Integrated Multi-Omics Analysis Reveals Photosynthetic Acclimation and Metabolic Reprogramming in Populus ussuriensis kom. Under Cold Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Cold Treatment
2.2. Full-Length cDNA Preparation and Sequencing
2.3. Metabolites Analysis
2.4. Integrated Metabolic Profiling and Transcriptional Analysis
2.5. Nutrient Metabolite Determinations
2.6. Leaf Gas Exchange and Photosynthetic Pigments Measurements
2.7. RNA Extraction and RT-qPCR Analysis
2.8. Statistical Analysis
2.9. Data Availability
3. Results
3.1. Transcriptome Sequencing and Differential Gene Annotation of Populus ussuriensis kom. Under Cold Stress
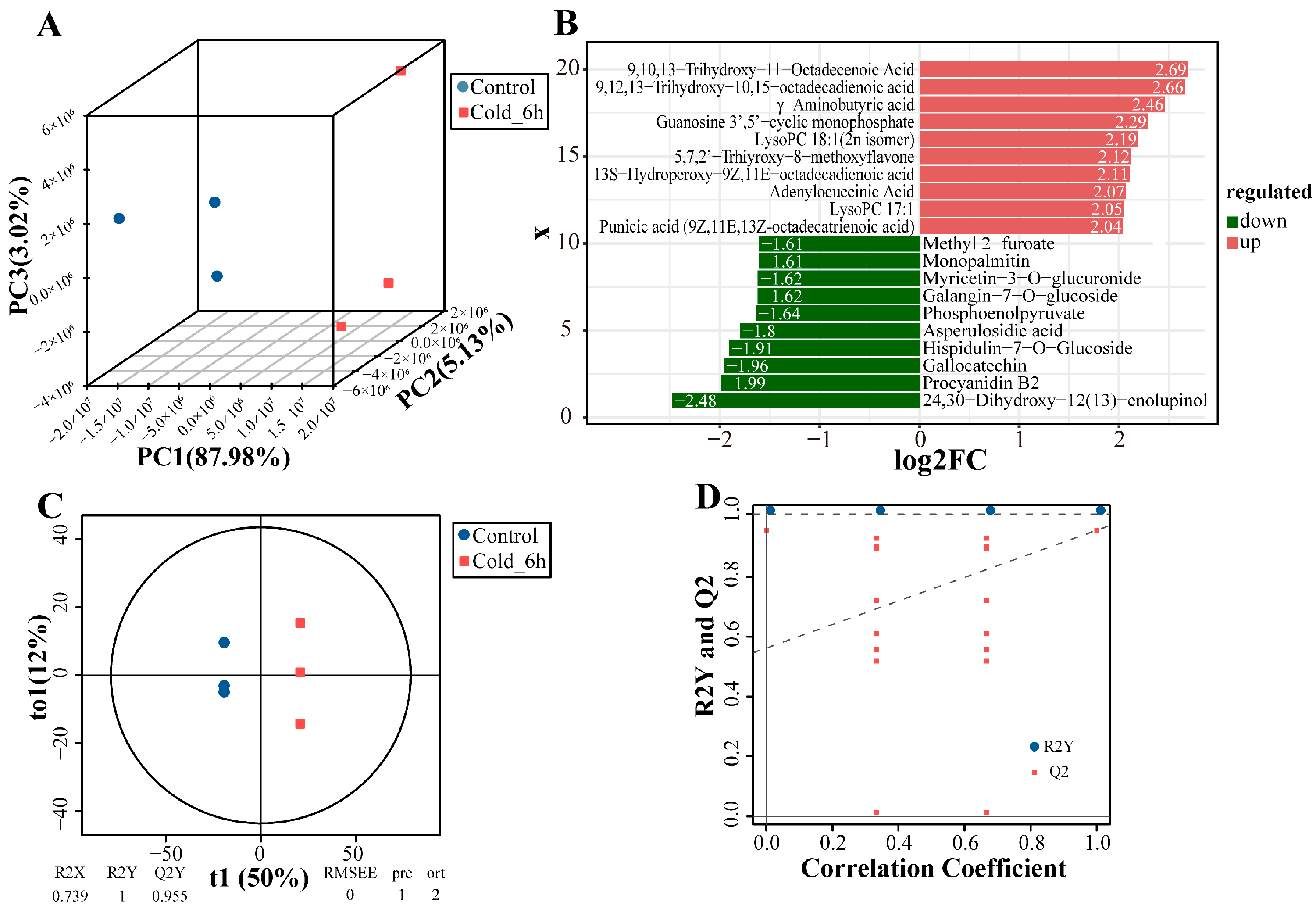
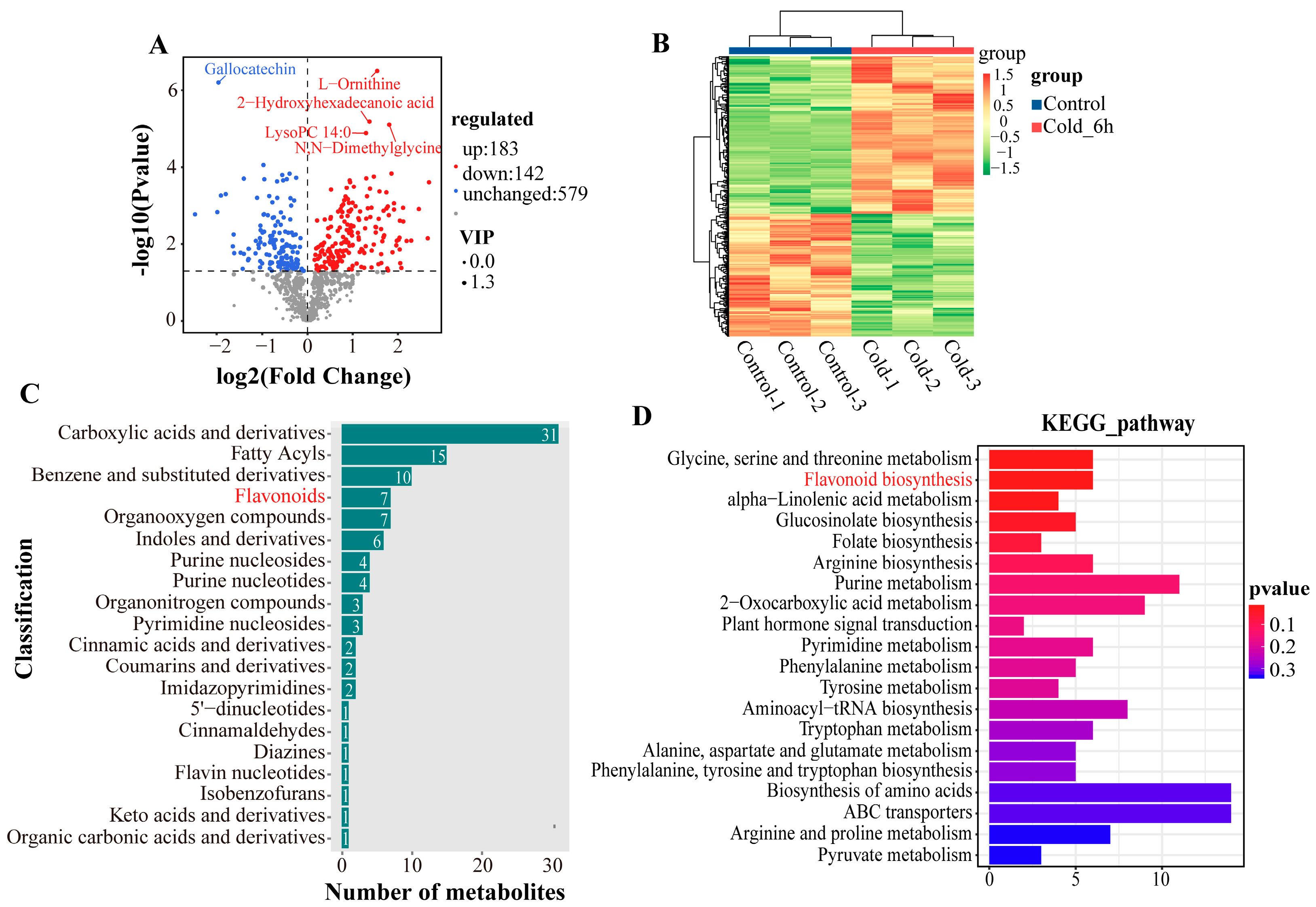
3.2. Metabolite Profiling and Differentially Changed Metabolites Identification of P. ussuriensis Under Cold Stress
3.3. Joint Analysis of Transcriptome and Metabolome
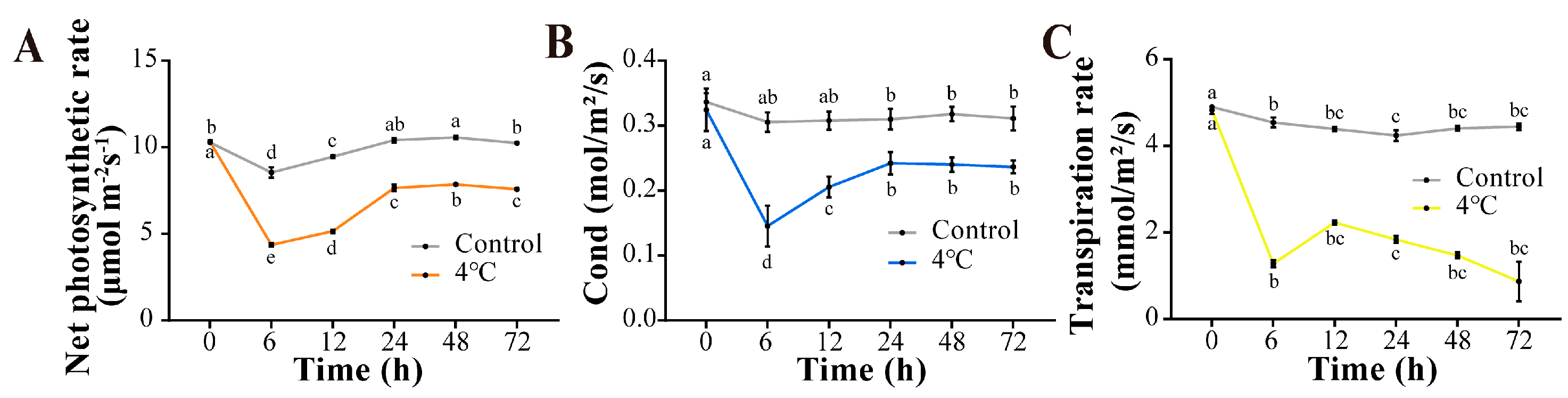
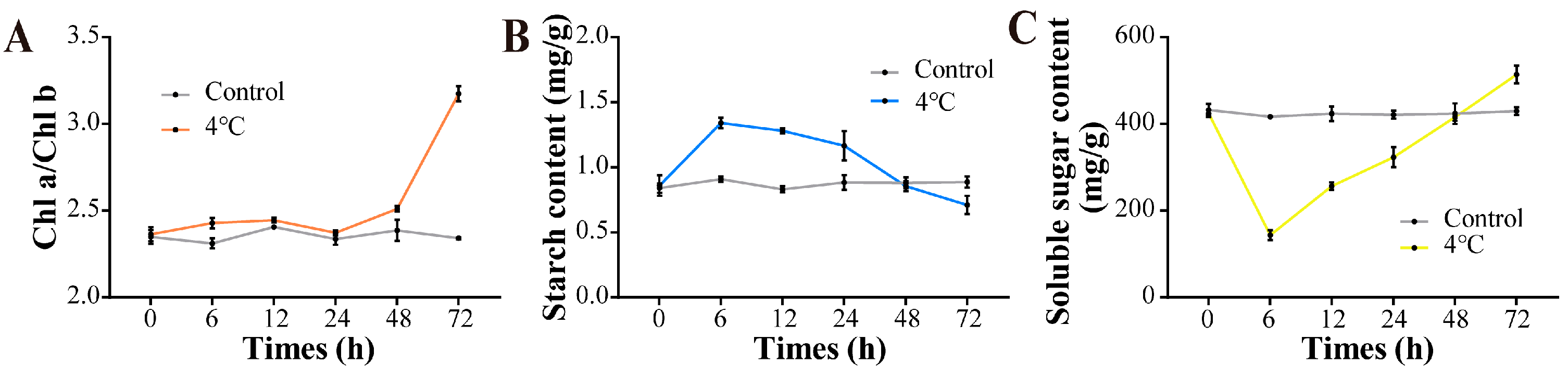
3.4. Chlorophyll Content and Gas Exchange
3.5. Starch Content and Soluble Sugars
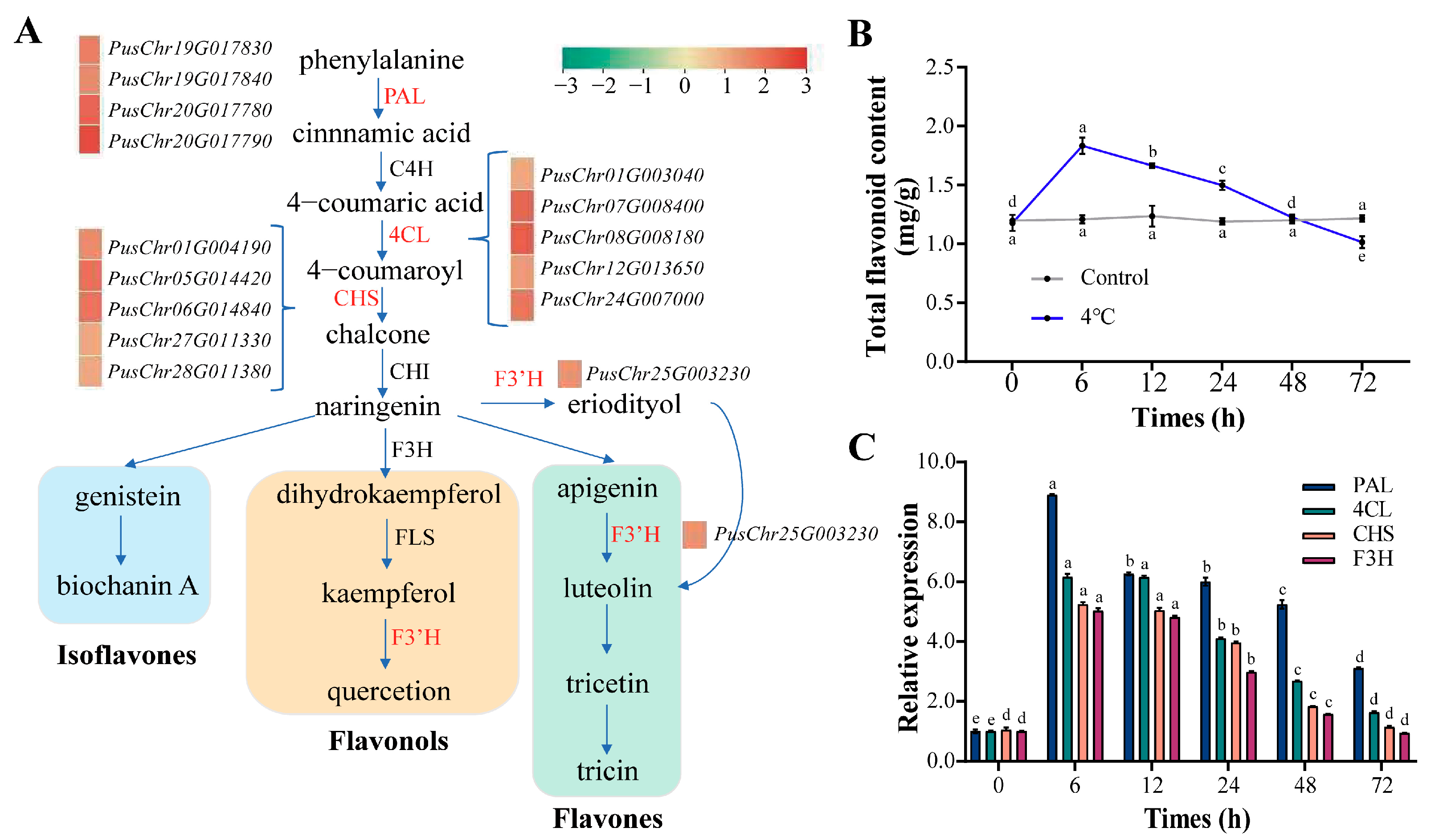
3.6. Flavonoid Biosynthetic Genes and Content
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paredes, M.; Quiles, M.J. The effects of cold stress on photosynthesis in Hibiscus plants. PLoS ONE 2015, 10, e0137472. [Google Scholar] [CrossRef] [PubMed]
- Yamori, W.; Noguchi, K.; Terashima, I. Temperature acclimation of photosynthesis in spinach leaves: Analyses of photosynthetic components and temperature dependencies of photosynthetic partial reactions. Plant Cell Environ. 2005, 28, 536–547. [Google Scholar] [CrossRef]
