Identification of the 2AP Regulatory Gene CnProDH in Aromatic Coconut and Screening of Its Regulatory Factors
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Extraction of RNA and Synthesis of cDNA
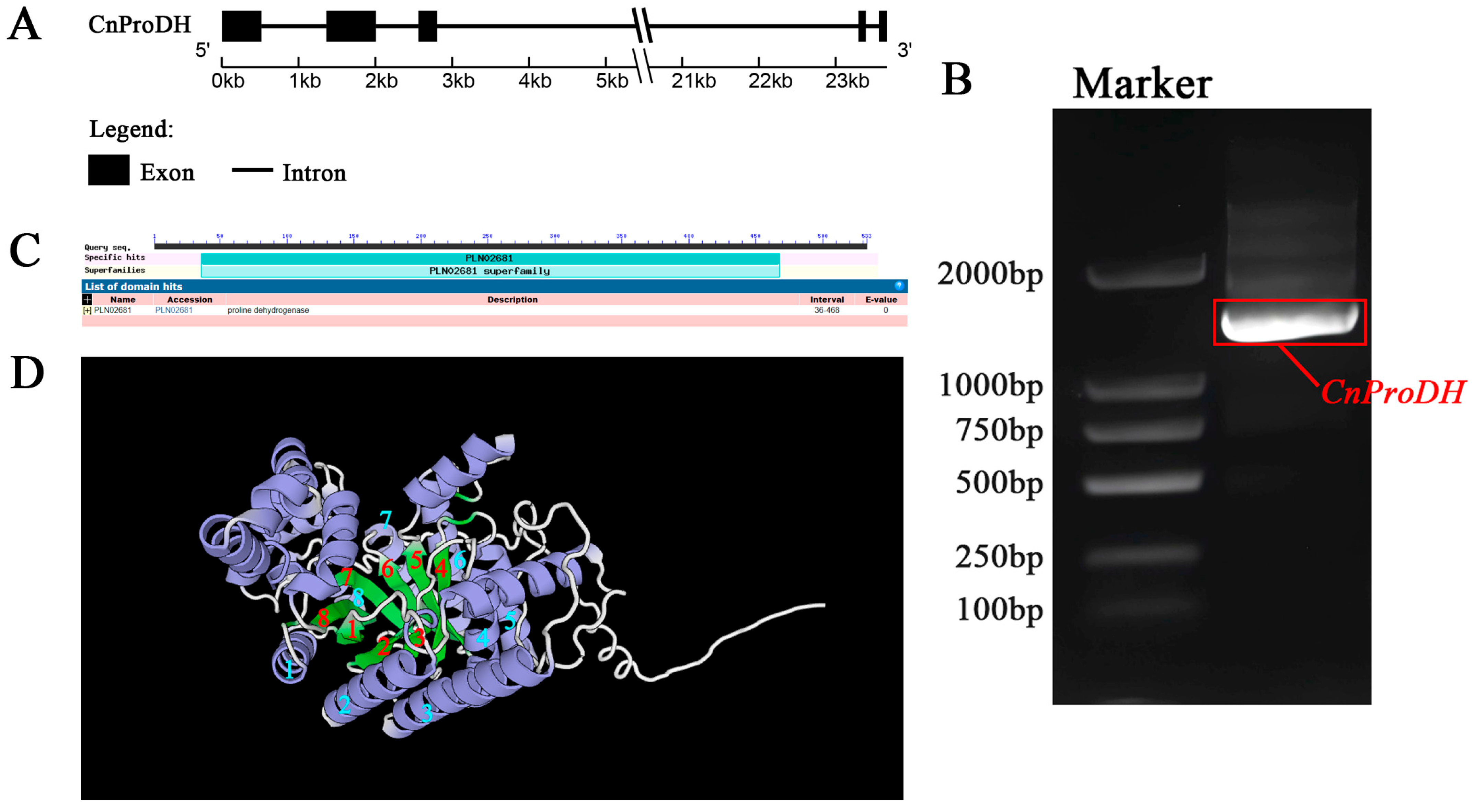
2.3. Gene Cloning and Identification
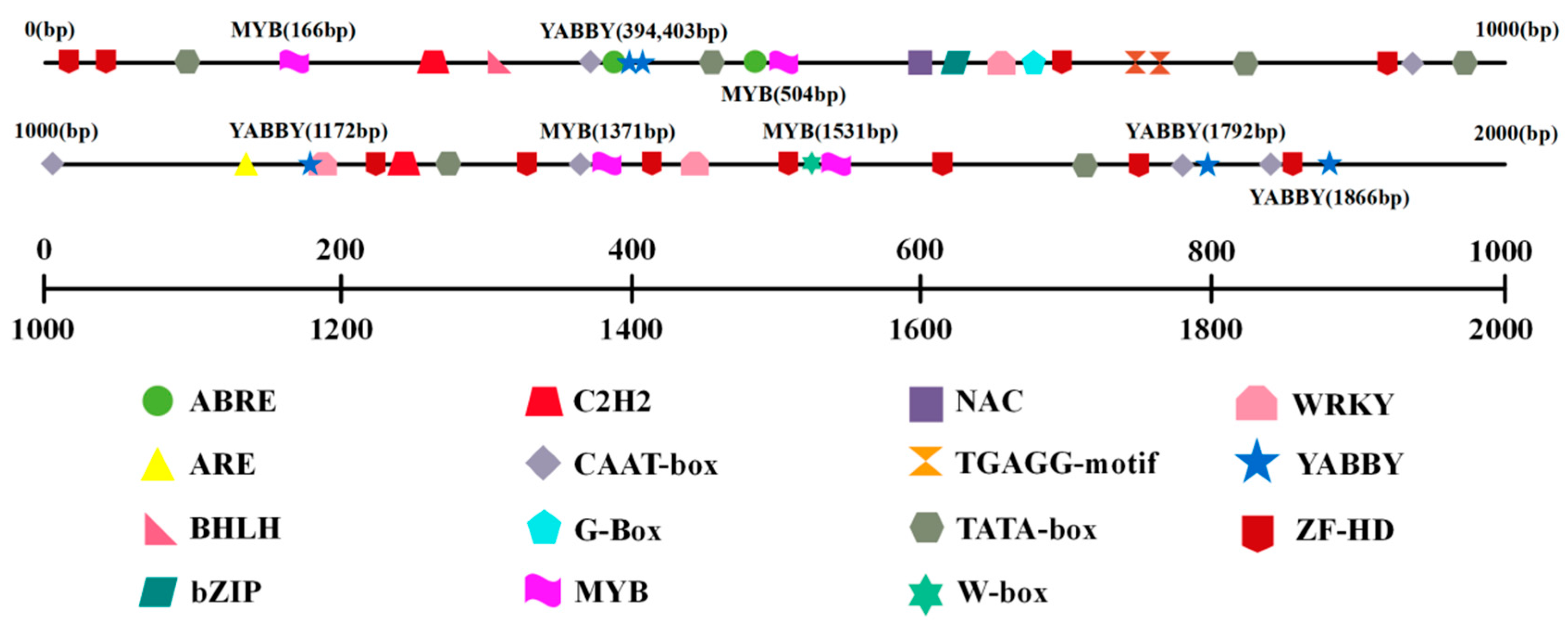
2.4. Prediction of Cis-Acting Elements
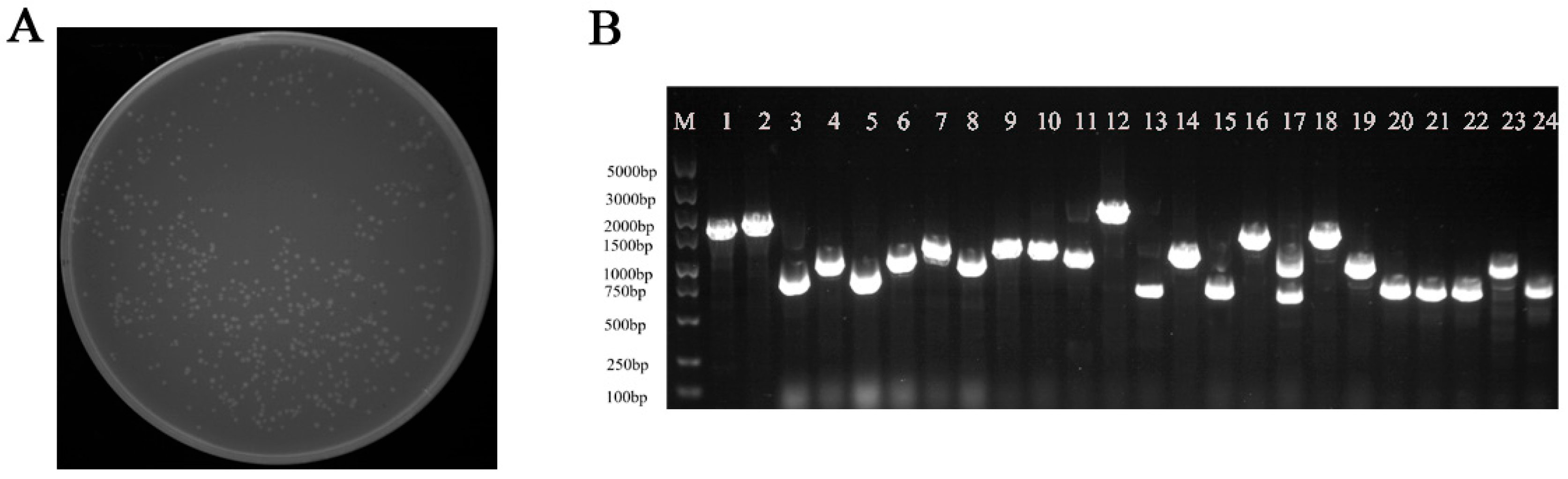
2.5. Construction of Y1H Library
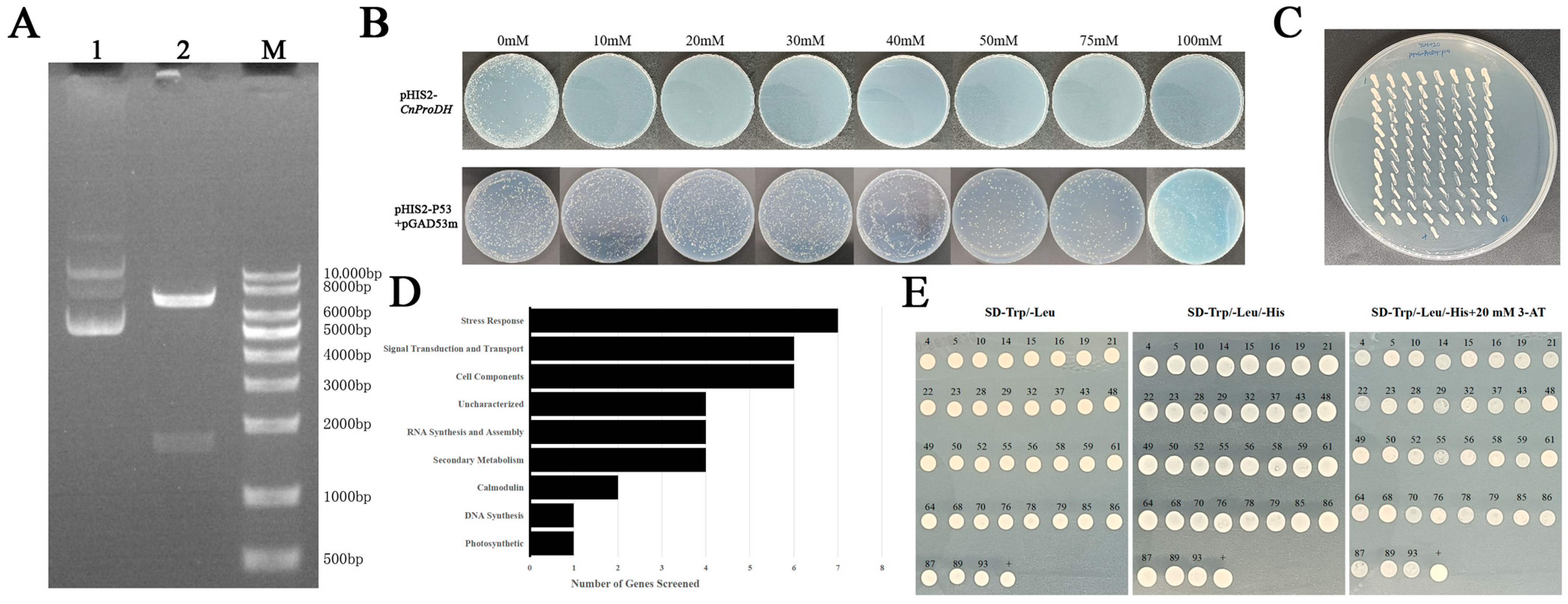
2.6. Construction of Bait Vector and Self-Activation Detection
2.7. Screening of Library
2.8. Identification and Verification of Positive Clones
3. Results
3.1. Cloning and Identification of the CnProDH Gene
3.2. Prediction of Cis-Acting Elements in the CnProDH Promoter
3.3. Quality Detection of Y1H Library
3.4. Bait Vector Construction and Self-Activation Detection
3.5. Obtainment, Identification, and Verification of Positive Clones
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chan, E.; Elevitch, C.R. Cocos nucifera (coconut). In Species Profifiles for Pacifific Island Agroforestry; Permanent Agriculture Resources: Holualoa, HI, USA, 2006; Volume 2, pp. 1–27. Available online: http://www.traditionaltree.org (accessed on 3 September 2025).
- Dumhai, R.; Wanchana, S.; Saensukc, C.; Choowongkomon, K.; Mahatheeranontf, S.; Kraithongg, T.; Toojindab, T.; Vanavichitc, A.; Arikitc, S. Discovery of a novel CnAMADH2 allele associated with higher levels of 2-acetyl-1-pyrroline (2AP) in yellow dwarf coconut (Cocos nucifera L.). Sci. Hortic. 2019, 243, 490–497. [Google Scholar] [CrossRef]
- Buttery, R.G.; Turnbaugh, J.G.; Ling, L.C. Contribution of volatiles to rice aroma. J. Agric. Food Chem. 1988, 36, 1006–1009. [Google Scholar] [CrossRef]
