.; Marčiulynienė, D.; Marčiulynas, A.; Lynikienė, J.; Gedminas, A.; Prylutsky, O.; Menkis, A. Biogeography of fungal communities associated with Pinus sylvestris L. and Picea abies (L.) H. Karst. along the latitudinal gradient in Europe. J. Fungi 2023, 9, 829. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Magurran, A.E. Ecological Diversity and Its Measurement; Princeton University Press: Princeton, NJ, USA, 1988; p. 192. [Google Scholar]
- ter Braak, C.J.F.; Smilauer, P. Canoco Reference Manual and User’s Guide to Canoco for Windows: Software for Canonical Community Ordination, Version 4; Microcomputer Power: Ithaca, NY, USA, 1998; p. 351. [Google Scholar]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0 contributors. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Xu, J. Fungal DNA barcoding. Genome 2016, 59, 913–932. [Google Scholar] [CrossRef]
- Tedersoo, L.; Anslan, S.; Bahram, M.; Drenkhan, R.; Pritsch, K.; Buegger, F.; Padari, A. Regional scale in depth analysis of soil fungal diversity reveals strong pH and plant species effects in Northern Europe. Front. Microb. 2020, 11, 1953. [Google Scholar] [CrossRef]
- Baldrian, P. Forest microbiome: Diversity, complexity, and dynamics. FEMS Microbiol. Rev. 2017, 41, 109–130. [Google Scholar] [CrossRef]
- Nguyen, D.; Boberg, J.; Ihrmark, K.; Stenström, E.; Stenlid, J. Do foliar fungal communities of Norway spruce shift along a tree species diversity gradient in mature European forests? Fungal Ecol. 2016, 23, 97–108. [Google Scholar] [CrossRef]
- Marčiulynas, A.; Marčiulynienė, D.; Mishcherikova, V.; Franić, I.; Lynikienė, J.; Gedminas, A.; Menkis, A. High Variability of Fungal Communities Associated with the Functional Tissues and Rhizosphere Soil of Picea abies in the Southern Baltics. Forests 2022, 13, 1103. [Google Scholar] [CrossRef]
- Bálint, M.; Tiffin, P.; Hallström, B.; O’Hara, R.B.; Olson, M.S.; Fankhauser, J.D.; Piepenbring, M.; Schmitt, I. Host Genotype Shapes the Foliar Fungal Microbiome of Balsam Poplar (Populus balsamifera). PLoS ONE 2013, 8, e53987. [Google Scholar] [CrossRef]
- Rodriguez, R.J.; Redman, R.S. More than 400 million years of evolution and some plants still can’t make it on their own: Plant stress tolerance via fungal symbiosis. J. Exp. Bot. 2008, 59, 1109–1114. [Google Scholar] [CrossRef]
- Siber-Canavesi, F.; Sieber, T.N. Successional patterns of fungal communities in needles of European silver fir (Abies alba Mill.). New Phytol. 1993, 25, 149–161. [Google Scholar] [CrossRef]
- Pusz, W.; Baturo-Ciesniewska, A.; Kaczmarek-Pienczewska, A.; Zwijacz-Kozica, T.; Patejuk, K. The mycobiota of needles and shoots of silver fir (Abies alba Mill.) with symptoms of Herpotrichia needle browning in the Tatra Mts. (Poland). Ann. For. Res. 2020, 63, 45–56. [Google Scholar] [CrossRef]
