Population Structure and Genetic Diversity of Castanea sativa Mill. Genotypes in the Republic of Croatia
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area and Plant Material Collection
2.2. DNA Isolation
2.3. SSR Primer Screening and PCR Amplification and Analysis
2.4. Genetic Analyses
3. Results
3.1. Genetic Diversity Analysis of Loci and Populations
3.2. Genetic Differentiation Between Chestnut Populations: PET, HRK, and BAC
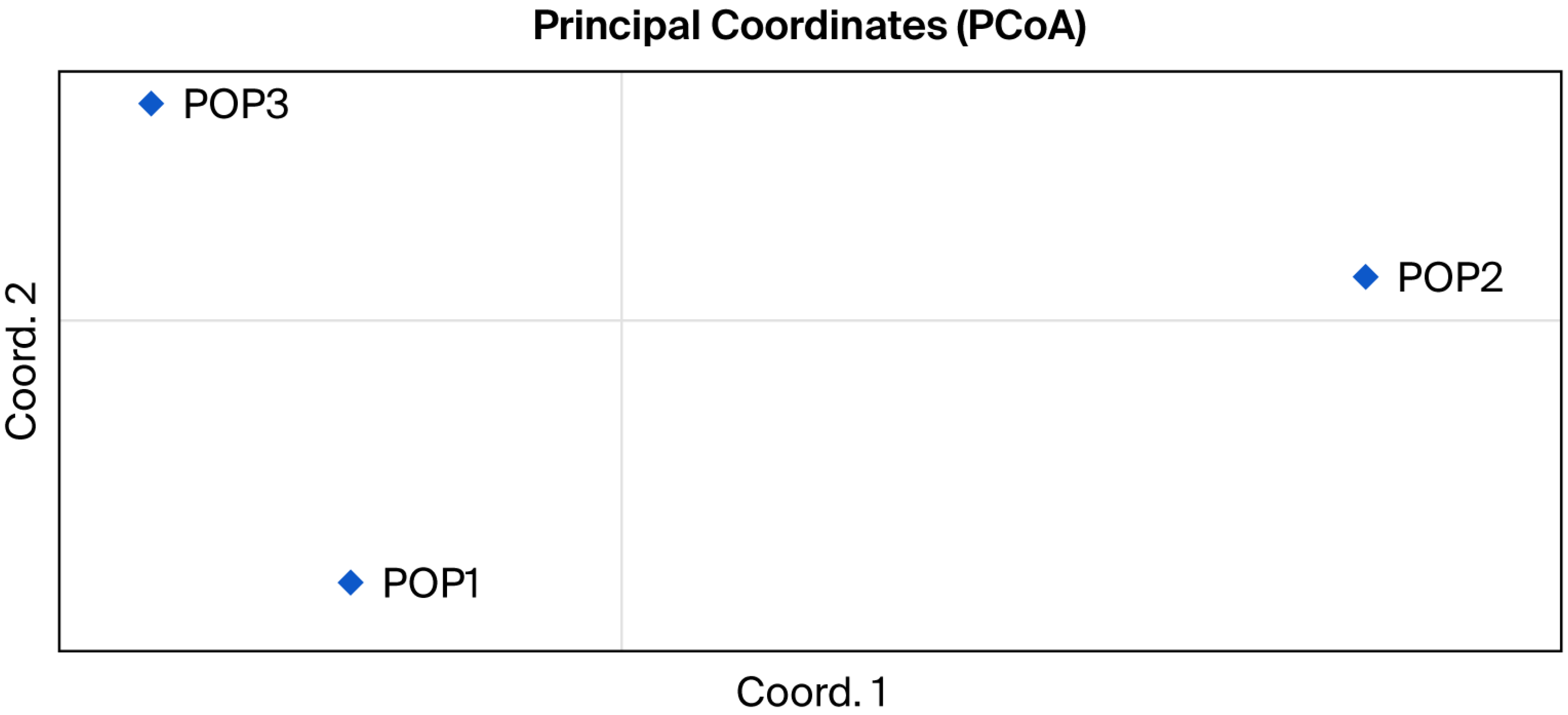
3.3. Population Genetic Structure Analysis
4. Discussion
4.1. Hypothesis and Objectives Evaluated
4.2. Genetic Diversity and Allelic Patterns
4.3. Population Differentiation and Gene Flow
4.4. Role of Mutation: Insights from R-Statistics
4.5. Population Structure Patterns
4.6. Methodological Context and Limitations
4.7. Implications for Conservation and Genetic Resource Management
4.8. Conservation Implications for European Chestnut
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Conedera, M.; Tinner, W.; Krebs, P.; de Rigo, D.; Caudullo, G. Castanea sativa in Europe: Distribution, habitat, usage and threats. In European Atlas of Forest Tree Species; San-Miguel-Ayanz, J., de Rigo, D., Caudullo, G., Houston Durrant, T., Mauri, A., Eds.; Publications Office of the European Union: Luxembourg, 2016; pp. 78–79. [Google Scholar]
- Fernández López, J.; Alía, R. EUFORGEN Technical Guidelines for Genetic Conservation and Use for European Chestnut (Castanea sativa); International Plant Genetic Resources Institute: Rome, Italy, 2003; p. 6. [Google Scholar]
- Conedera, M.; Manetti, M.C.; Giudici, F.; Amorini, E. Distribution and economic potential of the sweet chestnut (Castanea sativa Mill.) in Europe. Ecol. Mediterr. 2004, 30, 179–193. [Google Scholar] [CrossRef]
- Tumpa, T.; Islam, M.T.; Chowdhury, M.S.H.; Nath, T.K. Coppicing behavior and carbon sequestration potential of sweet chestnut (Castanea sativa): A review. Sustainability 2022, 14, 925. [Google Scholar] [CrossRef]
- Metreveli, V.; Gavashelishvili, A.; de Rigo, D.; Caudullo, G. Potential distribution and suitable habitat for chestnut in Georgia. Forests 2023, 14, 2076. [Google Scholar] [CrossRef]
- Marques, T.; Pina-Martins, F.; Silva, D.N.; Ribeiro, J.; Matos, J. Current biological insights of Castanea sativa Mill. to improve crop resilience to heat and drought. Plants 2025, 14, 335. [Google Scholar] [CrossRef] [PubMed]
- Rigling, D.; Prospero, S. Cryphonectria parasitica, the causal agent of chestnut blight: Invasion history, population biology and disease ecology. Mol. Plant Pathol. 2018, 19, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Camisón, Á.; Brotons, J.; Miranda-Fontaíña, M.E.; Aguín, O.; Pintos, C.; Mansilla, J.P.; Rey, L. Increased tolerance to Phytophthora cinnamomi in offspring of ink-diseased chestnut trees. Ann. For. Sci. 2019, 76, 119. [Google Scholar] [CrossRef]
- Marcolin, E.; Battipaglia, G.; Dalla Valle, C.; Sitzia, T.; Mosca, E.; Paletto, A. Impact of the Asian gall wasp Dryocosmus kuriphilus on the radial growth of European chestnut. J. Appl. Ecol. 2021, 58, 1212–1224. [Google Scholar] [CrossRef]