- Sharma, A.; Kumar, V.; Shahzad, B.; Ramakrishnan, M.; Preet Singh Sidhu, G.; Shreeya Bali, A.; Handa, N.; Kapoor, D.; Yadav, P.; Khanna, K.; et al. Photosynthetic Response of Plants Under Different Abiotic Stresses: A Review. J. Plant Growth Regul. 2020, 39, 509–531. [Google Scholar] [CrossRef]
- Yamori, W.; Sakata, N.; Suzuki, Y.; Shikanai, T.; Makino, A. Cyclic electron flow around photosystem I via chloroplast NAD(P)H dehydrogenase (NDH) complex performs a significant physiological role during photosynthesis and plant growth at low temperature in rice. Plant J. 2011, 68, 966–976. [Google Scholar] [CrossRef]
- Simkin, A.J.; Kapoor, L.; Doss, C.G.P.; Hofmann, T.A.; Lawson, T.; Ramamoorthy, S. The role of photosynthesis related pigments in light harvesting, photoprotection and enhancement of photosynthetic yield in planta. Photosynth. Res. 2022, 152, 23–42. [Google Scholar] [CrossRef]
- Agati, G.; Brunetti, C.; Fini, A.; Gori, A.; Guidi, L.; Landi, M.; Sebastiani, F.; Tattini, M. Are Flavonoids Effective Antioxidants in Plants? Twenty Years of Our Investigation. Antioxidants 2020, 9, 1098. [Google Scholar] [CrossRef]
- Di Ferdinando, M.; Brunetti, C.; Fini, A.; Tattini, M. Flavonoids as Antioxidants in Plants Under Abiotic Stresses. In Abiotic Stress Responses in Plants; Ahmad, P., Prasad, M., Eds.; Springer: New York, NY, USA, 2012. [Google Scholar]
- Nabavi, S.M.; Šamec, D.; Tomczyk, M.; Milella, L.; Russo, D.; Habtemariam, S.; Suntar, I.; Rastrelli, L.; Daglia, M.; Xiao, J.; et al. Flavonoid biosynthetic pathways in plants: Versatile targets for metabolic engineering. Biotechnol Adv. 2020, 38, 107316. [Google Scholar] [CrossRef]