- Buttery, R.G.; Ling, L.C.; Juliano, B.O. Cooked rice aroma and 2-acetyl-1-pyrroline. J. Agric. Food Chem. 1983, 31, 823–826. [Google Scholar] [CrossRef]
- Wongpornchai, S.; Sriseadka, T.; Choonvisase, S. Identification and quantitation of the rice aroma compound, 2-acetyl-1-pyrroline, in bread flowers (Vallaris glabra Ktze). J. Agric. Food Chem. 2003, 51, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Zhang, A.; Qin, X.; Yu, H.; Ji, X.; He, S.; Zong, Y.; Gu, C.; Feng, Z.; Hu, L.; et al. Changes in key volatile components associated with leaf quality of Pandanus amaryllifolius Roxb. alongside growth duration. Food chem. X 2024, 25, 102126. [Google Scholar] [CrossRef]
- Yundaeng, C.; Somta, P.; Tangphatsornruang, S.; Chankaew, S.; Srinives, P. A single base substitution in BADH/AMADH is responsible for fragrance in cucumber (Cucumis sativus L.), and development of SNAP markers for the fragrance. Theor. Appl. Genet. 2015, 128, 1881–1892. [Google Scholar] [CrossRef]
- Qian, L.; Jin, H.; Yang, Q.; Zhu, L.; Yu, X.; Fu, X.; Zhao, M.; Yuan, F. A Sequence Variation in GmBADH2 Enhances Soybean Aroma and Is a Functional Marker for Improving Soybean Flavor. Int. J. Mol. Sci. 2022, 23, 4116. [Google Scholar] [CrossRef]
- Yundaeng, C.; Somta, P.; Tangphatsornruang, S.; Wongpornchai, S.; Srinives, P. Gene discovery and functional marker development for fragrance in sorghum (Sorghum bicolor (L.) Moench). Theor. Appl. Genet. 2013, 126, 2897–2906. [Google Scholar] [CrossRef]
