Elevational Patterns of Plant Species Richness: Insights from Western Himalayas
Abstract
1. Introduction
2. Methodology
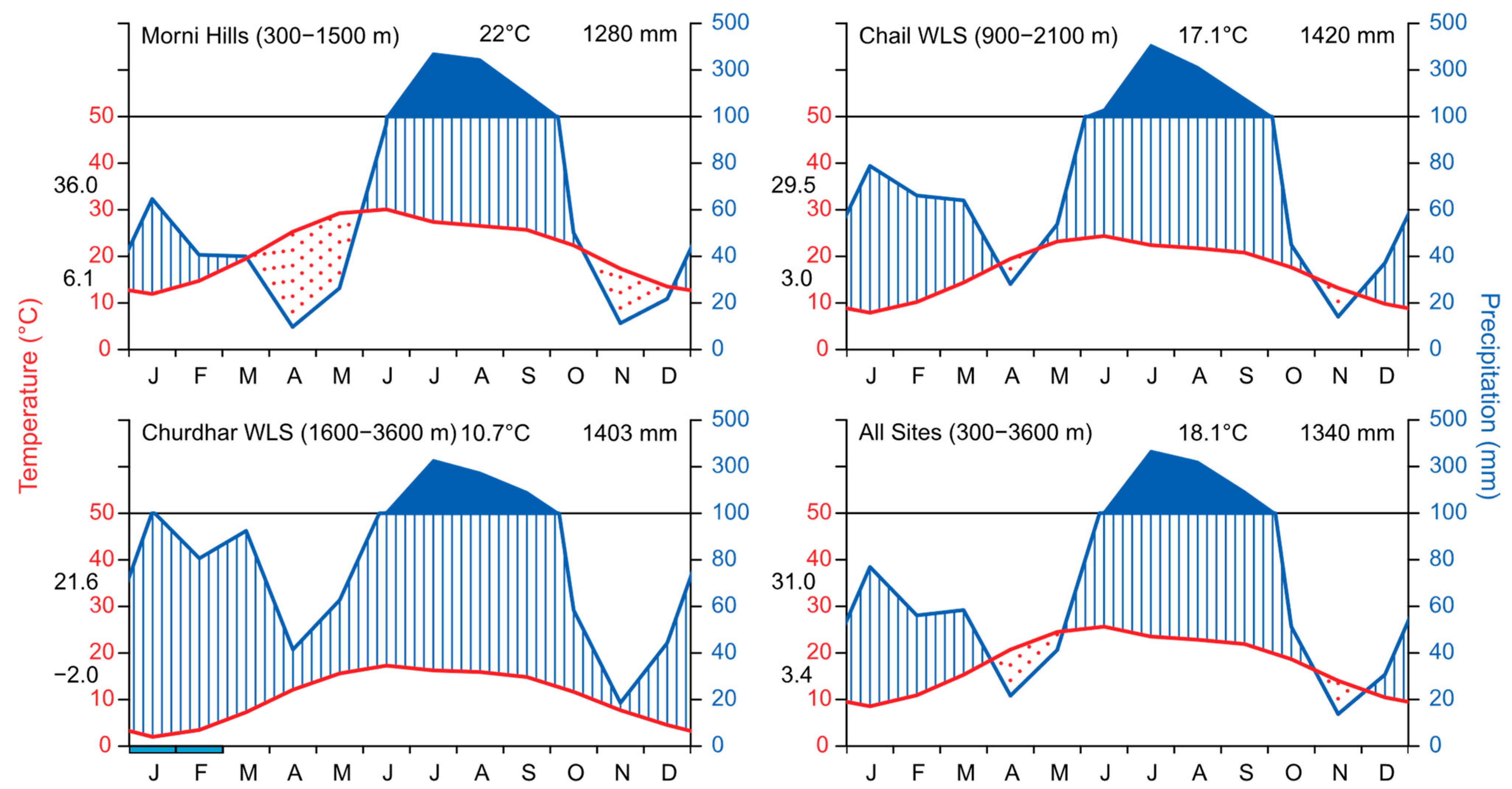
2.1. Study Sites
2.2. Species Check-List
2.3. Distribution Ranges
2.4. Species Richness
2.5. Data Analysis
3. Results
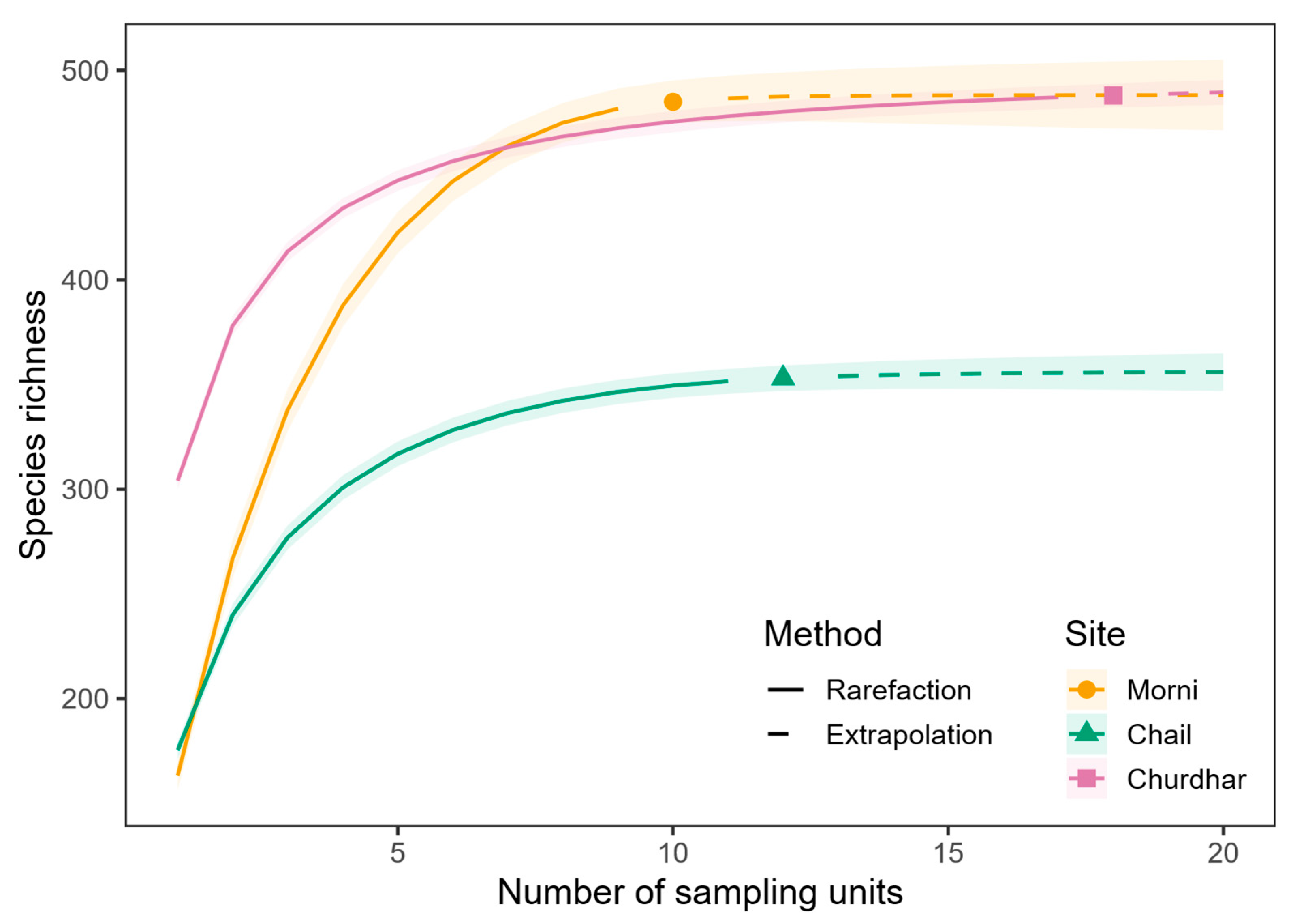
3.1. Species Richness Across Sites
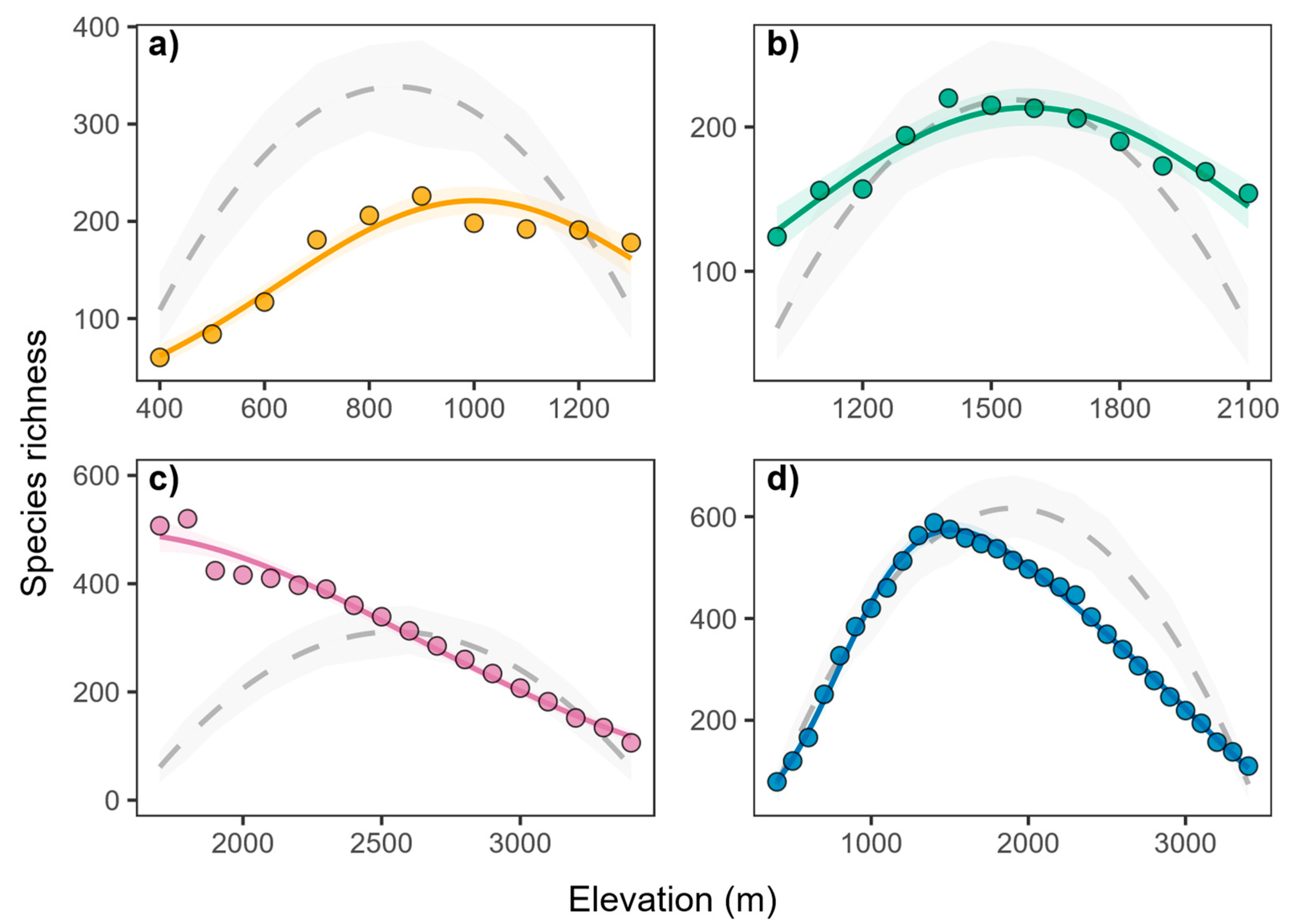
3.2. Elevational Patterns of Species Richness
3.3. Comparison with the Null Model
3.4. Elevational Patterns of Residual Species Richness
3.5. Influence of Observed Species
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AIC | Akaike’s Information Criterion |
| AICc | Akaike’s Information Criterion corrected for small sample size |
| APG IV | Angiosperm Phylogeny Group IV |
| GBIF | Global Biodiversity Information Facility |
| GLM | Generalised linear model |
| MDE | Mid-domain effect |
| POWO | Plants of the World Online |
| WCVP | World Checklist of Vascular Plants |
| WLS | Wildlife Sanctuary |
References
- Bánki, O.; Roskov, Y.; Döring, M.; Ower, G.; Vandepitte, L.; Hobern, D.; Remsen, D.; Schalk, P.; DeWalt, R.E.; Keping, M.; et al. Catalogue of Life Checklist (Annual Checklist 2023); Catalogue of Life: Leiden, The Netherlands, 2023. [Google Scholar] [CrossRef]
- Rahbek, C.; Borregaard, M.K.; Colwell, R.K.; Dalsgaard, B.; Holt, B.G.; Morueta-Holme, N.; Nogues-Bravo, D.; Whittaker, R.J.; Fjeldså, J. Humboldt’s Enigma: What Causes Global Patterns of Mountain Biodiversity? Science 2019, 365, 1108–1113. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Kelt, D.A.; Sun, Z.; Liu, H.; Hu, L.; Ren, H.; Wen, J. Global Variation in Elevational Diversity Patterns. Sci. Rep. 2013, 3, 3007. [Google Scholar] [CrossRef] [PubMed]
- Rahbek, C. The Elevational Gradient of Species Richness: A Uniform Pattern? Ecography 1995, 18, 200–205. [Google Scholar] [CrossRef]
- Lomolino, M.V. Elevation Gradients of Species-Density: Historical and Prospective Views. Glob. Ecol. Biogeogr. 2001, 10, 3–13. [Google Scholar] [CrossRef]
- Steinbauer, M.J.; Grytnes, J.-A.; Jurasinski, G.; Kulonen, A.; Lenoir, J.; Pauli, H.; Rixen, C.; Winkler, M.; Bardy-Durchhalter, M.; Barni, E.; et al. Accelerated Increase in Plant Species Richness on Mountain Summits is Linked to Warming. Nature 2018, 556, 231–234. [Google Scholar] [CrossRef]
- Von Humboldt, A.; Bonpland, A. Essai sur la Géographie des Plantes; Chez Levrault, Schoell et Compagnie: Paris, France, 1805; p. 274. [Google Scholar] [CrossRef]
- Stevens, G.C. The Elevational Gradient in Altitudinal Range: An Extension of Rapoport’s Latitudinal Rule to Altitude. Am. Nat. 1992, 140, 893–911. [Google Scholar] [CrossRef] [PubMed]
- Rahbek, C. The Role of Spatial Scale and the Perception of Large-Scale Species-Richness Patterns. Ecol. Lett. 2005, 8, 224–239. [Google Scholar] [CrossRef]
- McCain, C.M.; Grytnes, J.A. Elevational Gradients in Species Richness. In Encyclopedia of Life Sciences (eLS); Wiley: Chichester, UK, 2010. [Google Scholar] [CrossRef]
- Bryant, J.A.; Lamanna, C.; Morlon, H.; Kerkhoff, A.J.; Enquist, B.J.; Green, J.L. Microbes on Mountainsides: Contrasting Elevational Patterns of Bacterial and Plant Diversity. Proc. Natl. Acad. Sci. USA 2008, 105, 11505–11511. [Google Scholar] [CrossRef]
- McCain, C.M. Global Analysis of Bird Elevational Diversity. Glob. Ecol. Biogeogr. 2009, 18, 346–360. [Google Scholar] [CrossRef]
