Genome-Wide Identification of Rubber Tree LRR-RLK Genes and Functional Characterization of HbPSKR2 (HbLRR-RLK174)
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome-Wide Identification of LRR-RLKs in Rubber Tree (Hevea brasiliensis)
2.2. Phylogenetic Analysis
2.3. Chromosomal Localization, Conserved Motif, and Gene Structure Analysis
2.4. HbLRR-RLK Duplication and Synteny Analysis
2.5. Analysis of Cis-Regulatory Elements HbLRR-RLKs
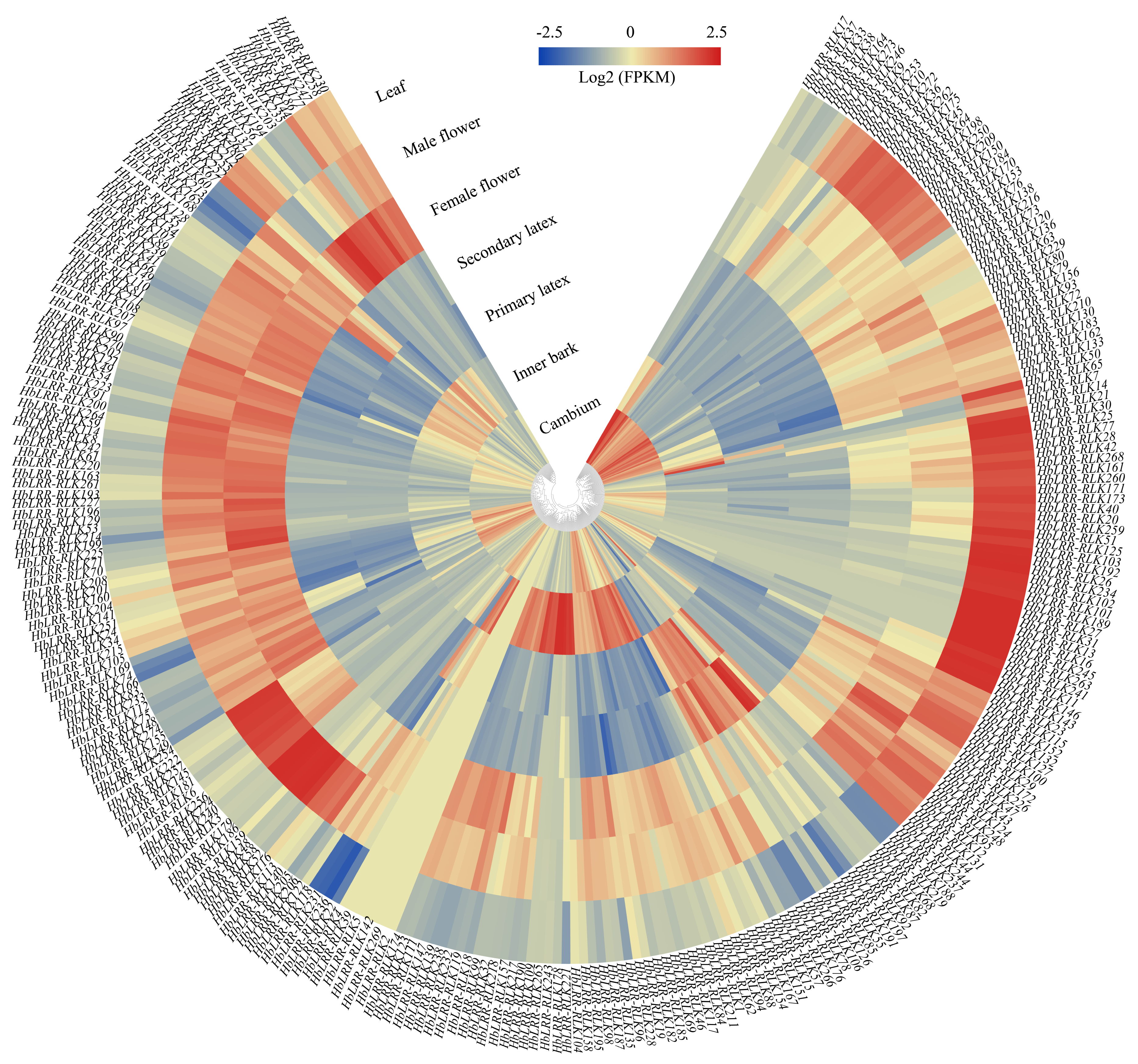
2.6. Tissue-Specific HbLRR-RLKs Expression Patterns
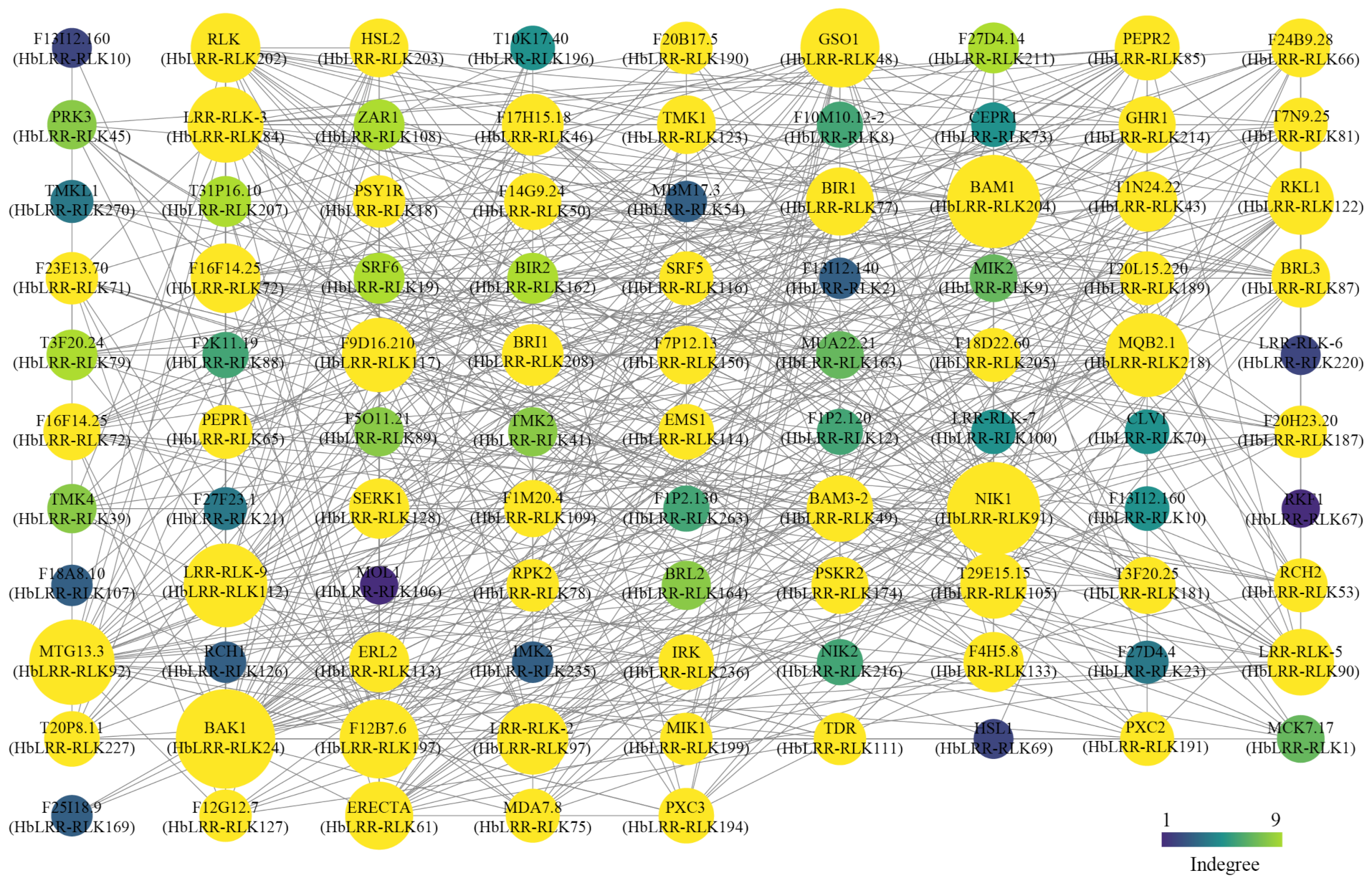
2.7. HbLRR-RLK Protein–Protein Interactions
2.8. Subcellular Localization of HbPSKR2
2.9. Overexpression of HbPSKR2 in Arabidopsis
2.10. Quantitative Real-Time PCR (qRT-PCR) Assay of the HbPSKR2 Gene
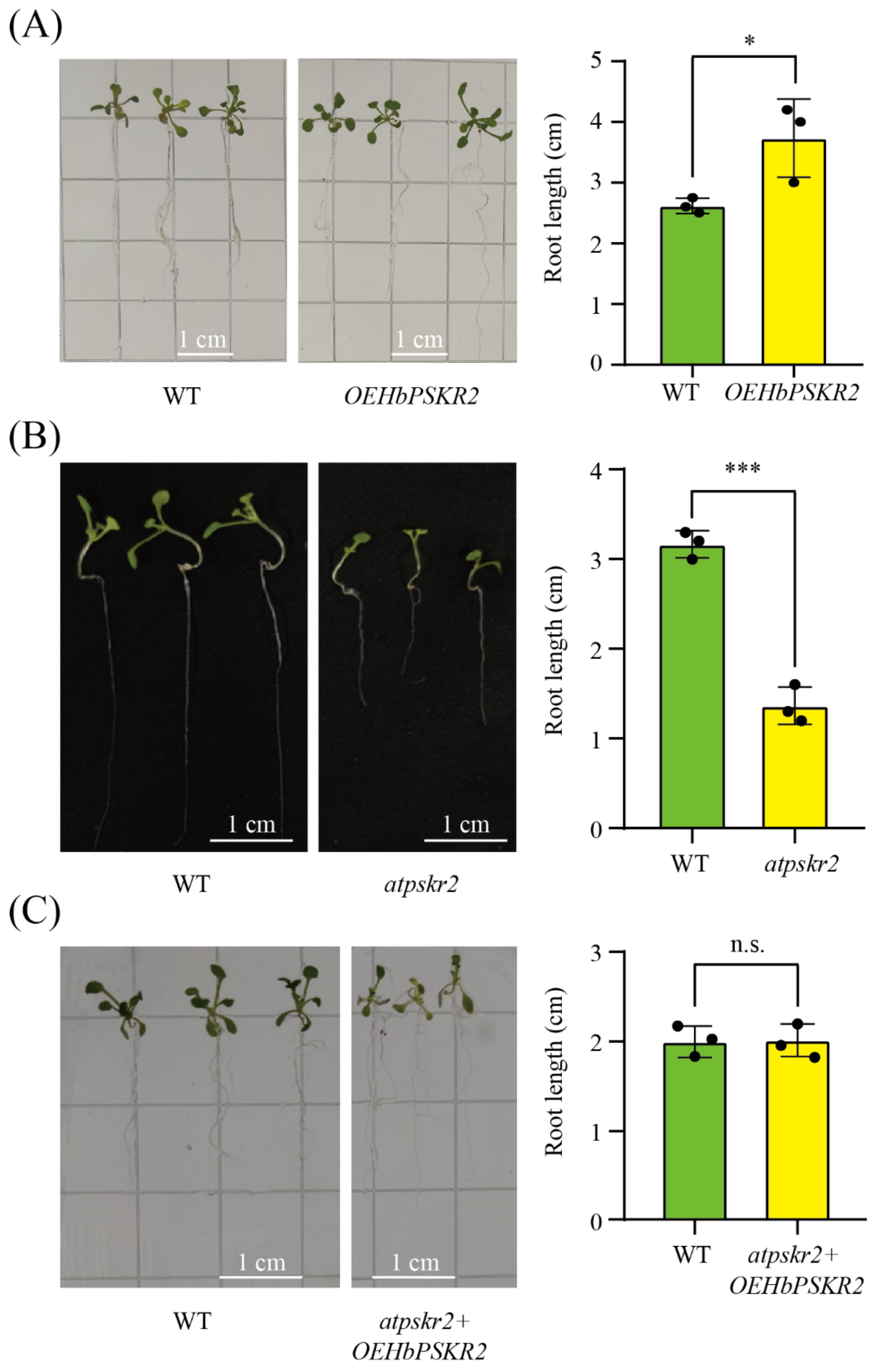
2.11. Phenotypic Analysis of Transgenic Arabidopsis
2.12. Statistical Analysis
3. Results
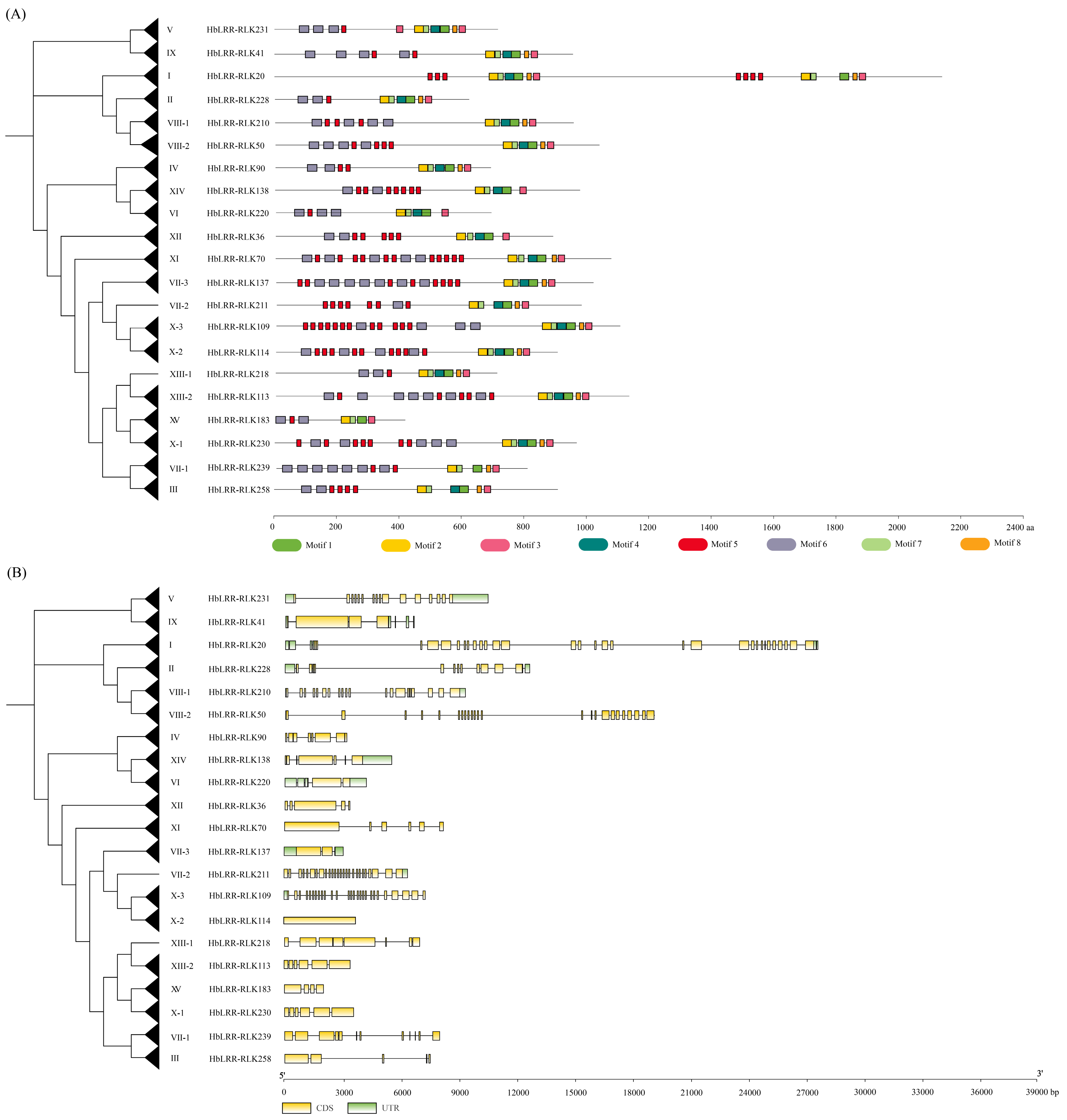
3.1. Characterization of LRR-RLKs in Rubber Tree
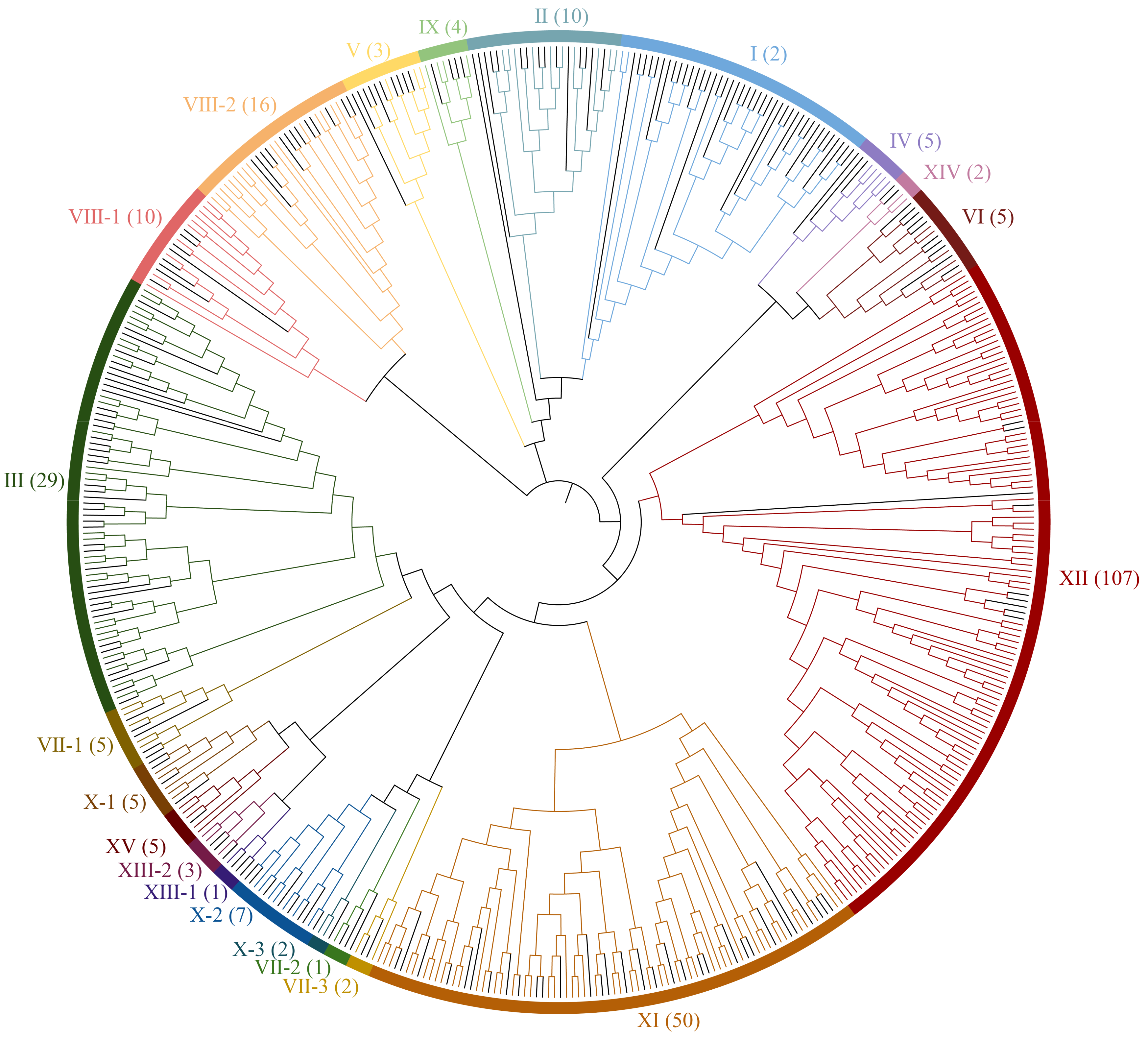
3.2. Phylogenetic Analysis of Rubber Tree HbLRR-RLKs
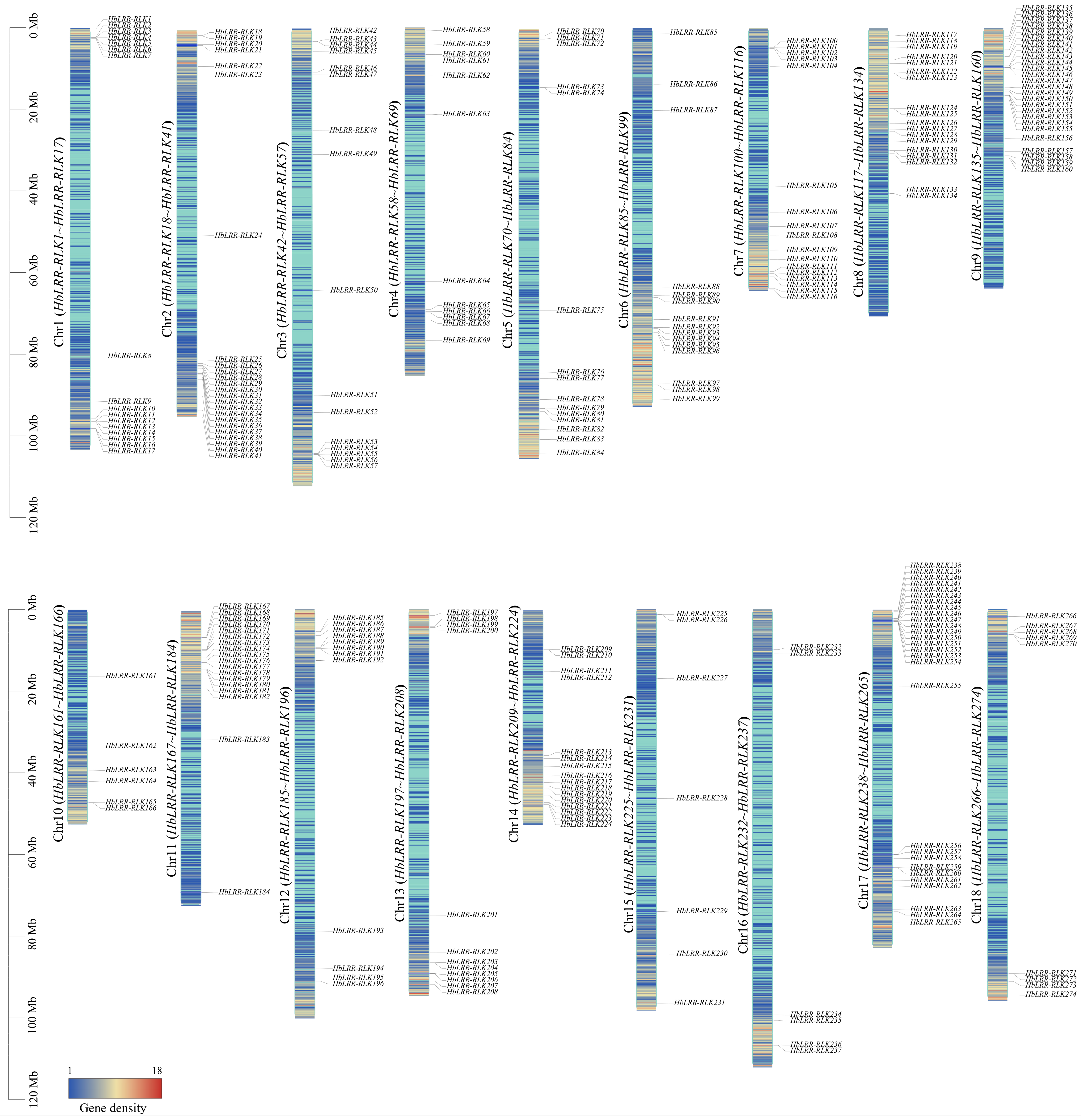
3.3. HbLRR-RLK Gene Duplication and Synteny Analysis
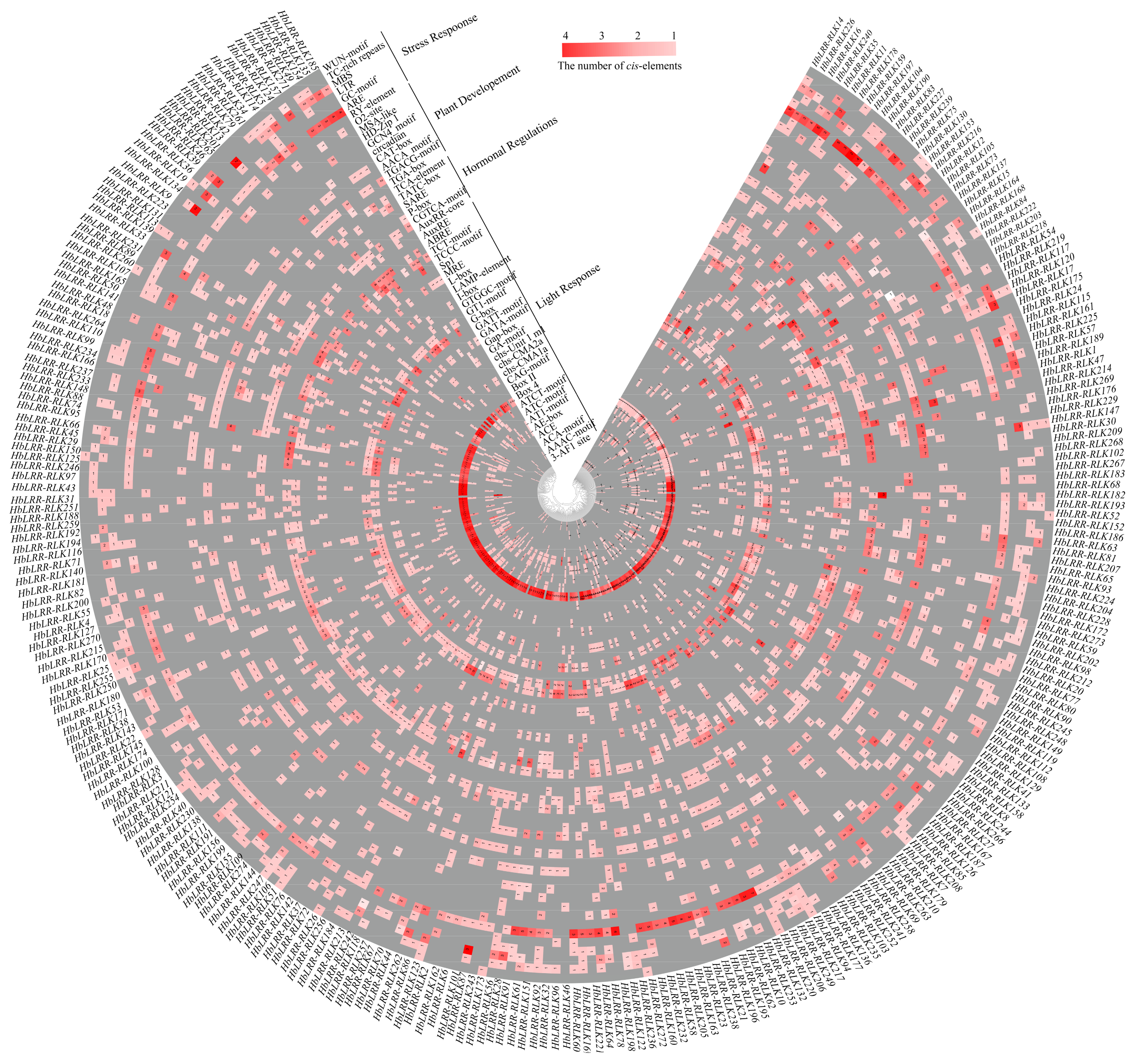
3.4. HbLRR-RLKs Cis-Regulatory Element Analysis
3.5. HbLRR-RLKs Tissue-Specific Expression Patterns
3.6. Prediction of the HbLRR-RLK Protein–Protein Interaction Network
3.7. Subcellular Localization of HbPSKR2
3.8. Functional Identification of HbPSKR2 in Arabidopsis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| LRR-RLK | Leucine-Rich Repeat Receptor-like Kinase |
| ECD | Extracellular receptor domain |
| TM | Transmembrane domain |
| KD | Kinase domain |
| WAK | Wall-associated kinase |
