Comparative Analysis of Endophytic Bacterial Microbiomes in Healthy and Phytoplasma-Infected European Blueberry Plants
Abstract
1. Introduction
2. Materials and Methods
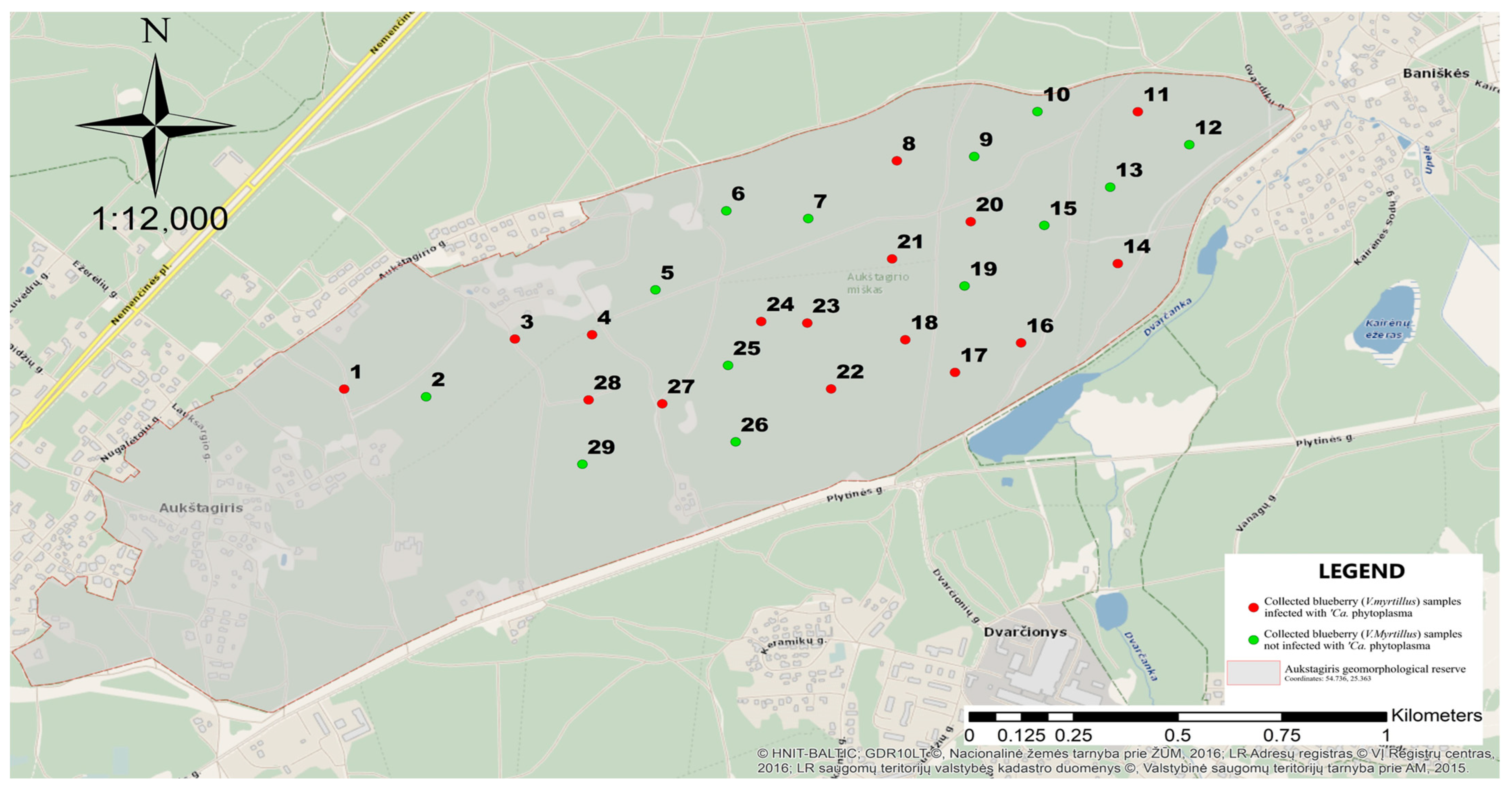
2.1. Sample Collection
2.2. DNA Extraction, Phytoplasma Detection and Identification
2.3. Bacterial DNA Amplification, Library Preparation and Sequencing
2.4. Bioinformatic Analysis
3. Results and Discussion
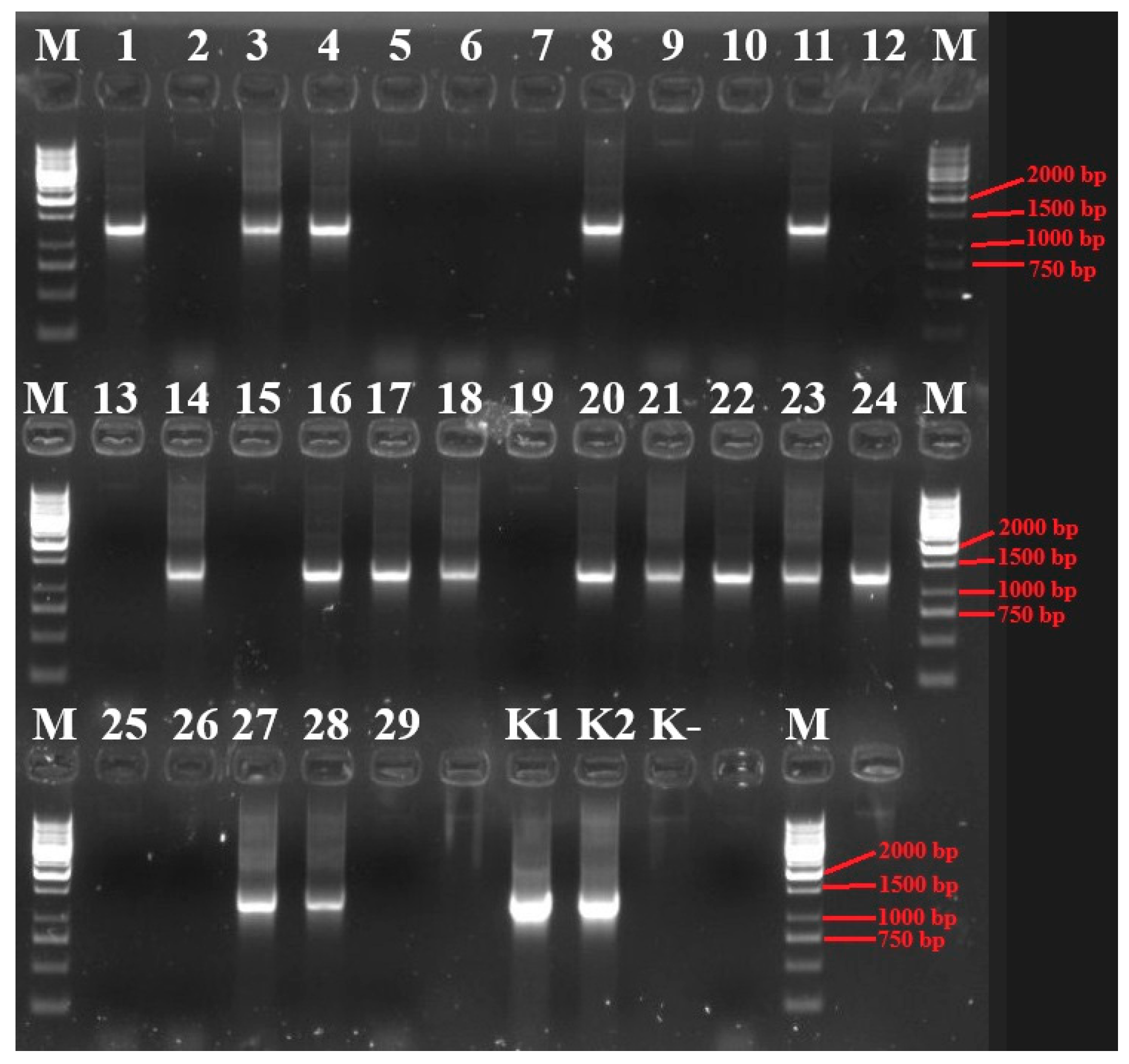
3.1. Molecular Identification of Phytoplasma in Blueberry Plants
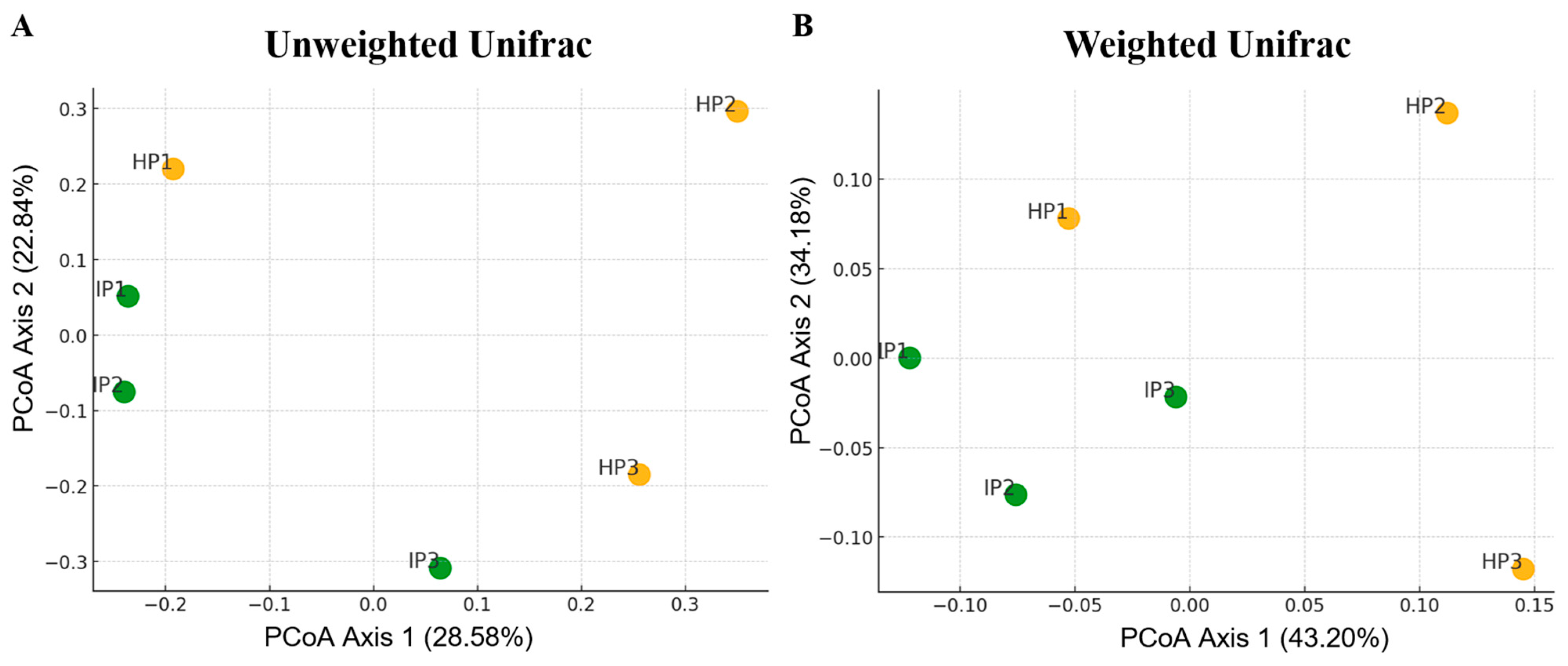
3.2. Diversity and Richness of Bacterial Microbiota of Healthy and Infected with ‘Candidatus Phytoplasma’ Vaccinium Myrtillus Plants
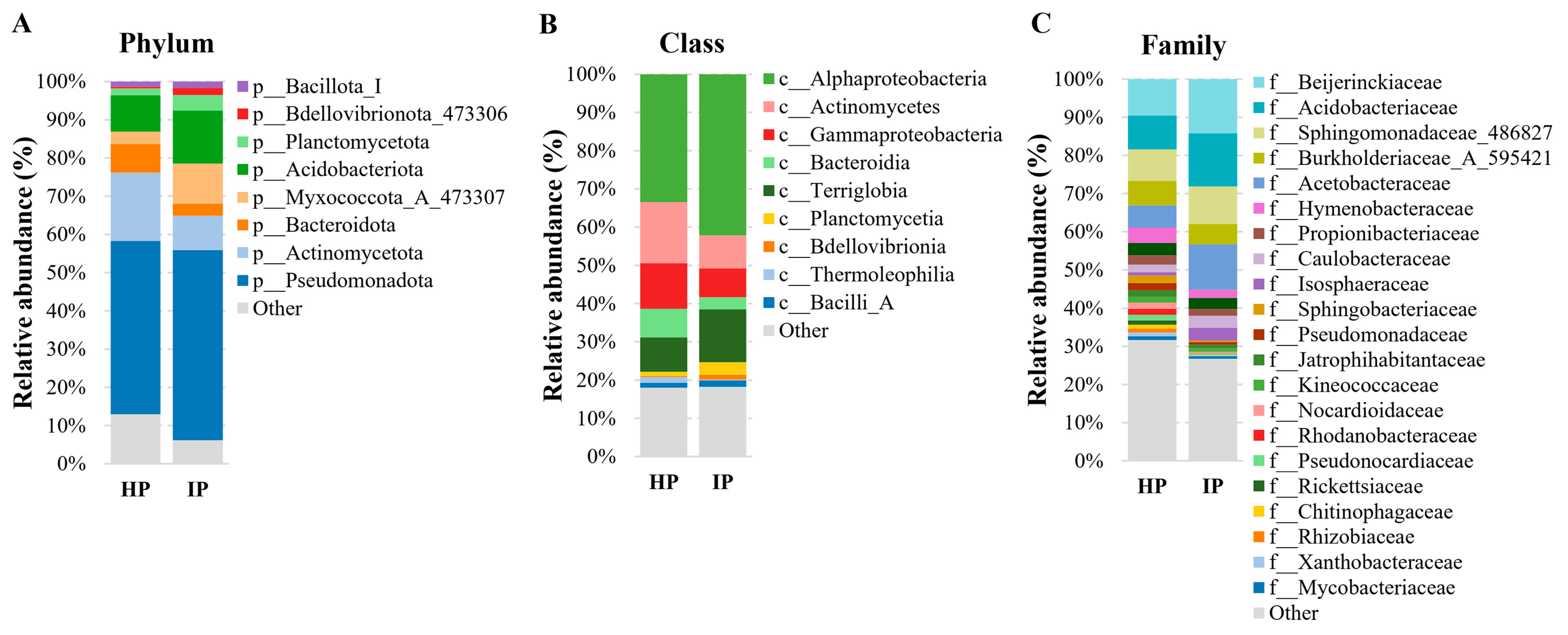
3.3. Bacterial Community Profiling of Healthy and Infected with Phytoplasma V. myrtillus Plants
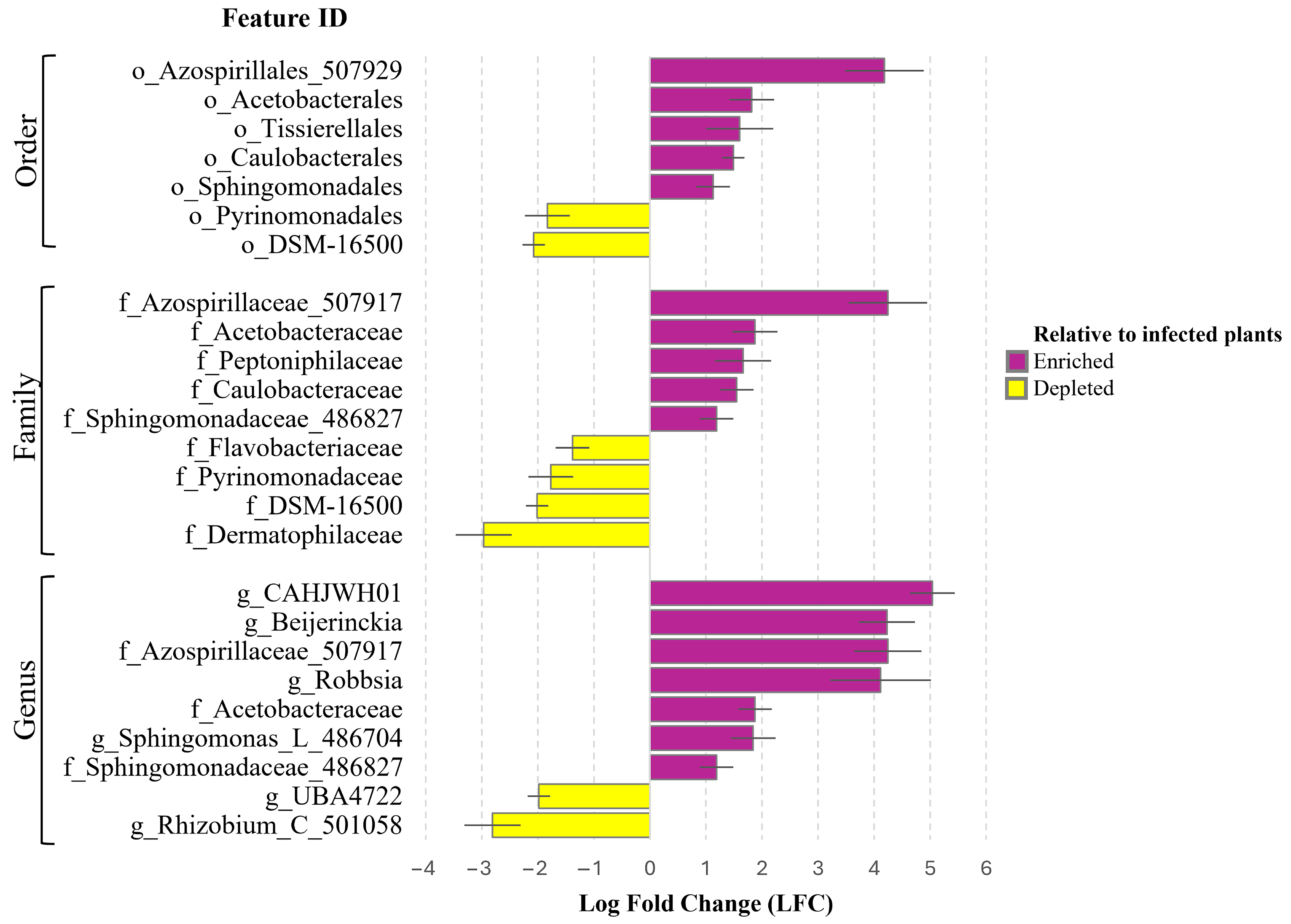
3.4. ANCOM-BC Analysis for Identifying Significant Taxonomic Differences and Potential Biomarkers
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Edger, P.P.; Iorizzo, M.; Bassil, N.V.; Benevenuto, J.; Ferrão, L.F.V.; Giongo, L.; Hummer, K.; Lawas, L.M.F.; Leisner, C.P.; Li, C.; et al. There and back again; historical perspective and future directions for Vaccinium breeding and research studies. Hortic. Res. 2022, 9, uhac083. [Google Scholar] [CrossRef] [PubMed]
- Cignarella, A.; Nastasi, M.; Cavalli, E.; Puglisi, L. Novel lipid-lowering properties of vaccinium myrtillus L. leaves, a traditional antidiabetic treatment, in several models of rat dyslipidaemia: A comparison with ciprofibrate. Thromb. Res. 1996, 84, 311–322. [Google Scholar] [CrossRef]
- Kopystecka, A.; Kozioł, I.; Radomska, D.; Bielawski, K.; Bielawska, A.; Wujec, M. Vaccinium uliginosum and Vaccinium myrtillus—Two Species—One Used as a Functional Food. Nutrients 2023, 15, 4119. [Google Scholar] [CrossRef] [PubMed]
- Shamilov, A.A.; Olennikov, D.N.; Pozdnyakov, D.I.; Bubenchikova, V.N.; Garsiya, E.R.; Larskii, M.V. Caucasian blueberry: Comparative study of phenolic compounds and neuroprotective and antioxidant potential of Vaccinium myrtillus and Vaccinium arctostaphylos Leaves. Life 2022, 12, 2079. [Google Scholar] [CrossRef] [PubMed]
- Azari, H.; Morovati, A.; Gargari, B.P.; Sarbakhsh, P. Beneficial effects of blueberry supplementation on the components of metabolic syndrome: A systematic review and meta-analysis. Food Funct. 2022, 13, 4875–4900. [Google Scholar] [CrossRef]