- Heiniger, U.; Rigling, D. Biological control of chestnut blight in Europe. Annu. Rev. Phytopathol. 1994, 32, 581–599. [Google Scholar] [CrossRef]
- EFSA Panel on Plant Health. Risk assessment of Dryocosmus kuriphilus for the EU territory. EFSA J. 2010, 8, 1619. [Google Scholar] [CrossRef]
- Vettraino, A.M.; Natili, G.; Anselmi, N.; Vannini, A. Recovery and pathogenicity of Phytophthora species associated with ink disease of chestnut in Italy. Plant Pathol. 2001, 50, 90–96. [Google Scholar] [CrossRef]
- Mattioni, C.; Martin, M.A.; Pollegioni, P.; Cherubini, M.; Villani, F. Microsatellite markers reveal a strong geographical structure in European populations of Castanea sativa (Fagaceae): Evidence for multiple glacial refugia. Am. J. Bot. 2013, 100, 951–961. [Google Scholar] [CrossRef]
- Bouffartigue, C.; Debille, S.; Fabreguettes, O.; Lheureux, F.; Trontin, J.-F.; Villani, F.; Zampini, A.; Marinoni, D.; Ciancolini, A.; Mattioni, C.; et al. Two main genetic clusters with high admixture between forest and cultivated chestnut (Castanea sativa Mill.) in France. Ann. For. Sci. 2020, 77, 74. [Google Scholar] [CrossRef]
- Mattioni, C.; Martin, M.A.; Chiocchini, F.; Cherubini, M.; Gaudet, M.; Pollegioni, P.; Velichkov, I.; Jarman, R.; Chambers, F.M.; Paule, L.; et al. Landscape genetics structure of European sweet chestnut (Castanea sativa Mill.): Indications for conservation priorities. Tree Genet. Genomes 2017, 13, 39. [Google Scholar] [CrossRef]
- Novak Agbaba, S.; Liović, B.; Medak, J.; Slade, D. Chestnut research in Croatia. Acta Hortic. 2005, 693, 49–54. [Google Scholar] [CrossRef]
- Horvat, I. Biljnosociološka istraživanja šuma u Hrvatskoj. Glas. šum. pokuse 1938, 6, 127–279. [Google Scholar]
- Medak, J. Šume pitomog kestena s prasećim zeljem (Aposeri foetidae-Castanetum sativae ass. nova) u Hrvatskoj. Sum. list 2011, 135, 5–24. [Google Scholar]
- Anić, M. Pitomi kesten na Cresu. Glas. šum. pokuse 1953, 11, 321–356. [Google Scholar]
- Marinček, L.; Župančič, M. Nomenklaturna revizija acidofilnih bukovih in gradnovih gozdov zahodnega območja ilirske florne province. Hladnikia 1995, 4, 29–35. [Google Scholar]
- Medak, J. Šumske Zajednice i Staništa Pitomog Kestena (Castanea sativa Mill.) u Hrvatskoj [Forest Communities and Habitats of Sweet Chestnut (Castanea sativa Mill.) in Croatia]. Ph.D. Dissertation, Faculty of Forestry and Wood Technology, University of Zagreb, Zagreb, Croatia, 18 December 2009. [Google Scholar]
- Lazanin, S. Border-crossings and migration in the Croatian and Slavonian Military Frontiers in the early modern period. Hist. Flux 2021, 3, 57–74. [Google Scholar] [CrossRef]
- Hruševar, D.; Bakrač, K.; Miko, S.; Ilijanić, N.; Šparica Miko, M.; Hasan, O.; Mitić, B. Vegetation history in central Croatia from ~10,000 cal BC to the beginning of the Common Era—Filling the palaeoecological gap for the Western Balkans. Diversity 2023, 15, 235. [Google Scholar] [CrossRef]
- Ježić, M.; Nuskern, L.; Peranić, K.; Popović, M.; Ćurković-Perica, M.; Mendaš, O.; Škegro, I.; Poljak, I.; Vidaković, A.; Idžojtić, M. Regional variability of chestnut (Castanea sativa) tolerance toward blight disease. Plants 2024, 13, 3060. [Google Scholar] [CrossRef] [PubMed]