| Duf26 | Domain of unknown function 26 |
| LysM | Lysin motif |
| PSK | Phytosulfokine |
| PSKR | Phytosulfokine receptor |
| NR | Natural rubber |
| NJ | Neighbor-joining |
| Hb | Hevea brasiliensis |
| At | Arabidopsis thaliana |
| Pt | Populus trichocarpa |
| Me | Manihot esculenta |
| BAK | Brassinosteroid-insensitive 1-associated receptor kinase |
| NAA | Nα-acetyltransferase |
| Y2H | Yeast 2 hybrid |
References
- Hubbard, S.R.; Miller, W.T. Receptor tyrosine kinases: Mechanisms of activation and signaling. Curr. Opin. Cell Biol. 2007, 19, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Zhou, J.M. Receptor-like cytoplasmic kinases: Central players in plant receptor kinase-mediated signaling. Annu. Rev. Plant Biol. 2018, 69, 267–299. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.C.; Zhang, R. Relationship of a putative receptor protein kinase from maize to the S-locus glycoproteins of brassica. Nature 1990, 345, 743–746. [Google Scholar] [CrossRef]
- Dievart, A.; Clark, S.E. Using mutant alleles to determine the structure and function of leucine-rich repeat receptor-like kinases. Curr. Opin. Plant Biol. 2003, 6, 507–516. [Google Scholar] [CrossRef]
- Gou, X.; He, K.; Yang, H.; Yuan, T.; Lin, H.; Clouse, S.D.; Li, J. Genome-wide cloning and sequence analysis of leucine-rich repeat receptor-like protein kinase genes in Arabidopsis thaliana. BMC Genom. 2010, 11, 19. [Google Scholar] [CrossRef]
- Afzal, A.J.; Wood, A.J.; Lightfoot, D.A. Plant receptor-like serine threonine kinases: Roles in signaling and plant defense. Mol. Plant Microbe Interact. 2008, 21, 507–517. [Google Scholar] [CrossRef]
- Ngou, B.P.M.; Wyler, M.; Schmid, M.W.; Kadota, Y.; Shirasu, K. Evolutionary trajectory of pattern recognition receptors in plants. Nat. Commun. 2024, 15, 308. [Google Scholar] [CrossRef]