- Yadav, P.; Vaidya, E.; Rani, R.; Yadav, N.K.; Singh, B.K.; Rai, P.K.; Singh, D. Recent Perspective of Next Generation Sequencing: Applications in Molecular Plant Biology and Crop Improvement. Proc. Natl. Acad. Sci. India 2018, 88, 435–449. [Google Scholar] [CrossRef]
- Liao, J.; Tan, J.; Li, X.; Wang, L.; Zhu, L.; Chen, Z.; Gan, L.; Liu, W.; Jin, X. Combined transcriptome and metabolome analysis revealed the antimicrobial mechanism of Griseorhodin C against Methicillin-resistant Staphylococcus aureus. Sci. Rep. 2024, 14, 30242. [Google Scholar] [CrossRef]
- Wang, L.; Wu, L.M.; Greaves, I.K.; Zhu, A.; Dennis, E.S.; Peacock, W.J. PIF4-controlled auxin pathway contributes to hybrid vigor in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2017, 114, E3555–E3562. [Google Scholar] [CrossRef]
- Guo, H.; Zhang, C.; Wang, Y.; Zhang, Y.; Zhang, Y.; Wang, Y.; Wang, C. Expression profiles of genes regulated by BplMYB46 in Betula platyphylla. J. For. Res. 2019, 30, 2267–2276. [Google Scholar] [CrossRef]
- Wang, H.; Pak, S.; Yang, J.; Wu, Y.; Li, W.; Feng, H.; Yang, J.; Wei, H.; Li, C. Two high hierarchical regulators, PuMYB40 and PuWRKY75, control the low phosphorus driven adventitious root formation in Populus ussuriensis. Plant Biotechnol. J. 2022, 20, 1561–1577. [Google Scholar] [CrossRef]
- Pérez-Vicente, A.; Gil-Izquierdo, A.; García-Viguera, C. In vitro gastrointestinal digestion study of pomegranate juice phenolic compounds, anthocyanins, and vitamin C. J. Agric. Food Chem. 2002, 50, 2308–2312. [Google Scholar] [CrossRef]