- Grau, O.; Grytnes, J.A.; Birks, H.J.B. A Comparison of Altitudinal Species Richness Patterns of Bryophytes with Other Plant Groups in Nepal, Central Himalaya. J. Biogeogr. 2007, 34, 1907–1915. [Google Scholar] [CrossRef]
- Rodríguez-Quiel, E.E.; Kluge, J.; Mendieta-Leiva, G.; Bader, M.Y. Elevational Patterns in Tropical Bryophyte Diversity Differ Among Substrates: A Case Study on Baru Volcano, Panama. J. Veg. Sci. 2022, 33, e13136. [Google Scholar] [CrossRef]
- Bhattarai, K.R.; Vetaas, O.R.; Grytnes, J.A. Fern Species Richness Along a Central Himalayan Elevational Gradient, Nepal. J. Biogeogr. 2004, 31, 389–400. [Google Scholar] [CrossRef]
- Kessler, M.; Kluge, J.; Hemp, A.; Ohlemüller, R. A Global Comparative Analysis of Elevational Species Richness Patterns of Ferns. Glob. Ecol. Biogeogr. 2011, 20, 868–880. [Google Scholar] [CrossRef]
- Khine, P.K.; Fraser-Jenkins, C.; Lindsay, S.; Middleton, D.; Miehe, G.; Thomas, P.; Kluge, J. A Contribution Toward the Knowledge of Ferns and Lycophytes from Northern and Northwestern Myanmar. Am. Fern J. 2017, 107, 219–256. [Google Scholar] [CrossRef]
- Qian, H.; Kessler, M.; Vetaas, O.R. Pteridophyte Species Richness in the Central Himalaya Is Limited by Cold Climate Extremes at High Elevations and Rainfall Seasonality at Low Elevations. Ecol. Evol. 2022, 12, e8958. [Google Scholar] [CrossRef]
- Nowak, P.; Khine, P.K.; Homeier, J.; Leuschner, C.; Miehe, G.; Kluge, J. A Plot-Based Elevational Assessment of Species Densities, Life Forms and Leaf Traits of Seed Plants in the South-Eastern Himalayan Biodiversity Hotspot, North Myanmar. Plant Ecol. Divers. 2020, 13, 437–450. [Google Scholar] [CrossRef]
- Kumar, A. Climate-Driven Elevational Shifting of Plant Distributions in the Selected Sites of Siwaliks of North Western India. Ph.D. Thesis, Panjab University, Chandigarh, India, 2024; p. 259. Available online: http://hdl.handle.net/10603/607391 (accessed on 26 September 2025).
- Trigas, P.; Panitsa, M.; Tsiftsis, S. Elevational Gradient of Vascular Plant Species Richness and Endemism in Crete—The Effect of Post-Isolation Mountain Uplift on a Continental Island System. PLoS ONE 2013, 8, e59425. [Google Scholar] [CrossRef]
- Vetaas, O.R.; Grytnes, J.A. Distribution of Vascular Plant Species Richness and Endemic Richness Along the Himalayan Elevation Gradient in Nepal. Glob. Ecol. Biogeogr. 2002, 11, 291–301. [Google Scholar] [CrossRef]
- Acharya, B.K.; Chettri, B.; Vijayan, L. Distribution Pattern of Trees Along an Elevation Gradient of Eastern Himalaya, India. Acta Oecol. 2011, 37, 329–336. [Google Scholar] [CrossRef]
- Homeier, J.; Breckle, S.-W.; Günter, S.; Rollenbeck, R.T.; Leuschner, C. Tree Diversity, Forest Structure and Productivity Along Altitudinal and Topographical Gradients in a Species-Rich Ecuadorian Montane Rain Forest. Biotropica 2010, 42, 140–148. [Google Scholar] [CrossRef]
- Djordjević, V.; Tsiftsis, S.; Kindlmann, P.; Stevanović, V. Orchid Diversity Along an Altitudinal Gradient in the Central Balkans. Front. Ecol. Evol. 2022, 10, 929266. [Google Scholar] [CrossRef]
- Peters, M.K.; Hemp, A.; Appelhans, T.; Behler, C.; Classen, A.; Detsch, F.; Ensslin, A.; Ferger, S.W.; Frederiksen, S.B.; Gebert, F.; et al. Predictors of Elevational Biodiversity Gradients Change from Single Taxa to the Multi-Taxa Community Level. Nat. Commun. 2016, 7, 13736. [Google Scholar] [CrossRef]