- Bella, J.; Hindle, K.L.; McEwan, P.A.; Lovell, S.C. The leucine-rich repeat structure. Cell. Mol. Life Sci. 2008, 65, 2307–2333. [Google Scholar] [CrossRef]
- Kajava, A.V. Structural diversity of leucine-rich repeat proteins. J. Mol. Biol. 1998, 277, 519–527. [Google Scholar] [CrossRef]
- Magalhães, D.M.; Scholte, L.L.; Silva, N.V.; Oliveira, G.C.; Zipfel, C.; Takita, M.A.; De Souza, A.A. LRR-RLK family from two Citrus species: Genome-wide identification and evolutionary aspects. BMC Genom. 2016, 17, 623. [Google Scholar] [CrossRef]
- Guo, Y.; Han, L.; Hymes, M.; Denver, R.; Clark, S.E. CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification. Plant J. 2010, 63, 889–900. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Zhang, R.; Gui, J.; Zhong, Y.; Li, L. The receptor-like kinase AtVRLK1 regulates secondary cell wall thickening. Plant Physiol. 2018, 177, 671–683. [Google Scholar] [CrossRef] [PubMed]
- Yue, L.; Zhong, M.; Kang, D.; Qin, H.; Li, H.; Chai, X.; Kang, Y.; Yang, X. Genome-wide identification and characterization of flavonol synthase (FLS) gene family in Brassica vegetables and their roles in response to biotic and abiotic stresses. Sci. Hortic. 2024, 331, 113168. [Google Scholar] [CrossRef]
- Matsubayashi, Y.; Sakagami, Y. 120- and 160-kDa receptors for endogenous mitogenic peptide, phytosulfokine-α, in rice plasma membranes. J. Biol. Chem. 2000, 275, 15520–15525. [Google Scholar] [CrossRef]
- Wang, J.; Li, H.; Han, Z.; Zhang, H.; Wang, T.; Lin, G.; Chang, J.; Yang, W.; Chai, J. Allosteric receptor activation by the plant peptide hormone phytosulfokine. Nature 2015, 525, 265–268. [Google Scholar] [CrossRef]