- Ristaino, J.B.; Anderson, P.K.; Bebber, D.P.; Brauman, K.A.; Cunniffe, N.J.; Fedoroff, N.V.; Finegold, C.; Garrett, K.A.; Gilligan, C.A.; Jones, C.M.; et al. The persistent threat of emerging plant disease pandemics to global food security. Proc. Natl. Acad. Sci. USA 2021, 118, e2022239118. [Google Scholar] [CrossRef]
- Bertaccini, A. Plants and phytoplasmas: When bacteria modify plants. Plants 2022, 11, 1425. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant–microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef]
- Noman, M.; Ahmed, T.; Ijaz, U.; Shahid, M.; Azizullah, N.; Li, D.; Manzoor, I.; Song, F. Plant–microbiome crosstalk: Dawning from composition and assembly of microbial community to improvement of disease resilience in plants. Int. J. Mol. Sci. 2021, 22, 6852. [Google Scholar] [CrossRef]
- Fitzpatrick, C.R.; Copeland, J.; Wang, P.W.; Guttman, D.S.; Kotanen, P.M.; Johnson, M.T.J. Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl. Acad. Sci. USA 2018, 115, E1157–E1165. [Google Scholar] [CrossRef]
- Kim, H.; Lee, K.K.; Jeon, J.; Harris, W.A.; Lee, Y. Domestication of Oryza species eco-evolutionarily shapes bacterial and fungal communities in rice seed. Microbiome 2020, 8, 20. [Google Scholar] [CrossRef]
- Wei, W.; Zhao, Y. Phytoplasma taxonomy: Nomenclature, classification, and identification. Biology 2022, 11, 1119. [Google Scholar] [CrossRef] [PubMed]
- Trivellone, V.; Wei, W.; Filippin, L.; Dietrich, C.H. Screening potential insect vectors in a museum biorepository reveals undiscovered diversity of plant pathogens in natural areas. Ecol. Evol. 2021, 11, 6493–6503. [Google Scholar] [CrossRef] [PubMed]
- Pérez-López, E.; Luna-Rodríguez, M.; Olivier, C.; Dumonceaux, T. The underestimated diversity of phytoplasmas in Latin America. Int. J. Syst. Evol. Microbiol. 2016, 66, 492–513. [Google Scholar] [CrossRef] [PubMed]
- Schneider, B.; Seemueller, E.; Smart, C.D.; Kirkpatrick, B.C. Phylogenetic classification of plant pathogenic mycoplasma-like organisms or phytoplasmas. Mol. Diagn. Proced. Mycoplasmol. 1995, 1, 369–380. [Google Scholar] [CrossRef]