- Ellegren, H. Microsatellites: Simple sequences with complex evolution. Nat. Rev. Genet. 2004, 5, 435–445. [Google Scholar] [CrossRef]
- Smith, D.N.; Devey, M.E. Occurrence and inheritance of microsatellites in Pinus radiata. Genome 1994, 37, 977–983. [Google Scholar] [CrossRef] [PubMed]
- Dow, B.D.; Ashley, M.V.; Howe, H.F. Characterization of highly variable (GA/CT)_n microsatellites in the bur oak, Quercus macrocarpa. Theor. Appl. Genet. 1995, 91, 137–141. [Google Scholar] [CrossRef]
- Dow, B.D.; Ashley, M.V. Microsatellite analysis of seed dispersal and parentage of saplings in bur oak, Quercus macrocarpa. Mol. Ecol. 1996, 5, 615–627. [Google Scholar] [CrossRef]
- Steinkellner, H.; Fluch, S.; Turetschek, E.; Lexer, C.; Streiff, R.; Kremer, A.; Burg, K.; Glössl, J. Identification and characterization of (GA/CT)_n microsatellite loci from Quercus petraea. Plant Mol. Biol. Rep. 1997, 15, 295–303. [Google Scholar] [CrossRef]
- Kampfer, S.; Lexer, C.; Glössl, J.; Steinkellner, H. Characterization of (GA)_n microsatellite loci from Quercus robur. Hereditas 1998, 129, 183–186. [Google Scholar] [CrossRef]
- Lexer, C.; Heinze, B.; Gerber, S.; Macalka-Kampfer, S.; Steinkellner, H.; Kremer, A.; Glössl, J. Microsatellite analysis of maternal half-sib families of Quercus robur (pedunculate oak): II. Inferring the number of pollen donors from the offspring. Theor. Appl. Genet. 2000, 100, 858–865. [Google Scholar] [CrossRef]
- Aldrich, P.R.; Michler, C.H.; Sun, W.; Romero-Severson, J. Microsatellite markers for northern red oak (Fagaceae: Quercus rubra). Mol. Ecol. Notes 2002, 2, 472–474. [Google Scholar] [CrossRef]
- Buck, E.J.; Hadonou, M.; James, C.J.; Blakesley, D.; Russell, K. Isolation and characterization of polymorphic microsatellites in European chestnut (Castanea sativa Mill.). Mol. Ecol. Notes 2003, 3, 239–241. [Google Scholar] [CrossRef]
- Marinoni, D.; Akkak, A.; Bounous, G.; Edwards, K.J.; Botta, R. Development and characterization of microsatellite markers in Castanea sativa (Mill.). Mol. Breed. 2003, 11, 127–136. [Google Scholar] [CrossRef]
- Mattioni, C.; Cherubini, M.; Micheli, E.; Villani, F.; Bucci, G. Role of domestication in shaping Castanea sativa genetic variation in Europe. Tree Genet. Genomes 2008, 4, 563–574. [Google Scholar] [CrossRef]
- Martín, M.A.; Mattioni, C.; Cherubini, M.; Taurchini, D.; Villani, F. Genetic diversity in European chestnut populations by means of genomic and genic microsatellite markers. Tree Genet. Genomes 2010, 6, 735–744. [Google Scholar] [CrossRef]
- Pereira-Lorenzo, S.; Lourenço Costa, R.M.; Ramos-Cabrer, A.M.; Marques Ribeiro, C.A.; Serra da Silva, M.F.; Manzano, G.; Barreneche, T. Variation in grafted European chestnut and hybrids by microsatellites reveals two main origins in the Iberian Peninsula. Tree Genet. Genomes 2010, 6, 701–715. [Google Scholar] [CrossRef]
- Alessandri, S.; Krznar, M.; Ajolfi, D.; Ramos Cabrer, A.M.; Pereira-Lorenzo, S.; Dondini, L. Genetic diversity of Castanea sativa Mill. accessions from the Tuscan-Emilian Apennines and Emilia Romagna region (Italy). Agronomy 2020, 10, 1319. [Google Scholar] [CrossRef]