- Bisht, M.; Sekar, K.C.; Mukherjee, S.; Thapliyal, N.; Bahukhandi, A.; Singh, D.; Bhojak, P.; Mehta, P.; Upadhyay, S.; Dey, D. Influence of Anthropogenic Pressure on the Plant Species Richness and Diversity Along the Elevation Gradients of Indian Himalayan High-Altitude Protected Areas. Front. Ecol. Evol. 2022, 10, 751989. [Google Scholar] [CrossRef]
- Khuroo, A.A.; Weber, E.; Malik, A.H.; Reshi, Z.A.; Dar, G.H. Altitudinal Distribution Patterns of the Native and Alien Woody Flora in Kashmir Himalaya, India. Environ. Res. 2011, 111, 967–977. [Google Scholar] [CrossRef]
- Li, L.; Xu, X.; Qian, H.; Huang, X.; Liu, P.; Landis, J.B.; Fu, Q.; Sun, L.; Wang, H.; Sun, H.; et al. Elevational Patterns of Phylogenetic Structure of Angiosperms in a Biodiversity Hotspot in Eastern Himalaya. Divers. Distrib. 2022, 28, 2534–2548. [Google Scholar] [CrossRef]
- Di Musciano, M.; Zannini, P.; Ferrara, C.; Spina, L.; Nascimbene, J.; Vetaas, O.R.; Bhatta, K.P.; D’Agostino, M.; Peruzzi, L.; Carta, A.; et al. Investigating Elevational Gradients of Species Richness in a Mediterranean Plant Hotspot Using a Published Flora. Front. Biogeogr. 2021, 13, e50007. [Google Scholar] [CrossRef]
- Grytnes, J.A.; Vetaas, O.R. Species Richness and Altitude: A Comparison Between Null Models and Interpolated Plant Species Richness Along the Himalayan Altitudinal Gradient, Nepal. Am. Nat. 2002, 159, 294–304. [Google Scholar] [CrossRef]
- Gaston, K.J. Global Patterns in Biodiversity. Nature 2000, 405, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Patil, M.; Kumar, P.; Singh, A.N. Determinants of Plant Species Richness Along Elevational Gradients: Insights with Climate, Energy and Water–Energy Dynamics. Ecol. Process. 2024, 13, 86. [Google Scholar] [CrossRef]
- Colwell, R.K.; Lees, D.C. The Mid-Domain Effect: Geometric Constraints on the Geography of Species Richness. Trends Ecol. Evol. 2000, 15, 70–76. [Google Scholar] [CrossRef]
- Colwell, R.K.; Rahbek, C.; Gotelli, N.J. The Mid-Domain Effect and Species Richness Patterns: What Have We Learned so Far? Am. Nat. 2004, 163, E1–E23. [Google Scholar] [CrossRef] [PubMed]
- Dunn, R.R.; McCain, C.M.; Sanders, N.J. When Does Diversity Fit Null Model Predictions? Scale and Range Size Mediate the Mid-Domain Effect. Glob. Ecol. Biogeogr. 2007, 16, 305–312. [Google Scholar] [CrossRef]
- Hu, W.; Wu, F.; Gao, J.; Yan, D.; Liu, L.; Yang, X. Influences of Interpolation of Species Ranges on Elevational Species Richness Gradients. Ecography 2016, 40, 1231–1241. [Google Scholar] [CrossRef]
- Rana, S.K.; Gross, K.; Price, T.D. Drivers of Elevational Richness Peaks, Evaluated for Trees in the East Himalaya. Ecology 2019, 100, e02548. [Google Scholar] [CrossRef]
- Chawla, A.; Rajkumar, S.; Singh, K.N.; Lal, B.; Singh, R.D.; Thukral, A.K. Plant Species Diversity Along an Altitudinal Gradient of Bhabha Valley in Western Himalaya. J. Mt. Sci. 2008, 5, 157–177. [Google Scholar] [CrossRef]
- Oommen, M.A.; Shanker, K. Elevational Species Richness Patterns Emerge from Multiple Local Mechanisms in Himalayan Woody Plants. Ecology 2005, 86, 3039–3047. [Google Scholar] [CrossRef]
- Beck, H.E.; Zimmermann, N.E.; McVicar, T.R.; Vergopolan, N.; Berg, A.; Wood, E.F. Present and Future Köppen-Geiger Climate Classification Maps at 1-Km Resolution. Sci. Data 2018, 5, 180214. [Google Scholar] [CrossRef]
- Champion, H.G.; Seth, S.K. A Revised Survey of the Forest Types of India; Government of India: Delhi, India, 1968; p. 404. [Google Scholar]
- IUCN. The IUCN Red List of Threatened Species; Version 2022-2. 2023. Available online: https://www.iucnredlist.org/ (accessed on 17 June 2023).