- Jaakola, L.; Pirttilä, A.M.; Halonen, M.; Hohtola, A. Isolation of high quality RNA from bilberry (Vaccinium myrtillus L.) fruit. Mol Biotechnol. 2001, 19, 201–203. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Delcher, A.L.; Mount, S.M.; Wortman, J.R.; Smith, R.K., Jr.; Hannick, L.I.; Maiti, R.; Ronning, C.M.; Rusch, D.B.; Town, C.D.; et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003, 31, 5654–5666. [Google Scholar] [CrossRef]
- Majoros, W.H.; Pertea, M.; Salzberg, S.L. TigrScan and GlimmerHMM: Two open source ab initio eukaryotic gene-finders. Bioinformatics 2004, 20, 2878–2879. [Google Scholar] [CrossRef]
- Stanke, M.; Steinkamp, R.; Waack, S.; Morgenstern, B. AUGUSTUS: A web server for gene finding in eukaryotes. Nucleic Acids Res. 2004, 32, W309–W312. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Fang, S.; Liu, C.; Zhao, L.; Cong, B.; Zhang, Z. Transcriptomics integrated with metabolomics reveal the effects of ultraviolet-b radiation on flavonoid biosynthesis in Antarctic moss. Front. Plant Sci. 2021, 12, 788377. [Google Scholar] [CrossRef]
- Xiong, Q.; Sun, C.; Li, A.; Zhang, J.; Shi, Q.; Zhang, Y.; Hu, J.; Zhou, N.; Wei, H.; Liu, B.; et al. Metabolomics and biochemical analyses revealed metabolites important for the antioxidant properties of purple glutinous rice. Food Chem. 2022, 389, 133080. [Google Scholar] [CrossRef]
- Durand, M.; Porcheron, B.; Hennion, N.; Maurousset, L.; Lemoine, R.; Pourtau, N. Water deficit enhances C export to the roots in Arabidopsis thaliana plants with contribution of sucrose transporters in both shoot and roots. Plant Physiol. 2016, 170, 1460–1479. [Google Scholar] [CrossRef]