- Saensuk, C.; Wanchana, S.; Choowongkomon, K.; Wongpornchai, S.; Kraithong, T.; Imsabai, W.; Chaichoompu, E.; Ruanjaichon, V.; Toojinda, T.; Vanavichit, A.; et al. De novo transcriptome assembly and identification of the gene conferring a “pandan-like” aroma in coconut (Cocos nucifera L.). Plant Sci. 2016, 252, 324–334. [Google Scholar] [CrossRef]
- Okpala, N.E.; Mo, Z.; Duan, M.; Tang, X. The genetics and biosynthesis of 2-acetyl-1-pyrroline in fragrant rice. Plant Physiol. Biochem. 2019, 135, 272–276. [Google Scholar] [CrossRef]
- Qamar, A.; Mysore, K.S.; Senthil-Kumar, M. Role of proline and pyrroline-5-carboxylate metabolism in plant defense against invading pathogens. Front. Plant Sci. 2015, 6, 503. [Google Scholar] [CrossRef]
- Verbruggen, N.; Hermans, C. Proline accumulation in plants: A review. Amino Acids 2008, 35, 753–759. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Nadaraia, S.; Gu, D.; Becker, D.F.; Tanner, J.J. Structure of the proline dehydrogenase domain of the multifunctional PutA flavoprotein. Nat. Struct. Biol. 2003, 10, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.C.; Huang, Y.W.; Hung, H.J.; Ho, C.T.; Wu, M.L. Delta1-pyrroline-5-carboxylic acid formed by proline dehydrogenase from the Bacillus subtilis ssp. natto expressed in Escherichia coli as a precursor for 2-acetyl-1-pyrroline. J. Agric. Food Chem. 2007, 55, 5097–5102. [Google Scholar] [CrossRef]
- Luo, H.; Zhang, T.; Zheng, A.; He, L.; Lai, R.; Liu, J.; Xing, P.; Tang, X. Exogenous proline induces regulation in 2-acetyl-1-pyrroline (2-AP) biosynthesis and quality characters in fragrant rice (Oryza sativa L.). Sci. Rep. 2020, 10, 13971. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.M.; Reed, R.R. Molecular cloning of the olfactory neuronal transcription factor Olf-1 by genetic selection in yeast. Nature 1993, 364, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Reece-Hoyes, J.S.; Marian Walhout, A.J. Yeast one-hybrid assays: A historical and technical perspective. Methods 2012, 57, 441–447. [Google Scholar] [CrossRef]