- Zhao, Y.; Wei, W.; Lee, I.; Shao, J.; Suo, X.; Davis, R. Construction of an interactive online phytoplasma classification tool, iPhyClassifier, and its application in analysis of the peach X-disease phytoplasma group (16SrIII). Int. J. Syst. Evol. Microbiol. 2009, 5, 5607–5612. [Google Scholar] [CrossRef]
- Wei, W.; Trivellone, V.; Dietrich, C.H.; Zhao, Y.; Bottner-Parker, K.D.; Ivanauskas, A. Identification of phytoplasmas representing multiple new genetic lineages from phloem-feeding leafhoppers highlights the diversity of phytoplasmas and their potential vectors. Pathogens 2021, 10, 352. [Google Scholar] [CrossRef]
- Wright, A.A.; Shires, M.; Molnar, C.; Bishop, G.; Johnson, A.; Frias, C.; Harper, S.J. Titer and Distribution of ‘Candidatus Phytoplasma Pruni’ in Prunus Avium. Phytopathology 2022, 112, 1406–1412. [Google Scholar] [CrossRef]
- Ivanauskas, A.; Valiūnas, D.; Jomantienė, R.; Picciau, L.; Davis, R.E. Possible insect vectors of ‘Candidatus Phytoplasma asteris’ and ‘Ca. Phytoplasma pruni’-related strains in Lithuania. Zemdirb.-Agric. 2014, 101, 313–320. [Google Scholar] [CrossRef]
- Lee, I.; Polashock, J.; Bottner-Parker, K.D.; Bagadia, P.G.; Rodriguez-Saona, C.; Zhao, Y.; Davis, R.E. New subgroup 16SrIII-Y phytoplasmas associated with false-blossom diseased cranberry (Vaccinium macrocarpon) plants and with known and potential insect vectors in New Jersey. Eur. J. Plant Pathol. 2014, 139, 399–406. [Google Scholar] [CrossRef]
- Dėlkus, M.; Žižytė-Eidetienė, M.; Ivanauskas, A.; Valiunas, D. First report of lingonberry stunted yellows disease of Vaccinium vitis-idaea associated with ‘Candidatus Phytoplasma trifolii’-related phytoplasma strain in Lithuania. Plant Dis. 2024, 108, 1391. [Google Scholar] [CrossRef] [PubMed]
- Pradit, N.; Mescher, M.C.; Wang, Y.; Vorsa, N.; Rodriguez-Saona, C. Phytoplasma infection of cranberries benefits non-vector phytophagous insects. Front. Ecol. Evol. 2019, 7, 181. [Google Scholar] [CrossRef]
- Valiunas, D.; Alminaite, A.; Jomantiene, R.; Davis, R.E.; Maas, J.L. Possible cause of European blueberry disease is related to North American milkweed yellows phytoplasma. J. Plant Pathol. 2004, 86, 135–140. [Google Scholar]
- Dėlkus, M.; Žižytė-Eidetienė, M.; Ivanauskas, A.; Valiunas, D. First report of ‘Candidatus Phytoplasma trifolii’-related strain associated with Vaccinium reddish witches’-broom disease of European blueberry in Lithuania. Plant Dis. 2025, 109, 709. [Google Scholar] [CrossRef]