- Chazaux, M.; Schiphorst, C.; Lazzari, G.; Caffarri, S. Precise estimation of chlorophyll a, b and carotenoid content by deconvolution of the absorption spectrum and new simultaneous equations for Chl determination. Plant J. 2022, 109, 1630–1648. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, J.; Li, W.; Chen, Y.; Lu, H.; Zhao, S.; Li, D.; Wei, M.; Li, C. PuHSFA4a Enhances Tolerance To Excess Zinc by Regulating Reactive Oxygen Species Production and Root Development in Populus. Plant Physiol. 2019, 180, 2254–2271. [Google Scholar] [CrossRef]
- Hasdai, M.; Weiss, B.; Levi, A.; Samach, R. Differential responses of Arabidopsis ecotypes to cold, chilling and freezing temperatures. Ann. Appl. Biol. 2006, 148, 113–120. [Google Scholar] [CrossRef]
- Gao, Y.; Thiele, W.; Saleh, O.; Scossa, F.; Arabi, F.; Zhang, H.; Sampathkumar, A.; Kühn, K.; Fernie, A.; Bock, R.; et al. Chloroplast translational regulation uncovers nonessential photosynthesis genes as key players in plant cold acclimation. Plant Cell 2022, 34, 2056–2079. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Molina, A.; Kleine, T.; Schneider, K.; Mühlhaus, T.; Lehmann, M.; Leister, D. Translational components contribute to acclimation responses to high light, heat, and cold in Arabidopsis. iScience 2020, 23, 101331. [Google Scholar] [CrossRef]
- Borisova-Mubarakshina, M.; Naydov, I.; Vetoshkina, D.; Kozuleva, M.; Vilyanen, D.; Rudenko, N.N.; Ivanov, B. Photosynthetic Antenna Size Regulation as an Essential Mechanism of Higher Plants Acclimation to Biotic and Abiotic Factors: The Role of the Chloroplast Plastoquinone Pool and Hydrogen Peroxide. In Vegetation Index and Dynamics; IntechOpen Ltd.: London, UK, 2021. [Google Scholar]