- Qiao, J.; Jiang, H.; Lin, Y.; Shang, L.; Wang, M.; Li, D.; Fu, X.; Geisler, M.; Qi, Y.; Gao, Z.; et al. A novel miR167a-OsARF6-OsAUX3 module regulates grain length and weight in rice. Mol. Plant 2021, 14, 1683–1698. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.; Kang, K.; Shim, Y.; Yoo, S.C.; Paek, N.C. Inactivating transcription factor OsWRKY5 enhances drought tolerance through abscisic acid signaling pathways. Plant Physiol. 2022, 188, 1900–1916. [Google Scholar] [CrossRef]
- Fu, D.; Chen, Y.; Gao, F. Yeast One-Hybrid Screening for Transcription Factors of IbbHLH2 in Purple-Fleshed Sweet Potato. Genes 2023, 14, 1042. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Xiao, Y.; Zhou, Z.W.; Yuan, J.; Guo, H.; Yang, Z.; Yang, J.; Sun, P.; Sun, L.; Deng, Y.; et al. High-quality reference genome sequences of two coconut cultivars provide insights into evolution of monocot chromosomes and differentiation of fiber content and plant height. Genome Biol. 2021, 22, 304. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Chow, C.N.; Yang, C.W.; Wu, N.Y.; Wang, H.T.; Tseng, K.C.; Chiu, Y.H.; Lee, T.Y.; Chang, W.C. PlantPAN 4.0: Updated database for identifying conserved non-coding sequences and exploring dynamic transcriptional regulation in plant promoters. Nucleic Acids Res. 2024, 52, D1569–D1578. [Google Scholar] [CrossRef]
- UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2025. Nucleic Acids Res. 2025, 53, D609–D617. [Google Scholar] [CrossRef]