- Yurgel, S.N.; Ajeethan, N.; Smertenko, A. Response of plant-associated microbiome to plant root colonization by exogenous bacterial endophyte in perennial crops. Front. Microbiol. 2022, 13, 863946. [Google Scholar] [CrossRef]
- Mažeikienė, I.; Frercks, B.; Burokienė, D.; Mačionienė, I.; Šalaševičienė, A. Endophytic community composition and genetic-enzymatic features of cultivable bacteria in Vaccinium myrtillus L. in forests of the Baltic-Nordic region. Forests 2021, 12, 1647. [Google Scholar] [CrossRef]
- Deng, S.; Hiruki, C. Amplification of 16S rRNA genes from culturable and nonculturable Mollicutes. J. Microbiol. Methods 1991, 14, 53–61. [Google Scholar] [CrossRef]
- Schneider, B.; Goldstein, H.W.; Smith, D.B. The ASA Framework: An Update. Pers. Psychol. 1995, 48, 747–773. [Google Scholar] [CrossRef]
- Gundersen, D.E.; Lee, I.M.; Schaff, D.A.; Harrison, N.A.; Chang, C.J.; Davis, R.E.; Kingsbury, D.T. Genomic diversity and differentiation among phytoplasma strains in 16S rRNA groups I (aster yellows and related phytoplasmas) and III (X-disease and related phytoplasmas). Int. J. Syst. Bacteriol. 1996, 46, 64–75. [Google Scholar] [CrossRef]
- Lee, I.; Gundersen-Rindal, D.E.; Davis, R.E.; Bartoszyk, I.M. Revised Classification scheme of phytoplasmas based on RFLP analyses of 16S rRNA and ribosomal protein gene Sequences. Int. J. Syst. Bacteriol. 1998, 48, 1153–1169. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 10 December 2024).
- Zhao, Y.; Wei, W.; Lee, I.M.; Shao, J.; Suo, X.; Davis, R.E. The iPhyClassifier, an Interactive Online Tool for Phytoplasma Classification and Taxonomic Assignment. In Phytoplasma; Dickinson, M., Hodgetts, J., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2013; Volume 938, pp. 329–338. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable, and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D.; Jiang, Y.; Balaban, M.; Cantrell, K.; Zhu, Q.; Gonzalez, A.; Morton, J.T.; Nicolaou, G.; Parks, D.H.; Karst, S.M.; et al. Greengenes2 unifies microbial data in a single reference tree. Nat. Biotechnol. 2024, 42, 715–718. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Aust. Ecol. 2001, 26, 32–46. [Google Scholar] [CrossRef]
- Vázquez-Baeza, Y.; Pirrung, M.; Gonzalez, A.; Knight, R. EMPeror: A tool for visualizing high-throughput microbial community data. GigaScience 2013, 2, 16. [Google Scholar] [CrossRef]