- Xie, S.; Nie, L.; Zheng, Y.; Wang, J.; Zhao, M.; Zhu, S.; Hou, J.; Chen, G.; Wang, C.; Yuan, L. Comparative proteomic analysis reveals that chlorophyll metabolism contributes to leaf color changes in Wucai (Brassica campestris L.) responding to cold acclimation. J. Proteome Res. 2019, 8, 2478–2492. [Google Scholar] [CrossRef] [PubMed]
- Reinbothe, C.; El Bakkouri, M.; Buhr, F.; Muraki, N.; Nomata, J.; Kurisu, G.; Fujita, Y.; Reinbothe, S. Chlorophyll biosynthesis: Spotlight on protochlorophyllide reduction. Trends Plant Sci. 2010, 15, 614–624. [Google Scholar] [CrossRef]
- Wang, P.; Grimm, B. Organization of chlorophyll biosynthesis and insertion of chlorophyll into the chlorophyll-binding proteins in chloroplasts. Photosynth Res. 2015, 126, 189–202. [Google Scholar] [CrossRef]
- Brodersen, C.R.; Vogelmann, T.C. Do changes in light direction affect absorption profiles in leaves? Funct. Plant Biol. 2010, 37, 403–412. [Google Scholar] [CrossRef]
- Garcia-Molina, A.; Leister, D. Accelerated relaxation of photoprotection impairs biomass accumulation in Arabidopsis. Nat. Plants 2020, 6, 9–12. [Google Scholar] [CrossRef]
- Zhang, J.; Lee, K.P.; Liu, Y.; Kim, C. Temperature-driven changes in membrane fluidity differentially impact FILAMENTATION TEMPERATURE-SENSITIVE H2-mediated photosystem II repair. Plant Cell 2024, 37, koae323. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, L.; Hu, H.; Yang, J.; Cui, J.; Wei, G.; Xu, J. Transcriptome and metabolome changes in Chinese cedar during cold acclimation reveal the roles of flavonoids in needle discoloration and cold resistance. Tree Physiol. 2022, 42, 1858–1875. [Google Scholar] [CrossRef]