- Lazarević, J.; Menkis, A. Fungal biodiversity in the phyllosphere of Pinus heldreichii H. Christ-an endemic and high-altitude pine of Mediterranean region. Diversity 2020, 12, 172. [Google Scholar] [CrossRef]
- Peay, K.G.; Kennedy, P.G.; Talbot, J.M. Dimensions of biodiversity in the Earth’s mycobiome. Nat. Rev. Microbiol. 2016, 14, 434–447. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Põlme, S. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef]
- Baldrian, P.; López-Mondéjar, R.; Kohout, P. Forest microbiome and global change. Nat. Rev. Microbiol. 2023, 21, 487–501. [Google Scholar] [CrossRef]
- Barbi, F.; Martinović, T.; Odriozola, I.; Machac, A.; Moravcová, A.; Algora, C.; Ballian, D.; Barthold, S.; Brabcová, V.; Awokunle Hollá, S.; et al. Disentangling drivers behind fungal diversity gradients along altitude and latitude. New Phytol. 2025, 247, 295–308. [Google Scholar] [CrossRef]
- Crane, P.E. Morphology, taxonomy, and nomenclature of the Chrysomyxa ledi complex and related rust fungi on spruce and Ericaceae in North America and Europe. Can. J. Bot. 2001, 79, 57–982. [Google Scholar] [CrossRef]
- Karadić, D. Forest Phytopathology; University of Belgrade, Faculty of Forestry: Belgrade, Serbia and University of Banja Luka, Faculty of Forestry: Banja Luka, Bosnia and Herzegovina, 2010; pp. 330–331. [Google Scholar]
- Talgo, V.; Chastanger, G.; Thomsen, I.M.; Cech, T.; Riley, K.; Lang, K.; Klemsdal, S.S. Sydowia polyspora associated with current season needle necrosis (CSNN) on true fir (Abies spp.). Fungal Biol. 2010, 114, 545. [Google Scholar] [CrossRef]
- Botella, L.; Diez, J.J. Phylogenetic diversity of fungal endophytes in Spanish stands of Pinus halepensis. Fungal Divers. 2011, 47, 9–18. [Google Scholar] [CrossRef]
- Buzzini, P.; Turchetti, B.; Yurkov, A. Extremophilic yeasts: The toughest yeasts around? Yeast 2018, 35, 487–497. [Google Scholar] [CrossRef]
- Bensch, K.; Braun, U.; Groenewald, J.Z.; Crous, P.W. The genus Cladosporium. Stud. Mycol. 2012, 72, 1–401. [Google Scholar] [CrossRef]
- Tennakoon, D.S.; Thambugala, K.M.; Jeewon, R.; Hongsanan, S.; Kuo, C.-H.; Hyde, K.D. Additions to Chaetothyriaceae (Chaetothyriales): Longihyalospora gen. nov. and Ceramothyrium longivolcaniforme, a new host record from decaying leaves of Ficus ampelas. MycoKeys 2019, 61, 91–109, ISSN 1314–4057. Available online: https://www.sciencedirect.com/science/article/pii/S1314405719000892 (accessed on 5 April 2025). [CrossRef]
- Wijesinghe, S.N.; Dayarathne, M.C.; Maharachchikumbura, S.S.N.; Wanasinghe, D.N.; Hyde, K.D. Ceramothyrium chiangraiense, a novel species of Chaetothyriales (Eurotiomycetes) from Ficus sp. in Thailand. Asian J. Mycol. 2019, 2, 269–280. [Google Scholar] [CrossRef]