- Lin, H.; Peddada, S.D. Analysis of compositions of microbiomes with bias correction. Nat. Commun. 2020, 11, 3514. [Google Scholar] [CrossRef]
- Hrovat, K.; Dutilh, B.E.; Medema, M.H.; Melkonian, C. Taxonomic resolution of different 16S rRNA variable regions varies strongly across plant-associated bacteria. ISME Commun. 2024, 4, ycae034. [Google Scholar] [CrossRef]
- Michalko, J.; Medo, J.; Ferus, P.; Konôpková, J.; Košútová, D.; Hoťka, P.; Barta, M. Changes of endophytic bacterial community in mature leaves of Prunus laurocerasus L. during the seasonal transition from winter dormancy to vegetative growth. Plants 2022, 11, 417. [Google Scholar] [CrossRef]
- Huo, D.; Malacrinò, A.; Lindsey, L.E.; Benitez, M. Subtle responses of soil bacterial communities to corn-soybean-wheat rotation. Phytobiomes J. 2023, 7, 392–400. [Google Scholar] [CrossRef]
- Vlasselaer, L.; Crauwels, S.; Lievens, B.; De Coninck, B. Unveiling the microbiome of hydroponically cultivated lettuce: Impact of Phytophthora cryptogea infection on plant-associated microorganisms. FEMS Microbiol. Ecol. 2024, 100, fiae010. [Google Scholar] [CrossRef] [PubMed]
- Kwak, M.; Jeong, H.; Madhaiyan, M.; Lee, Y.; Sa, T.; Oh, T.K.; Kim, J.F. Genome Information of Methylobacterium oryzae, a Plant-Probiotic Methylotroph in the Phyllosphere. PLoS ONE 2014, 9, e106704. [Google Scholar] [CrossRef]
- Knief, C.; Ramette, A.; Frances, L.; Alonso-Blanco, C.; Vorholt, J.A. Site and plant species are important determinants of the Methylobacterium community composition in the plant phyllosphere. ISME J. 2010, 4, 719–728. [Google Scholar] [CrossRef]
- Grossi, C.E.M.; Fantino, E.; Serral, F.; Zawoznik, M.S.; Porto, D.F.D.; Ulloa, R.M. Methylobacterium sp. 2A is a plant growth-promoting rhizobacteria that has the potential to improve potato crop yield under adverse conditions. Front. Plant Sci. 2020, 11, 71. [Google Scholar] [CrossRef] [PubMed]
- De Araújo Diniz, P.F.; De Oliveira, L.E.M.; Lopes, N.A.; Florentino, L.A.; De Carvalho, T.S.; De Souza Moreira, F.M. Bactérias diazotróficas em solos sob seringueira. Rev. Bras. De Ciência Do Solo 2012, 36, 1426–1433. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Gallardo, A.; Covelo, F.; Prado-Comesaña, A.; Ochoa, V.; Maestre, F.T. Differences in thallus chemistry are related to species-specific effects of biocrust-forming lichens on soil nutrients and microbial communities. Funct. Ecol. 2014, 29, 1087–1098. [Google Scholar] [CrossRef]