- Rahmani, A.R.; Islam, M.Z.; Kasambe, R.M. Important Bird and Biodiversity Areas in India: Priority Sites for Conservation (Revised and Updated), 2nd ed.; Bombay Natural History Society and BirdLife International: Mumbai, India; Cambridge, UK, 2016; Volume 1, p. 1992. [Google Scholar]
- Walter, H.; Lieth, H.H.F. Klimadiagramm-Weltatlas; G. Fischer: Jena, Germany, 1967. [Google Scholar]
- Fick, S.E.; Hijmans, R.J. WorldClim 2: New 1-Km Spatial Resolution Climate Surfaces for Global Land Areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- eFI. eFlora of India: Database of Plants of Indian Subcontinent. 2023. Available online: https://efloraofindia.com/ (accessed on 17 April 2023).
- FOI. Flowers of India. 2023. Available online: http://www.flowersofindia.net/ (accessed on 17 April 2023).
- Kumar, A.; Patil, M.; Kumar, P.; Kumar, M.; Singh, A.N. Plant Ecology in Indian Siwalik Range: A Systematic Map and Its Bibliometric Analysis. Trop. Ecol. 2022, 63, 338–350. [Google Scholar] [CrossRef]
- Balkrishna, A.; Srivastava, A.; Shukla, B.; Mishra, R.; Joshi, B. Medicinal Plants of Morni Hills, Shivalik Range, Panchkula, Haryana. J. Non-Timber For. Prod. 2018, 25, 1–14. [Google Scholar] [CrossRef]
- Balkrishna, A.; Joshi, B.; Srivastava, A.; Shukla, B. Phyto-Resources of Morni Hills, Panchkula, Haryana. J. Non-Timber For. Prod. 2018, 25, 91–98. [Google Scholar] [CrossRef]
- Dhiman, H.; Saharan, H.; Jakhar, S. Floristic Diversity Assessment and Vegetation Analysis of the Upper Altitudinal Ranges of Morni Hills, Panchkula, Haryana, India. Asian J. Conserv. Biol. 2020, 9, 134–142. [Google Scholar]
- Dhiman, H.; Saharan, H.; Jakhar, S. Study of Invasive Plants in Tropical Dry Deciduous Forests—Biological Spectrum, Phenology, and Diversity. For. Stud. 2021, 74, 58–71. [Google Scholar] [CrossRef]
- Singh, N.; Vashistha, B.D. Flowering Plant Diversity and Ethnobotany of Morni Hills, Siwalik Range, Haryana, India. Int. J. Pharma Bio Sci. 2014, 5, B214–B222. [Google Scholar]
- Bhardwaj, A.; Verma, R.K.; Rana, J.C.; Thakur, K.; Verma, J. Orchid diversity at Chail Wild life Sanctuary, Himachal Pradesh, Northwest Himalaya. J. Orchid Soc. India 2014, 28, 67–74. [Google Scholar]
- Bhardwaj, A. Study on Dynamics of Plant Bioresources in Chail Wildlife Sanctuary of Himachal Pradesh. Ph.D. Thesis, Forest Research Institute (Deemed) University, Dehradun, India, 2017; p. 342. Available online: http://hdl.handle.net/10603/175719 (accessed on 5 October 2019).
- Kumar, R. Studies on Plant Biodiversity of Chail Wildlife Sanctuary in Himachal Pradesh. Master’s Thesis, Dr Yashwant Singh Parmar University of Horticulture and Forestry, Solan, India, 2013; p. 119. Available online: http://krishikosh.egranth.ac.in/handle/1/91126 (accessed on 4 October 2019).
- Choudhary, A.K.; Punam; Sharma, P.K.; Chandel, S. Study on the Physiography and Biodiversity of Churdhar Wildlife Sanctuary of Himachal Himalayas, India. Tigerpaper 2007, 34, 27–32. [Google Scholar]
- Choudhary, R.K.; Lee, J. A Floristic Reconnaissance of Churdhar Wildlife Sanctuary of Himachal Pradesh, India. Manthan 2012, 13, 2–12. [Google Scholar]
- Gupta, H. Comparative Studies on the Medicinal and Aromatic Flora of Churdhar and Rohtang Areas of Himachal Pradesh. Master’s Thesis, Dr Yashwant Singh Parmar University of Horticulture and Forestry, Solan, India, 1998; p. 228. Available online: http://krishikosh.egranth.ac.in/handle/1/5810135063 (accessed on 11 August 2022).