- Matsubayashi, Y.; Ogawa, M.; Kihara, H.; Niwa, M.; Sakagami, Y. Disruption and overexpression of Arabidopsis phytosulfokine receptor gene affects cellular longevity and potential for growth. Plant Physiol. 2006, 142, 45–53. [Google Scholar] [CrossRef]
- Zhang, H.; Hu, Z.; Lei, C.; Zheng, C.; Wang, J.; Shao, S.; Li, X.; Xia, X.; Cai, X.; Zhou, J. A plant phytosulfokine peptide initiates auxin-dependent immunity through cytosolic Ca2+ signaling in tomato. Plant Cell 2018, 30, 652–667. [Google Scholar] [CrossRef] [PubMed]
- Nie, Z.; Kang, G.; Yan, D.; Qin, H.; Yang, L.; Zeng, R. Downregulation of HbFPS1 affects rubber biosynthesis of Hevea brasiliensis suffering from tapping panel dryness. Plant J. 2023, 113, 504–520. [Google Scholar] [CrossRef]
- Zhang, S.X.; Wu, S.H.; Chao, J.Q.; Yang, S.G.; Bao, J.; Tian, W.M. Genome-wide identification and expression analysis of MYC transcription factor family genes in rubber tree (Hevea brasiliensis Muell. Arg.). Forests 2022, 13, 531. [Google Scholar] [CrossRef]
- Yan, Y.; Liang, C.; Liu, X.; Tan, Y.; Lu, Y.; Zhang, Y.; Luo, H.; He, C.; Cao, J.; Tang, C.; et al. Genome-wide association study identifies candidate genes responsible for inorganic phosphorus and sucrose content in rubber tree latex. Trop. Plants 2023, 2, 24. [Google Scholar] [CrossRef]
- Wu, S.; Zhang, S.; Chao, J.; Deng, X.; Chen, Y.; Shi, M.; Tian, W.M. Transcriptome analysis of the signalling networks in coronatine-induced secondary laticifer differentiation from vascular cambia in rubber trees. Sci. Rep. 2016, 6, 36384. [Google Scholar] [CrossRef] [PubMed]
- Wichaita, W.; Promlok, D.; Sudjaipraparat, N.; Sripraphot, S.; Suteewong, T.; Tangboriboonrat, P. A concise review on design and control of structured natural rubber latex particles as engineering nanocomposites. Eur. Polym. J. 2021, 159, 110740. [Google Scholar] [CrossRef]