- Cernava, T.; Müller, H.; Aschenbrenner, I.A.; Grube, M.; Berg, G. Analyzing the antagonistic potential of the lichen microbiome against pathogens by bridging metagenomic with culture studies. Front. Microbiol. 2015, 6, 620. [Google Scholar] [CrossRef]
- Costa, O.Y.; Pijl, A.; Kuramae, E.E. Dynamics of active potential bacterial and fungal interactions in the assimilation of acidobacterial EPS in soil. Soil Biol. Biochem. 2020, 148, 107916. [Google Scholar] [CrossRef]
- De Tender, C.; Vandecasteele, B.; Verstraeten, B.; Ommeslag, S.; Kyndt, T.; Debode, J. Biochar-enhanced resistance to botrytis cinerea in strawberry fruits (But Not Leaves) is associated with changes in the rhizosphere microbiome. Front. Plant Sci. 2021, 12, 700479. [Google Scholar] [CrossRef]
- Shahid, M.J.; Al-Surhanee, A.A.; Kouadri, F.; Ali, S.; Nawaz, N.; Afzal, M.; Rizwan, M.; Ali, B.; Soliman, M.H. Role of microorganisms in the remediation of wastewater in floating treatment wetlands: A review. Sustainability 2020, 12, 5559. [Google Scholar] [CrossRef]
- Muñoz-Dorado, J.; Marcos-Torres, F.J.; García-Bravo, E.; Moraleda-Muñoz, A.; Pérez, J. Myxobacteria: Moving, killing, feeding, and surviving together. Front. Microbiol. 2016, 7, 781. [Google Scholar] [CrossRef]
- Murphy, C.L.; Yang, R.; Decker, T.; Cavalliere, C.; Andreev, V.; Bircher, N.; Cornell, J.; Dohmen, R.; Pratt, C.J.; Grinnell, A.; et al. Genomes of novel Myxococcota reveal severely curtailed machineries for predation and cellular differentiation. Appl. Environ. Microbiol. 2021, 87, e01706–e01721. [Google Scholar] [CrossRef] [PubMed]
- Pérez, J.; Moraleda-Muñoz, A.; Marcos-Torres, F.J.; Muñoz-Dorado, J. Bacterial predation: 75 years and counting! Environ. Microbiol. 2016, 18, 766–779. [Google Scholar] [CrossRef] [PubMed]
- Cassán, F.; Coniglio, A.; López, G.; Molina, R.; Nievas, S.; De Carlan, C.L.N.; Donadio, F.; Torres, D.; Rosas, S.; Pedrosa, F.O.; et al. Everything you must know about Azospirillum and its impact on agriculture and beyond. Biol. Fertil. Soils 2020, 56, 461–479. [Google Scholar] [CrossRef]