- Radha; Puri, S.; Chandel, K.; Pundir, A.; Thakur, M.S.; Chauhan, B.; Simer, K.; Dhiman, N.; Shivani; Thakur, Y.S.; et al. Diversity of Ethnomedicinal Plants in Churdhar Wildlife Sanctuary of District Sirmour of Himachal Pradesh, India. J. Appl. Pharm. Sci. 2019, 9, 48–53. [Google Scholar] [CrossRef]
- Subramani, S.P.; Kapoor, K.S.; Goraya, G.S. Additions to the Floral Wealth of Sirmaur District, Himachal Pradesh from Churdhar Wildlife Sanctuary. J. Threat. Taxa 2014, 6, 6427–6452. [Google Scholar] [CrossRef]
- Thakur, U.; Bisht, N.S.; Kumar, M.; Kumar, A. Influence of Altitude on Diversity and Distribution Pattern of Trees in Himalayan Temperate Forests of Churdhar Wildlife Sanctuary, India. Water Air Soil Pollut. 2021, 232, 205. [Google Scholar] [CrossRef]
- Govaerts, R.; Lughadha, E.N.; Black, N.; Turner, R.; Paton, A. The World Checklist of Vascular Plants, a Continuously Updated Resource for Exploring Global Plant Diversity. Sci. Data 2021, 8, 215. [Google Scholar] [CrossRef]
- Turland, N.; Wiersema, J.; Barrie, F.; Greuter, W.; Hawksworth, D.; Herendeen, P.; Knapp, S.; Kusber, W.-H.; Li, D.-Z.; Marhold, K.; et al. (Eds.) International Code of Nomenclature for Algae, Fungi, and Plants (Shenzhen Code); Regnum vegetabile; Koeltz Botanical Books: Glashütten, Germany, 2018; Volume 159, p. 254. ISBN 978-3-946583-16-5. [Google Scholar] [CrossRef]
- APG, I.V. An Update of the Angiosperm Phylogeny Group Classification for the Orders and Families of Flowering Plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar] [CrossRef]
- POWO. Plants of the World Online; Royal Botanic Gardens: Kew, UK, 2022; Available online: http://www.plantsoftheworldonline.org/ (accessed on 8 August 2022).
- Brown, M.J.M.; Walker, B.E.; Black, N.; Govaerts, R.H.A.; Ondo, I.; Turner, R.; Lughadha, E.N. rWCVP: A Companion R Package for the World Checklist of Vascular Plants. New Phytol. 2023, 240, 1355–1365. [Google Scholar] [CrossRef]
- Rana, S.K.; Rawat, G.S. Database of Himalayan Plants Based on Published Floras During a Century. Data 2017, 2, 36. [Google Scholar] [CrossRef]
- Rana, S.K.; Rawat, G.S. Database of Vascular Plants of Himalaya, version 1.6; Wildlife Institute of India: Dehradun, India, 2019. [Google Scholar] [CrossRef]
- Chowdhery, H.J.; Wadhwa, B.M. Flora of Himachal Pradesh; Botanical Survey of India: Calcutta, India, 1984; Volumes 1–3. [Google Scholar]
- Collett, H. Flora Simlensis; Thacker, Spink & Co.: Simla, India, 1902. [Google Scholar] [CrossRef]
- Duthie, J.F. Flora of the Upper Gangetic Plain, and of the Adjacent Siwalik and Sub-Himalayan Tracts; Superintendent of Government Printing: Calcutta, India, 1903–1915; Volumes 1–3. [Google Scholar] [CrossRef]
- Chamberlain, S.; Barve, V.; Mcglinn, D.; Oldoni, D.; Desmet, P.; Geffert, L.; Ram, K. rgbif: Interface to the Global Biodiversity Information Facility API. CRAN. 2025. Available online: https://cran.r-project.org/web/packages/rgbif/index.html (accessed on 13 October 2025).
- Rana, S.K.; Price, T.D.; Qian, H. Plant Species Richness Across the Himalaya Driven by Evolutionary History and Current Climate. Ecosphere 2019, 10, e02945. [Google Scholar] [CrossRef]
- Colwell, R.K.; Hurtt, G.C. Nonbiological Gradients in Species Richness and a Spurious Rapoport Effect. Am. Nat. 1994, 144, 570–595. [Google Scholar] [CrossRef]
- Hollister, J.; Shah, T.; Robitaille, A.L.; Beck, M.W.; Johnson, M. elevatr: Access Elevation Data from Various APIs; Zenodo: Geneva, Switzerland, 2021. [Google Scholar] [CrossRef]
- Hijmans, R.J. terra: Spatial Data Analysis. CRAN. 2025. Available online: https://cran.r-project.org/web/packages/terra/index.html (accessed on 13 October 2025).
- Manish, K.; Pandit, M.K.; Telwala, Y.; Nautiyal, D.C.; Koh, L.P.; Tiwari, S. Elevational Plant Species Richness Patterns and Their Drivers Across Non-Endemics, Endemics and Growth Forms in the Eastern Himalaya. J. Plant Res. 2017, 130, 829–844. [Google Scholar] [CrossRef]
- Gao, C.; Chen, C.; Akyol, T.; Dusa, A.; Yu, G.; Cao, B.; Cai, P. ggVennDiagram: Intuitive Venn Diagram Software Extended. iMeta 2024, 3, e177. [Google Scholar] [CrossRef]
- Chao, A.; Gotelli, N.J.; Hsieh, T.C.; Sander, E.L.; Ma, K.H.; Colwell, R.K.; Ellison, A.M. Rarefaction and Extrapolation with Hill Numbers: A Framework for Sampling and Estimation in Species Diversity Studies. Ecol. Monogr. 2014, 84, 45–67. [Google Scholar] [CrossRef]
- Colwell, R.K.; Mao, C.X.; Chang, J. Interpolating, Extrapolating, and Comparing Incidence-based Species Accumulation Curves. Ecology 2004, 85, 2717–2727. [Google Scholar] [CrossRef]
- Colwell, R.K.; Chao, A.; Gotelli, N.J.; Lin, S.-Y.; Mao, C.X.; Chazdon, R.L.; Longino, J.T. Models and Estimators Linking Individual-Based and Sample-Based Rarefaction, Extrapolation and Comparison of Assemblages. J. Plant Ecol. 2012, 5, 3–21. [Google Scholar] [CrossRef]
- Kumar, A.; Kumar, P.; Patil, M.; Hussain, S.; Yadav, R.; Sharma, S.; Tokas, D.; Singh, S.; Singh, A.N. Disturbance and Vegetational Structure in an Urban Forest of Indian Siwaliks: An Ecological Assessment. Environ. Monit. Assess. 2024, 196, 691. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2024; Available online: https://www.r-project.org/ (accessed on 31 October 2024).
- Hsieh, T.C.; Ma, K.H.; Chao, A. iNEXT: An R Package for Rarefaction and Extrapolation of Species Diversity (Hill Numbers). Methods Ecol. Evol. 2016, 7, 1451–1456. [Google Scholar] [CrossRef]
- Hilbe, J.M. Modeling Count Data; Cambridge University Press: Cambridge, UK, 2014. [Google Scholar] [CrossRef]
- Johnson, J.B.; Omland, K.S. Model Selection in Ecology and Evolution. Trends Ecol. Evol. 2004, 19, 101–108. [Google Scholar] [CrossRef]
- Burnham, K.P.; Anderson, D.R. (Eds.) Model Selection and Multimodel Inference, 2nd ed.; Springer: New York, NY, USA, 2002; p. 488. [Google Scholar] [CrossRef]
- Hurvich, C.M.; Tsai, C.-L. Regression and Time Series Model Selection in Small Samples. Biometrika 1989, 76, 297–307. [Google Scholar] [CrossRef]
- Dunn, P.K.; Smyth, G.K. Randomized Quantile Residuals. J. Comput. Graph. Stat. 1996, 5, 236–244. [Google Scholar] [CrossRef]
- Hartig, F. DHARMa: Residual Diagnostics for Hierarchical (Multi-Level/Mixed) Regression Models. CRAN. 2024. Available online: https://cran.r-project.org/web/packages/DHARMa/index.html (accessed on 13 October 2025).