- Laosombut, T.; Arreewichit, P.; Nirapathpongporn, K.; Traiperm, P.; Kongsawadworakul, P.; Viboonjun, U.; Narangajavana, J. Differential expression of methyl jasmonate-responsive genes correlates with laticifer vessel proliferation in phloem tissue of rubber tree (Hevea brasiliensis). J. Plant Growth Regul. 2016, 35, 1049–1063. [Google Scholar] [CrossRef]
- Chao, J.; Wu, S.; Shi, M.; Xu, X.; Gao, Q.; Du, H.; Tian, W.M. Genomic insight into domestication of rubber tree. Nat. Commun. 2023, 14, 4651. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Linster, E.; Layer, D.; Bienvenut, W.V.; Dinh, T.V.; Weyer, F.A.; Leemhuis, W.; Brünje, A.; Hoffrichter, M.; Miklankova, P.; Kopp, J.; et al. The Arabidopsis Nα-acetyltransferase NAA60 locates to the plasma membrane and is vital for the high salt stress response. New Phytol. 2020, 228, 554–569. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.; Feeney, M.; Ahmadi, M.; Lonoce, C.; Sajari, R.; Di Cola, A.; Frigerio, L. Subcellular localization and interactions among rubber particle proteins from Hevea brasiliensis. J. Exp. Bot. 2017, 68, 5045–5055. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Liu, W.; Zhou, Y.; Li, X.; Wang, X.; Dong, Y.; Wang, N.; Liu, X.; Chen, H.; Yao, N.; Cui, X.; et al. Tissue-Specific Regulation of Gma-miR396 Family on Coordinating Development and Low Water Availability Responses. Front. Plant Sci. 2017, 8, 1112. [Google Scholar] [CrossRef]
- Noman, M.; Jameel, A.; Qiang, W.D.; Ahmad, N.; Liu, W.C.; Wang, F.W.; Li, H.Y. Overexpression of GmCAMTA12 Enhanced Drought Tolerance in Arabidopsis and Soybean. Int. J. Mol. Sci. 2019, 20, 4849. [Google Scholar] [CrossRef]