- Trichomerium Dioscoreae. Available online: https://ontosight.ai/glossary/term/trichomerium-dioscoreae-fungus-species-overview--67a29eaec445bf945af37842 (accessed on 10 April 2025).
- Callan, B.E.; Carris, L.M. 7-Fungi on Living Plant Substrata, Including Fruits. In Biodiversity of Fungi; Mueller, M.D., Bills, G.F., Foster, M.S., Eds.; Academic Press: Cambridge, MA, USA, 2004; pp. 105–126. Available online: https://www.sciencedirect.com/science/article/pii/B9780125095518500106 (accessed on 5 April 2025). [CrossRef]
- Ottosson, E.; Norden, J.; Dahlberg, A.; Edman, M.; Jönsson, M.; Larsson, K.H.; Lindahl, B.D. Species associations during the succession of wood-inhabiting fungal communities. Fungal Ecol. 2015, 14, 27–38. [Google Scholar] [CrossRef]

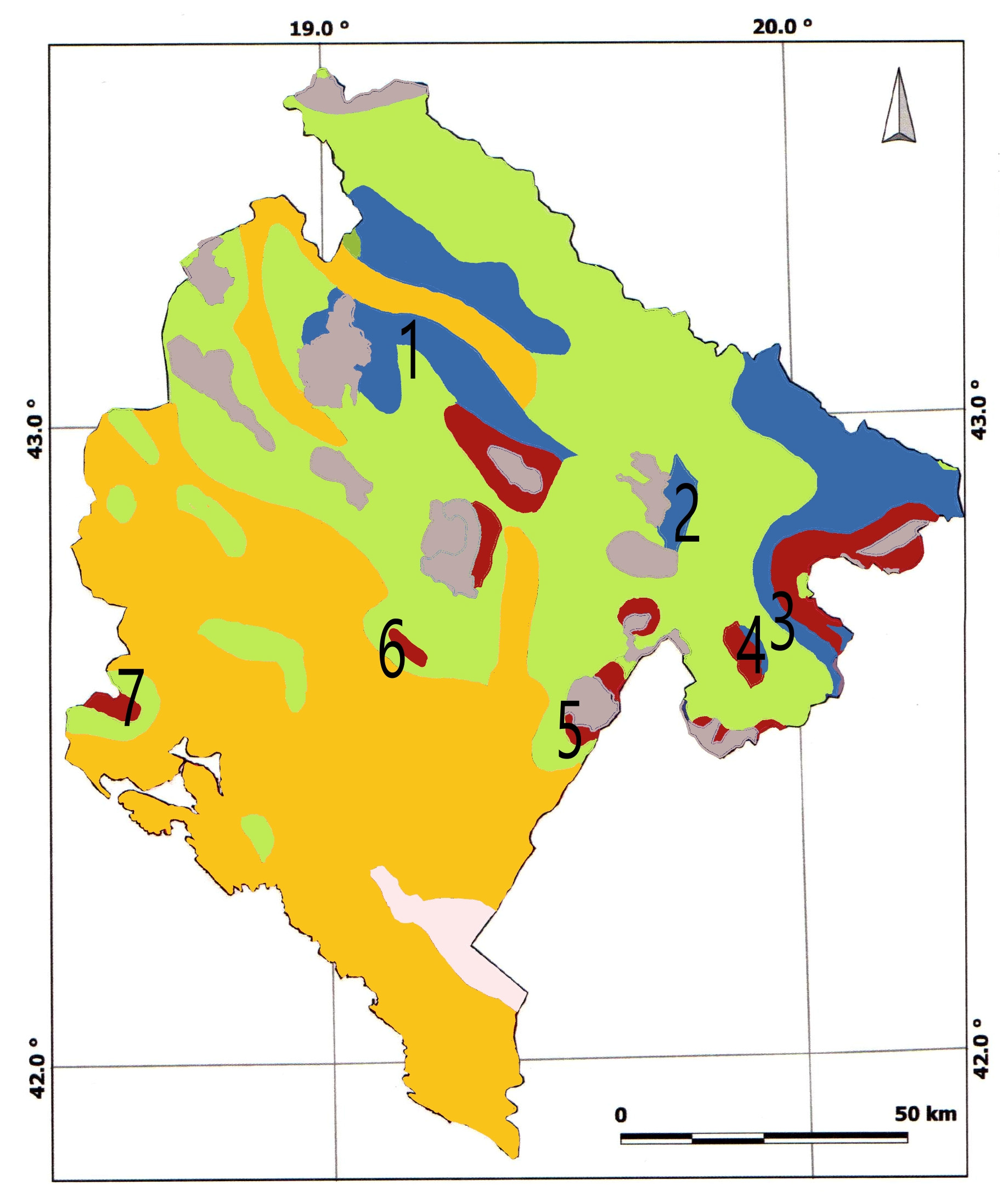

| Forest Vegetation [10] | Site | Geographical Position | Altitude (m) | Climate [12] | Soil Type [11] | Sampled |
|---|---|---|---|---|---|---|
| Abieti-Piceetum abietis | Mt. Durmitor | N 43.1455 E 19.0981 | 1350–1450 | Dfbx’’ | Kalkomelanosol | P. abies, A. alba |
| Abietinum albae subalpinum | Mt. Bjelasica | N 42.8748 E 19.6969 | 1700 | Dfbx’’ | Ranker | P. abies, A. alba |
| Abieti-Piceetum abietis | Mt. Prokletije | N 42.5964 E 20.0306 | 1400–1700 | Cfs’’b | Dystrict cambisol | P. abies, A. alba |
| Abieti-Piceetum abietis | Mt. Visitor | N 42.6146 E 19.8821 | 1700–1900- | Cfs’’b | Kakocambisol | A. alba |
| Pinetum heldreichii bertisceum | Mt. Kuči | N 44.8150 E 20.4347 | 1250–1700 | Cfws’’bx’’ | Leptosol/Molic leptosol | P. heldreichi |
| Pinetum heldreichii mediteraneo-montanum | Mt. Prekornica | N 42.6240 E 19.1995 | 1300 | Cfsb/Cfsb’’ | Leptosol/Molic leptosol | P. heldreichi |
| Pinetum heldreichii mediteraneo-montanum | Mt. Orjen | N 42.5436 E 18.5240 | 1200–1300 | Cfsb | Leptosol/Molic leptosol | P. heldreichii |
| Site | Picea abies | Abies alba | Pinus heldreichii | All | ||||
|---|---|---|---|---|---|---|---|---|
| Reads | OTUs | Reads | OTUs | Reads | OTUs | Reads | OTUs | |
| Mt. Durmitor | 529 | 80 | 1341 | 134 | - | - | 1870 | 158 |
| Mt. Bjelasica | 737 | 66 | 61 | 21 | - | - | 798 | 78 |
| Mt. Prokletije | 3138 | 168 | 982 | 137 | - | - | 4120 | 211 |
| Mt. Visitor | - | - | 232 | 59 | - | - | 232 | 59 |
| Mt. Kuči | - | - | - | - | 103 | 24 | 103 | 24 |
| Mt. Prekornica | - | - | - | - | 1091 | 71 | 1091 | 71 |
| Mt. Orjen | - | - | - | - | 1341 | 74 | 1341 | 74 |
| Total | 4404 | 226 | 2610 | 218 | 2535 | 111 | 9549 | 346 |
| OUT ID. | Taxon | Phylum * | Genbank Reference | Sequence Length, bp | Compared, bp | Similarity, % | Abies alba, % | Picea abies, % | Pinus heldreichii, % | All, % |