- Gotelli, N.J.; Anderson, M.J.; Arita, H.T.; Chao, A.; Colwell, R.K.; Connolly, S.R.; Currie, D.J.; Dunn, R.R.; Graves, G.R.; Green, J.L.; et al. Patterns and Causes of Species Richness: A General Simulation Model for Macroecology. Ecol. Lett. 2009, 12, 873–886. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Manish, K. Species Richness, Phylogenetic Diversity and Phylogenetic Structure Patterns of Exotic and Native Plants Along an Elevational Gradient in the Himalaya. Ecol. Process. 2021, 10, 64. [Google Scholar] [CrossRef]
- Thorne, J.H.; Choe, H.; Dorji, L.; Yangden, K.; Wangdi, D.; Phuntsho, Y.; Beardsley, K. Species Richness and Turnover Patterns for Tropical and Temperate Plants on the Elevation Gradient of the Eastern Himalayan Mountains. Front. Ecol. Evol. 2022, 10, 942759. [Google Scholar] [CrossRef]
- Fischer, H. A History of the Central Limit Theorem: From Classical to Modern Probability Theory; Springer: New York, NY, USA, 2011; p. 402. [Google Scholar] [CrossRef]

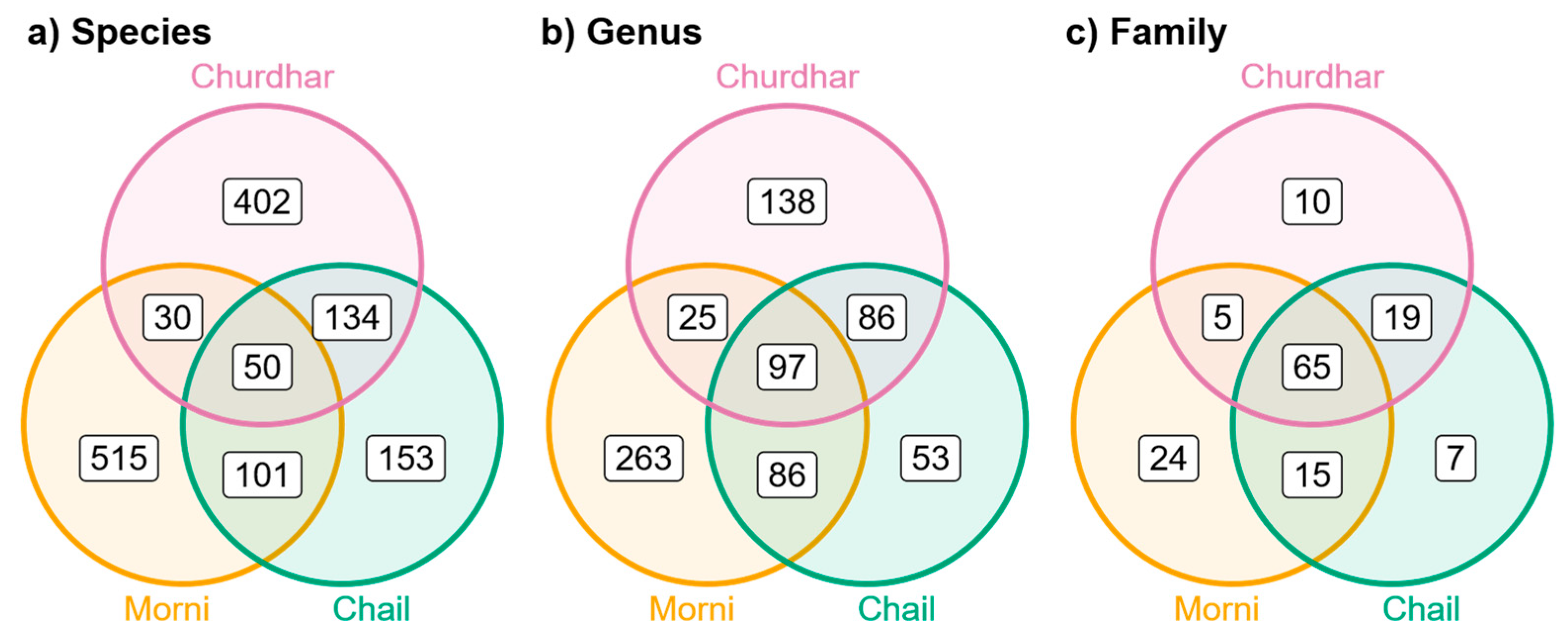