- Sun, S.; Fang, J.; Lin, M.; Hu, C.; Qi, X.; Chen, J.; Zhong, Y.; Muhammad, A.; Li, Z.; Li, Y. Comparative metabolomic and transcriptomic studies reveal key metabolism pathways contributing to freezing tolerance under cold stress in kiwifruit. Front. Plant Sci. 2021, 12, 628969. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 2019, 24, 2452. [Google Scholar] [CrossRef]
- Genzel, F.; Dicke, M.D.; Junker-Frohn, L.V.; Neuwohner, A.; Thiele, B.; Putz, A.; Usadel, B.; Wormit, A.; Wiese-Klinkenberg, A. Impact of moderate cold and salt stress on the accumulation of antioxidant flavonoids in the leaves of two Capsicum Cultivars. J. Agric. Food Chem. 2021, 69, 6431–6443. [Google Scholar] [CrossRef]
- Bilger, W.; Johnsen, T.; Schreiber, U. UV-excited chlorophyll fluorescence as a tool for the assessment of UV-protection by the epidermis of plants. J. Exp. Bot. 2001, 52, 2007–2014. [Google Scholar] [CrossRef] [PubMed]
- Döll, S.; Lippmann, R.; Mock, H.P. Proteomic approaches to identify cold-regulated soluble proteins. Methods Mol. Biol. 2014, 1166, 139–158. [Google Scholar]
- Dong, S.; Beckles, D.M. Dynamic changes in the starch-sugar interconversion within plant source and sink tissues promote a better abiotic stress response. J. Plant Physiol. 2019, 234–235, 80–93. [Google Scholar] [CrossRef]
- Kitashova, A.; Adler, S.O.; Richter, A.S.; Eberlein, S.; Dziubek, D.; Klipp, E.; Nägele, T. Limitation of sucrose biosynthesis shapes carbon partitioning during plant cold acclimation. Plant Cell Environ. 2023, 46, 464–478. [Google Scholar] [CrossRef]