- Giri, J.; Dansana, P.K.; Kothari, K.S.; Sharma, G.; Vij, S.; Tyagi, A.K. SAPs as novel regulators of abiotic stress response in plants. BioEssays 2013, 35, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, H.; He, D.; Damaris, R.N.; Yang, P. A stress-associated protein OsSAP8 modulates gibberellic acid biosynthesis by reducing the promotive effect of transcription factor OsbZIP58 on OsKO2. J. Exp. Bot. 2022, 73, 2420–2433. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Li, C.; Li, D.; Liu, Y.; Yang, X. Roles of YABBY transcription factors in the modulation of morphogenesis, development, and phytohormone and stress responses in plants. J. Plant Res. 2020, 133, 751–763. [Google Scholar] [CrossRef]
- Hayashi, F.; Ichino, T.; Osanai, M.; Wada, K. Oscillation and regulation of proline content by P5CS and ProDH gene expressions in the light/dark cycles in Arabidopsis thaliana L. Plant Cell Physiol. 2000, 41, 1096–1101. [Google Scholar] [CrossRef]
- Pandey, R.; Müller, A.; Napoli, C.A.; Selinger, D.A.; Pikaard, C.S.; Richards, E.J.; Bender, J.; Mount, D.W.; Jorgensen, R.A. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002, 30, 5036–5055. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Bolinger, A.A.; Zhou, J.; Tian, B. Bromodomain-containing protein 4 (BRD4): A key player in inflammatory bowel disease and potential to inspire epigenetic therapeutics. Expert Opin. Ther. Targets 2023, 27, 1–7. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Gene ID | Amino Acids | Molecular Mass/kDa | Isoelectric Point | Subcellular Localization 1 | ||||
|---|---|---|---|---|---|---|---|---|---|
| Chlo | Golg | Mito | Nucl | Plas | |||||