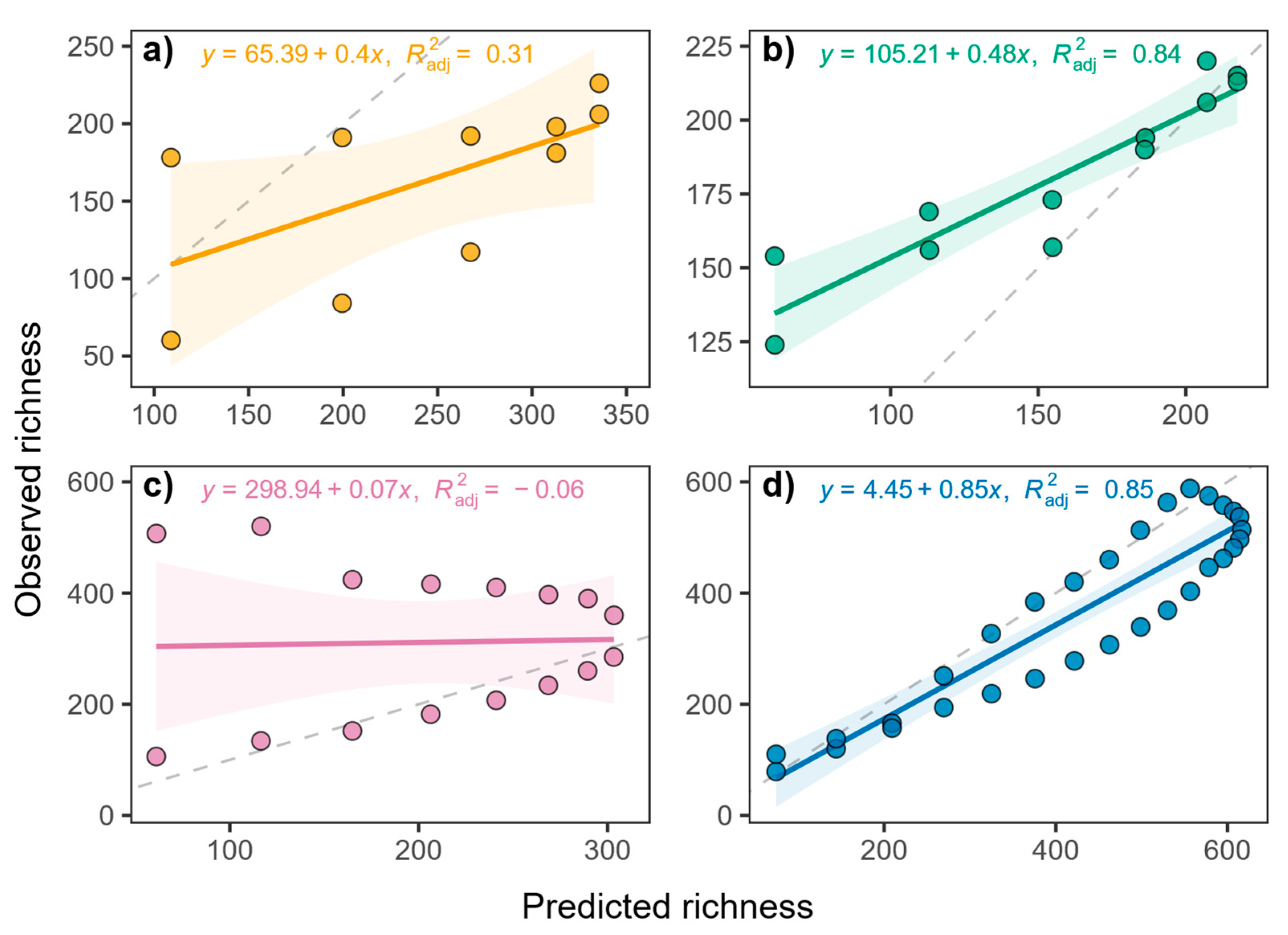
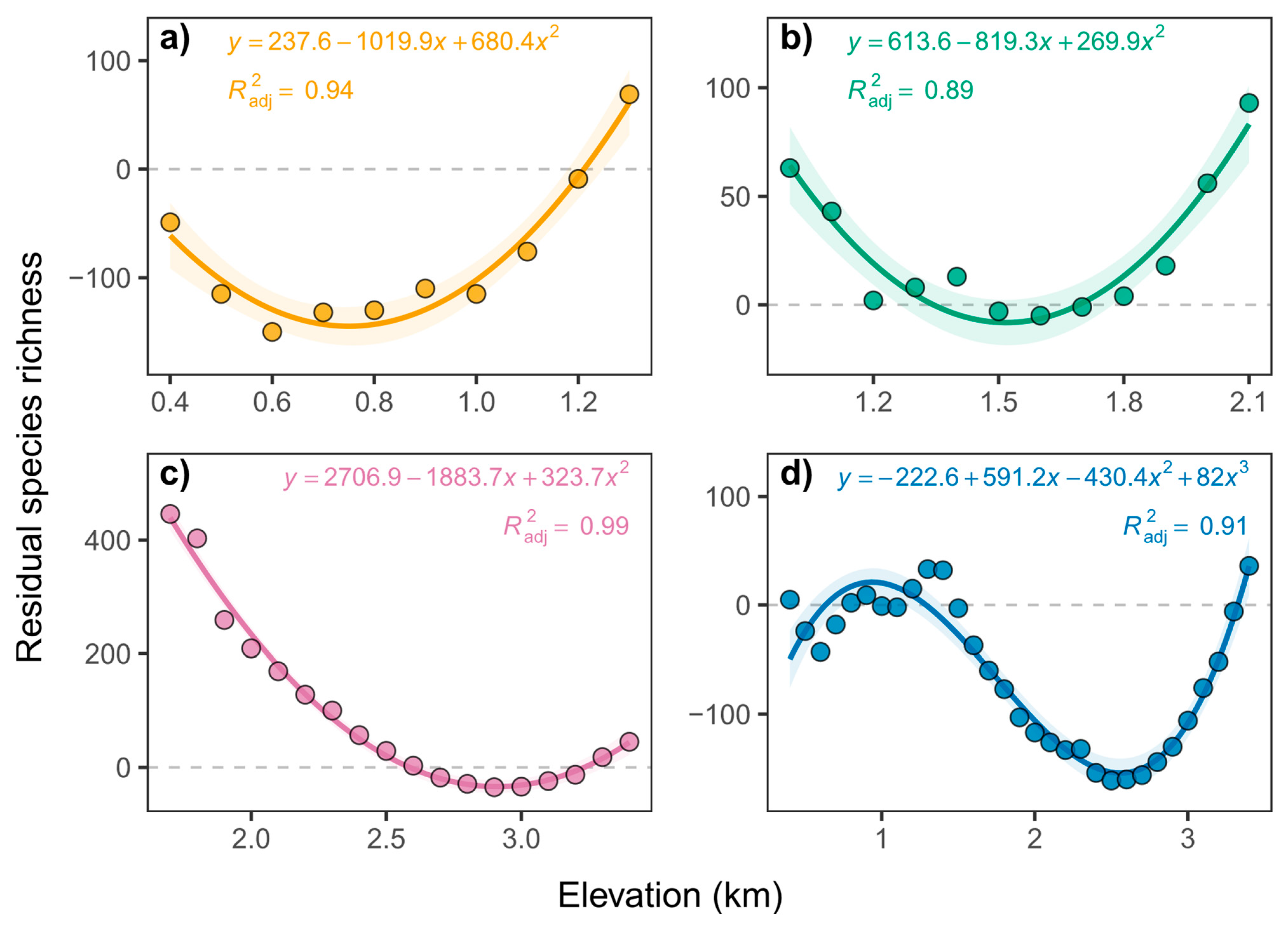
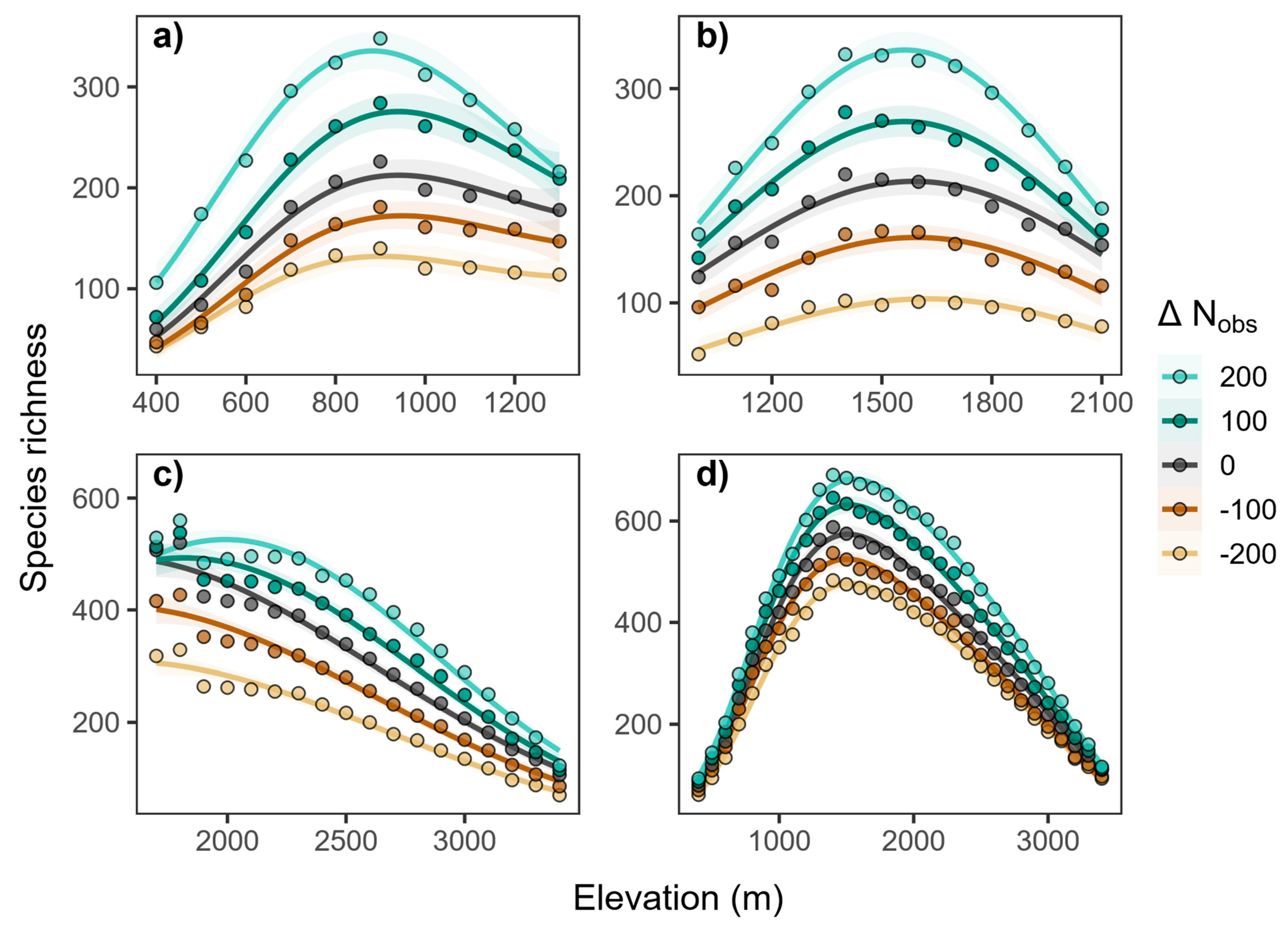
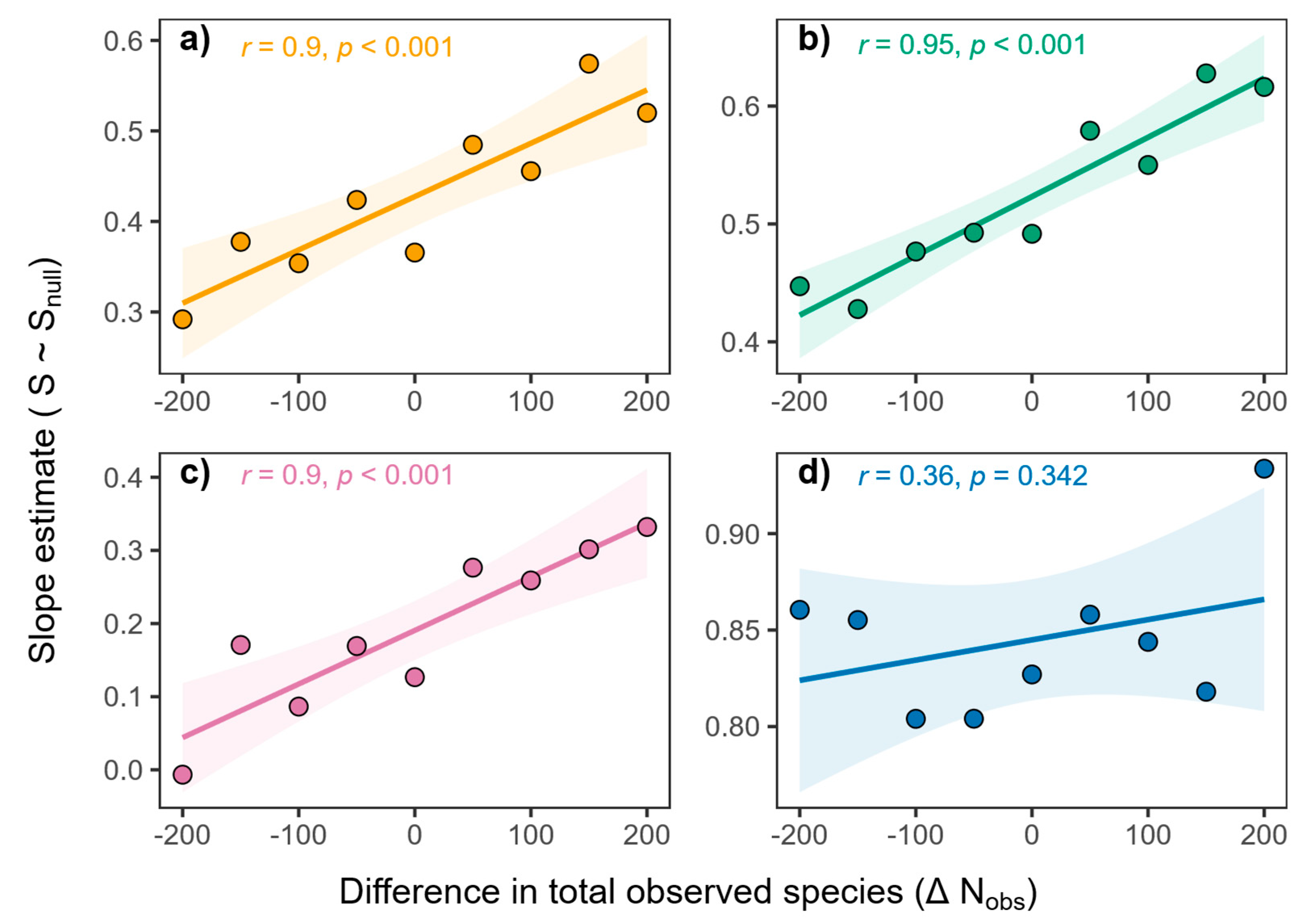
| Taxa | Morni | Chail | Churdhar | Total |
|---|---|---|---|---|
| Species | 696 | 438 | 616 | 1385 |
| Genus | 471 | 322 | 346 | 748 |
| Family | 109 | 106 | 99 | 145 |
| Introduced | 120 | 45 | 16 | 142 |
| Native | 576 | 393 | 600 | 1243 |
| Site | Model | D2 | D2adj | AICc | Dresid | dfresid | p | |
|---|---|---|---|---|---|---|---|---|
| Morni | S ~ Elev + Elev2 + Elev3 | 0.97 | 0.94 | 1.07 | 91.00 | 6.39 | 6 | 0.381 |
| Chail | S ~ Elev + Elev2 | 0.91 | 0.88 | 0.57 | 98.39 | 5.11 | 9 | 0.824 |
| Churdhar | S ~ Elev + Elev2 | 0.98 | 0.98 | 0.94 | 156.64 | 14.06 | 15 | 0.521 |
| All | S ~ Elev + Elev2 + Elev3 + Elev4 | 1.00 | 1.00 | 0.36 | 257.54 | 9.42 | 26 | 0.999 |
| Site | Intercept | Elev | Elev2 | Elev3 | Elev4 |
|---|---|---|---|---|---|
| Morni | −0.15 ± 0.92 | 14.95 ± 3.46 *** | −13.1 ± 4.12 ** | 3.65 ± 1.56 * | NA |
| Chail | 1.65 ± 0.51 ** | 4.68 ± 0.67 *** | −1.47 ± 0.21 *** | NA | NA |