- Cassan, F.D.; Coniglio, A.; Amavizca, E.; Maroniche, G.; Cascales, E.; Bashan, Y.; de-Bashan, L.E. The Azospirillum brasilense type VI secretion system promotes cell aggregation, biocontrol protection against phytopathogens and attachment to the microalgae Chlorella sorokiniana. Environ. Microbiol. 2021, 23, 6257–6274. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A Review on the Plant Microbiome: Ecology, Functions, and Emerging Trends in Microbial Application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef]
- Asaf, S.; Numan, M.; Khan, A.L.; Al-Harrasi, A. Sphingomonas: From diversity and genomics to functional role in environmental remediation and plant growth. Crit. Rev. Biotechnol. 2020, 40, 138–152. [Google Scholar] [CrossRef]
- Wilhelm, R.C. Following the terrestrial tracks of Caulobacter—Redefining the ecology of a reputed aquatic oligotroph. ISME J. 2018, 12, 3025–3037. [Google Scholar] [CrossRef]
- Spaepen, S.; Vanderleyden, J.; Remans, R. Indole-3-Acetic Acid in Microbial and Microorganism-Plant Signaling. FEMS Microbiol. Rev. 2007, 31, 425–448. [Google Scholar] [CrossRef]
- Sun, W.; Shahrajabian, M.H.; Wang, N. A Study of the Different Strains of the Genus Azospirillum Spp. on Increasing Productivity and Stress Resilience in Plants. Plants 2025, 14, 267. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wei, Y.; Yan, T.; Wang, C.; Chao, Y.; Jia, M.; An, L.; Sheng, H. Sphingomonas sp. Hbc-6 alters physiological metabolism and recruits beneficial rhizosphere bacteria to improve plant growth and drought tolerance. Front. Plant Sci. 2022, 13, 1002772. [Google Scholar] [CrossRef] [PubMed]
- Msomi, N.N.; Padayachee, T.; Nzuza, N.; Syed, P.R.; Kryś, J.D.; Chen, W.; Gront, D.; Nelson, D.R.; Syed, K. In Silico analysis of P450s and their role in secondary metabolism in the bacterial class gammaproteobacteria. Molecules 2021, 26, 1538. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Liu, Y.; Pan, J.; Cron, B.R.; Toner, B.M.; Anantharaman, K.; Breier, J.A.; Dick, G.J.; Li, M. Gammaproteobacteria mediating utilization of methyl-, sulfur- and petroleum organic compounds in deep ocean hydrothermal plumes. ISME J. 2020, 14, 3136–3148. [Google Scholar] [CrossRef]
- Benhizia, Y.; Benhizia, H.; Benguedouar, A.; Muresu, R.; Giacomini, A.; Squartini, A. Gammaproteobacteria can modulate legumes of the genus hedysarum. Syst. Appl. Microbiol. 2004, 27, 462–468. [Google Scholar] [CrossRef]