- Soltabayeva, A.; Dauletova, N.; Serik, S.; Sandybek, M.; Omondi, J.O.; Kurmanbayeva, A.; Srivastava, S. Receptor-like kinases (LRR-RLKs) in response of plants to biotic and abiotic stresses. Plants 2022, 11, 2660. [Google Scholar] [CrossRef]
- Matsushima, N.; Miyashita, H. Leucine-rich repeat (LRR) domains containing intervening motifs in plants. Biomolecules 2012, 2, 288–311. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Wang, S.; Ma, D.; Liu, C. Genome-wide analysis of LRR-RLK gene family in four Gossypium species and expression analysis during cotton development and stress responses. Genes 2018, 9, 592. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Wang, Y.; Yin, H.; Gao, M.; Zhang, Q.; Chen, Y. Genome-wide identification and characterization of the LRR-RLK gene family in two Vernicia species. Int. J. Genom. 2015, 2015, 823427. [Google Scholar] [CrossRef]
- Su, Y.; Peng, X.; Shen, S. Identification of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in paper mulberry and their potential roles in response to cold stress. Comput. Biol. Chem. 2022, 97, 107622. [Google Scholar] [CrossRef]
- Zhou, F.; Guo, Y.; Qiu, L.J. Genome-wide identification and evolutionary analysis of leucine-rich repeat receptor-like protein kinase genes in soybean. BMC Plant Biol. 2016, 16, 58. [Google Scholar] [CrossRef]
- Liu, S.; Lei, J.; Zhang, J.; Liu, H.; Ye, Z.; Yang, J.; Lu, Q.; Liu, P.; Chen, J.; Yang, J. Genome-wide identification and analysis of wheat LRR-RLK family genes following Chinese wheat mosaic virus infection. Front. Plant Sci. 2023, 13, 1109845. [Google Scholar] [CrossRef]