| Churdhar | 5.32 ± 0.36 *** | 1.19 ± 0.3 *** | −0.4 ± 0.06 *** | NA | NA |
| All | 1.8 ± 0.23 *** | 8.49 ± 0.61 *** | −5.69 ± 0.56 *** | 1.65 ± 0.21 *** | −0.19 ± 0.03 *** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, A.; Patil, M.; Kumar, P.; Singh, A.N. Elevational Patterns of Plant Species Richness: Insights from Western Himalayas. Forests 2025, 16, 1591. https://doi.org/10.3390/f16101591
Kumar A, Patil M, Kumar P, Singh AN. Elevational Patterns of Plant Species Richness: Insights from Western Himalayas. Forests. 2025; 16(10):1591. https://doi.org/10.3390/f16101591
Chicago/Turabian StyleKumar, Abhishek, Meenu Patil, Pardeep Kumar, and Anand Narain Singh. 2025. "Elevational Patterns of Plant Species Richness: Insights from Western Himalayas" Forests 16, no. 10: 1591. https://doi.org/10.3390/f16101591
APA StyleKumar, A., Patil, M., Kumar, P., & Singh, A. N. (2025). Elevational Patterns of Plant Species Richness: Insights from Western Himalayas. Forests, 16(10), 1591. https://doi.org/10.3390/f16101591