- Baldwin, S.A.; Khoshnoodi, M.; Rezadehbashi, M.; Taupp, M.; Hallam, S.; Mattes, A.; Sanei, H. The microbial community of a passive biochemical reactor treating arsenic, zinc, and Sulfate-Rich seepage. Front. Bioeng. Biotechnol. 2015, 3, 00027. [Google Scholar] [CrossRef]
- Lacombe-Harvey, M.; Brzezinski, R.; Beaulieu, C. Chitinolytic functions in actinobacteria: Ecology, enzymes, and evolution. Appl. Microbiol. Biotechnol. 2018, 102, 7219–7230. [Google Scholar] [CrossRef]
- Brescini, L.; Fioriti, S.; Morroni, G.; Barchiesi, F. Antifungal combinations in dermatophytes. J. Fungi 2021, 7, 727. [Google Scholar] [CrossRef]
- Pérez-Jaramillo, J.E.; Carrión, V.J.; De Hollander, M.; Raaijmakers, J.M. The wild side of plant microbiomes. Microbiome 2018, 6, 143. [Google Scholar] [CrossRef]
- Matse, D.T.; Huang, C.; Huang, Y.; Yen, M. Effects of coinoculation of Rhizobium with plant growth promoting rhizobacteria on the nitrogen fixation and nutrient uptake of Trifolium repens in low phosphorus soil. J. Plant Nutr. 2019, 43, 739–752. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dėlkus, M.; Lukša-Žebelovič, J.; Žižytė-Eidetienė, M.; Ivanauskas, A.; Valiūnas, D.; Servienė, E. Comparative Analysis of Endophytic Bacterial Microbiomes in Healthy and Phytoplasma-Infected European Blueberry Plants. Forests 2025, 16, 758. https://doi.org/10.3390/f16050758
Dėlkus M, Lukša-Žebelovič J, Žižytė-Eidetienė M, Ivanauskas A, Valiūnas D, Servienė E. Comparative Analysis of Endophytic Bacterial Microbiomes in Healthy and Phytoplasma-Infected European Blueberry Plants. Forests. 2025; 16(5):758. https://doi.org/10.3390/f16050758
Chicago/Turabian StyleDėlkus, Martynas, Juliana Lukša-Žebelovič, Marija Žižytė-Eidetienė, Algirdas Ivanauskas, Deividas Valiūnas, and Elena Servienė. 2025. "Comparative Analysis of Endophytic Bacterial Microbiomes in Healthy and Phytoplasma-Infected European Blueberry Plants" Forests 16, no. 5: 758. https://doi.org/10.3390/f16050758
APA StyleDėlkus, M., Lukša-Žebelovič, J., Žižytė-Eidetienė, M., Ivanauskas, A., Valiūnas, D., & Servienė, E. (2025). Comparative Analysis of Endophytic Bacterial Microbiomes in Healthy and Phytoplasma-Infected European Blueberry Plants. Forests, 16(5), 758. https://doi.org/10.3390/f16050758