- Zan, Y.; Ji, Y.; Zhang, Y.; Yang, S.; Song, Y.; Wang, J. Genome-wide identification, characterization and expression analysis of Populus leucine-rich repeat receptor-like protein kinase genes. BMC Genom. 2013, 14, 318. [Google Scholar] [CrossRef]
- Oren, M.; Barela Hudgell, M.A.; D’Allura, B.; Agronin, J.; Gross, A.; Podini, D.; Smith, L.C. Short tandem repeats, segmental duplications, gene deletion, and genomic instability in a rapidly diversified immune gene family. BMC Genom. 2016, 17, 900. [Google Scholar] [CrossRef]
- Li, X.; Salman, A.; Guo, C.; Yu, J.; Cao, S.; Gao, X.; Li, W.; Li, H.; Guo, Y. Identification and Characterization of LRR-RLK Family Genes in Potato Reveal Their Involvement in Peptide Signaling of Cell Fate Decisions and Biotic/Abiotic Stress Responses. Cells 2018, 7, 120. [Google Scholar] [CrossRef]
- Lespinet, O.; Wolf, Y.I.; Koonin, E.V.; Aravind, L. The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res. 2002, 12, 1048. [Google Scholar] [CrossRef]
- Ding, H.; Feng, X.; Yuan, Y.; Wang, B.; Wang, Y.; Zhang, J. Genomic investigation of duplication, functional conservation, and divergence in the LRR-RLK Family of Saccharum. BMC Genom. 2024, 25, 165. [Google Scholar] [CrossRef]
- Wei, Z.; Wang, J.; Yang, S.; Song, Y. Identification and expression analysis of the LRR-RLK gene family in tomato (Solanum lycopersicum) Heinz 1706. Genome 2015, 58, 121–134. [Google Scholar] [CrossRef]
- Hou, S.; Liu, D.; Huang, S.; Luo, D.; Liu, Z.; Xiang, Q.; Wang, P.; Mu, R.; Han, Z.; Chen, S.; et al. The Arabidopsis MIK2 receptor elicits immunity by sensing a conserved signature from phytocytokines and microbes. Nat. Commun. 2021, 12, 5494. [Google Scholar] [CrossRef]
- Song, X.; Li, J.; Lyu, M.; Kong, X.; Hu, S.; Song, Q.; Zuo, K. CALMODULIN-LIKE-38 and PEP1 RECEPTOR 2 integrate nitrate and brassinosteroid signals to regulate root growth. Plant Physiol. 2021, 187, 1779–1794. [Google Scholar] [CrossRef]