| CnProDH | AZ02G0033300.1 | 533 | 58076.63 | 7.11 | 8 | 2 | 1 | 2 | 1 |
| Gene Name/Gene ID | Number | Protein Name | Function | Possible Binding Sites |
|---|---|---|---|---|
| CnSAP8/ AZ05G0105480.1 | 23 | Stress-associated protein 8 | Involved in abiotic stress response. | MYB (ACGTG) |
| CnYABBY2/ AZ04G0074960.2 | 61 | YABBY2 | Involved in establishing dorsoventral polarity, morphogenesis and development, as well as phytohormone signaling and stress response. | YABBY (ATCAT; ATGAT) |
| CnBRD3/ AZ05G0120960.3 | 50 | Bromodomain- containing protein 3 | Involved in the regulation of plant growth, development, and stress responses as a coactivator. | Lysine residues |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, X.; Zhou, L.; Li, J.; Yin, J.; Ding, H.; Liu, X.; Yang, Y. Identification of the 2AP Regulatory Gene CnProDH in Aromatic Coconut and Screening of Its Regulatory Factors. Forests 2025, 16, 1707. https://doi.org/10.3390/f16111707
Sun X, Zhou L, Li J, Yin J, Ding H, Liu X, Yang Y. Identification of the 2AP Regulatory Gene CnProDH in Aromatic Coconut and Screening of Its Regulatory Factors. Forests. 2025; 16(11):1707. https://doi.org/10.3390/f16111707
Chicago/Turabian StyleSun, Xiwei, Lixia Zhou, Jing Li, Jinyao Yin, Hao Ding, Xiaomei Liu, and Yaodong Yang. 2025. "Identification of the 2AP Regulatory Gene CnProDH in Aromatic Coconut and Screening of Its Regulatory Factors" Forests 16, no. 11: 1707. https://doi.org/10.3390/f16111707
APA StyleSun, X., Zhou, L., Li, J., Yin, J., Ding, H., Liu, X., & Yang, Y. (2025). Identification of the 2AP Regulatory Gene CnProDH in Aromatic Coconut and Screening of Its Regulatory Factors. Forests, 16(11), 1707. https://doi.org/10.3390/f16111707