- Ren, R.; Wan, Z.; Chen, H.; Zhang, Z. The effect of inter-varietal variation in sugar hydrolysis and transport on sugar content and photosynthesis in Vitis vinifera L. leaves. Plant Physiol. Biochem. 2022, 189, 1–13. [Google Scholar] [CrossRef]
- McCormick, A.J.; Cramer, M.D.; Watt, D.A. Regulation of photosynthesis by sugars in sugarcane leaves. J. Plant Physiol. 2008, 165, 1817–1829. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Li, W.; Li, Z.; Yang, L.; Zhuang, W.; Zhang, J.; Zhang, Z.; Fan, Z.; Wang, F.; Zhao, S.; et al. Integrated Multi-Omics Analysis Reveals Photosynthetic Acclimation and Metabolic Reprogramming in Populus ussuriensis kom. Under Cold Stress. Forests 2025, 16, 660. https://doi.org/10.3390/f16040660
Li J, Li W, Li Z, Yang L, Zhuang W, Zhang J, Zhang Z, Fan Z, Wang F, Zhao S, et al. Integrated Multi-Omics Analysis Reveals Photosynthetic Acclimation and Metabolic Reprogramming in Populus ussuriensis kom. Under Cold Stress. Forests. 2025; 16(4):660. https://doi.org/10.3390/f16040660
Chicago/Turabian StyleLi, Jingjing, Wanxin Li, Zhuolong Li, Lu Yang, Wenhui Zhuang, Jingjing Zhang, Zhaohan Zhang, Zihan Fan, Fude Wang, Shicheng Zhao, and et al. 2025. "Integrated Multi-Omics Analysis Reveals Photosynthetic Acclimation and Metabolic Reprogramming in Populus ussuriensis kom. Under Cold Stress" Forests 16, no. 4: 660. https://doi.org/10.3390/f16040660
APA StyleLi, J., Li, W., Li, Z., Yang, L., Zhuang, W., Zhang, J., Zhang, Z., Fan, Z., Wang, F., Zhao, S., & Yang, J. (2025). Integrated Multi-Omics Analysis Reveals Photosynthetic Acclimation and Metabolic Reprogramming in Populus ussuriensis kom. Under Cold Stress. Forests, 16(4), 660. https://doi.org/10.3390/f16040660