- Du, X.; Zhao, Y.; Zhang, S.; Tian, W.; Chao, J. Cloning and Interacted Protein Identification of HbPSKR2 Gene from Rubber Tree. Chin. J. Trop. Crops 2024, 45, 653–662. [Google Scholar] [CrossRef]
- Ding, S.; Lv, J.; Hu, Z.; Wang, J.; Wang, P.; Yu, J.; Foyer, C.H.; Shi, K. Phytosulfokine peptide optimizes plant growth and defense via glutamine synthetase GS2 phosphorylation in tomato. EMBO J. 2023, 42, e111858. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wen, J.; Lease, K.A.; Doke, J.T.; Tax, F.E.; Walker, J.C. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 2002, 110, 213–222. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, X.; Jin, J.; Wu, S.; Deng, X.; Yang, S.; Shi, M.; Chao, J. Genome-Wide Identification of Rubber Tree LRR-RLK Genes and Functional Characterization of HbPSKR2 (HbLRR-RLK174). Forests 2025, 16, 552. https://doi.org/10.3390/f16030552
Du X, Jin J, Wu S, Deng X, Yang S, Shi M, Chao J. Genome-Wide Identification of Rubber Tree LRR-RLK Genes and Functional Characterization of HbPSKR2 (HbLRR-RLK174). Forests. 2025; 16(3):552. https://doi.org/10.3390/f16030552
Chicago/Turabian StyleDu, Xiaoyu, Jie Jin, Shaohua Wu, Xiaomin Deng, Shuguang Yang, Minjing Shi, and Jinquan Chao. 2025. "Genome-Wide Identification of Rubber Tree LRR-RLK Genes and Functional Characterization of HbPSKR2 (HbLRR-RLK174)" Forests 16, no. 3: 552. https://doi.org/10.3390/f16030552
APA StyleDu, X., Jin, J., Wu, S., Deng, X., Yang, S., Shi, M., & Chao, J. (2025). Genome-Wide Identification of Rubber Tree LRR-RLK Genes and Functional Characterization of HbPSKR2 (HbLRR-RLK174). Forests, 16(3), 552. https://doi.org/10.3390/f16030552