|---|---|---|---|---|---|---|---|---|---|---|
| 3598_3 | Unidentified sp. | A | KX220267 | 257 | 257/257 | 100% | 3.8 | 6.4 | 25.2 | 10.7 |
| 3598_2 | Unidentified sp. | A | MN902478 | 240 | 240/240 | 100% | 2.2 | 15.7 | - | 7.8 |
| 3598_4 | Chrysomyxa nagodhii | B | GU049432 | 317 | 312/317 | 98% | - | 12.6 | - | 5.8 |
| 3598_7 | Pseudeurotiaceae sp. | A | ON865419 | 240 | 239/240 | 99% | 2.0 | 4.5 | 2.2 | 3.2 |
| 3598_24 | Unidentified sp. | A | MT241934 | 259 | 259/259 | 100% | - | - | 11.6 | 3.1 |
| 3598_12 | Herpotrichiellaceae sp. | A | JF449676 | 258 | 257/258 | 99% | 5.0 | 3.3 | - | 2.9 |
| 3598_10 | Vishniacozyma victoriae | B | MF927673 | 234 | 234/234 | 100% | 1.3 | 5.4 | - | 2.8 |
| 3598_20 | Cladosporium sp. | A | LC317546 | 243 | 243/243 | 100% | 1.3 | 2.7 | 2.7 | 2.3 |
| 3598_28 | Unidentified sp. | A | MZ983694 | 242 | 242/242 | 100% | 4.1 | 0.7 | 3.4 | 2.3 |
| 3598_38 | Mycosphaerellaceae sp. | A | KJ406801 | 240 | 217/223 | 97% | 0.8 | 0.2 | 7.1 | 2.2 |
| 3598_41 | Unidentified sp. | A | MN903736 | 246 | 246/246 | 100% | 4.3 | 0.6 | 2.4 | 2.1 |
| 3598_27 | Botryosphaeriales sp. | A | PP759557 | 244 | 244/244 | 100% | 4.7 | 1.5 | - | 2.0 |
| 3598_15 | Phaeosphaeria sp. | A | KR909136 | 249 | 244/250 | 98% | 7.1 | 0.0 | - | 2.0 |
| 3598_45 | Ceramothyrium sp. | A | KC978733 | 252 | 246/253 | 97% | 3.1 | 2.4 | - | 1.9 |
| 3598_35 | Leotiomycetes sp. | A | OQ066890 | 240 | 240/240 | 100% | 0.5 | 3.6 | 0.0 | 1.8 |
| 3598_37 | Sydowia polyspora | A | KY659505 | 256 | 256/256 | 100% | 1.8 | 1.0 | 2.7 | 1.7 |
| 3598_39 | Trichomerium dioscoreae | A | NR_137946 | 265 | 261/265 | 98% | 3.6 | 1.1 | - | 1.5 |
| 3598_58 | Pseudeurotiaceae sp. | A | KR267039 | 243 | 243/243 | 100% | 1.3 | 2.3 | - | 1.4 |
| 3598_59 | Exobasidium bisporum | B | AB180368 | 281 | 266/268 | 99% | 2.1 | 0.5 | 1.3 | 1.2 |
| 3598_49 | Unidentified sp. | A | MN902540 | 240 | 235/240 | 98% | 3.1 | 0.6 | - | 1.1 |
| Total of the 20 taxa | 62.9 | 56.5 | 56.0 | 59.9 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lazarević, J.; Menkis, A. Exploring Fungal Communities in the Needles of Marginal Conifer Tree Populations. Forests 2025, 16, 968. https://doi.org/10.3390/f16060968
Lazarević J, Menkis A. Exploring Fungal Communities in the Needles of Marginal Conifer Tree Populations. Forests. 2025; 16(6):968. https://doi.org/10.3390/f16060968
Chicago/Turabian StyleLazarević, Jelena, and Audrius Menkis. 2025. "Exploring Fungal Communities in the Needles of Marginal Conifer Tree Populations" Forests 16, no. 6: 968. https://doi.org/10.3390/f16060968
APA StyleLazarević, J., & Menkis, A. (2025). Exploring Fungal Communities in the Needles of Marginal Conifer Tree Populations. Forests, 16(6), 968. https://doi.org/10.3390/f16060968