- Mattioni, C.; Ranzino, L.; Cherubini, M.; Leonardi, L.; La Mantia, T.; Castellana, S.; Villani, F.; Simeone, M.C. Monuments unveiled: Genetic characterization of large old chestnut (Castanea sativa Mill.) trees using comparative nuclear and chloroplast DNA analysis. Forests 2020, 11, 1118. [Google Scholar] [CrossRef]
- Caré, O.; Kuchma, O.; Hosius, B.; Voth, W.; Thurm, E.A.; Leinemann, L. Patterns of genetic variation and the potential origin of sweet chestnut (Castanea sativa Mill.) stands far from its natural northern distribution edge. Silvae Genet. 2024, 72, 200–210. [Google Scholar] [CrossRef]
- Idžojtić, M.; Zebec, M.; Mlinarec, J.; Franić, R. Genetic characterization of Lovran Marron chestnut (Castanea sativa Mill.) in Croatia. Šum. List 2012, 136, 283–291. [Google Scholar]
- Prgomet, S.; Poljak, I.; Jukić, D.; Idžojtić, M.; Hanzer, V. Genetic diversity of chestnut populations from Istria and Gorski Kotar. Šum. List 2014, 138, 415–422. [Google Scholar]
- Poljak, I.; Idžojtić, M.; Šatović, Z.; Ježić, M.; Ćurković Perica, M.; Simovski, B.; Acevski, J.; Liber, Z. Genetic diversity of the sweet chestnut (Castanea sativa Mill.) in Central Europe and the western Balkans, and evidence of Marron genotype introgression into wild populations. Tree Genet. Genomes 2017, 13, 18. [Google Scholar] [CrossRef]
- Croatian Forest Research Institute. Protection of Sweet Chestnut Forests, 2006–2010. Available online: https://www.sumins.hr/projekti/zastita-suma-pitomog-kestena/ (accessed on 5 August 2025).
- Croatian Forest Research Institute. Experimental Chestnut Grove, 2013–2015. Available online: https://www.sumins.hr/projekti/pokusni-nasad-pitomog-kestena-gornja-bacuga/ (accessed on 1 August 2025).
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987. [Google Scholar]
- Kimura, M.; Crow, J.F. The number of alleles that can be maintained in a finite population. Genetics 1964, 49, 725–738. [Google Scholar] [CrossRef]
- Shannon, C.E.; Weaver, W. The Mathematical Theory of Communication; University of Illinois Press: Urbana, IL, USA, 1949. [Google Scholar]
- Frankham, R.; Ballou, J.D.; Briscoe, D.A. Introduction to Conservation Genetics, 2nd ed.; Cambridge University Press: Cambridge, UK, 2010. [Google Scholar] [CrossRef]
- Kalinowski, S.T. Counting alleles with rarefaction: Private alleles and hierarchical sampling designs. Conserv. Genet. 2004, 5, 539–543. [Google Scholar] [CrossRef]
- Nei, M.; Chesser, R.K. Estimation of fixation indices and gene diversities. Ann. Hum. Genet. 1983, 47, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Wright, S. The genetical structure of populations. Ann. Eugen. 1951, 15, 323–354. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Slatkin, M. A measure of population subdivision based on microsatellite allele frequencies. Genetics 1995, 139, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Slatkin, M.; Barton, N.H. A comparison of three indirect methods for estimating average levels of gene flow. Evolution 1989, 43, 1349–1368. [Google Scholar] [CrossRef]
- Nei, M. Genetic distance between populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Coltman, D.W.; Pilkington, J.G.; Smith, J.A.; Pemberton, J.M. Parasite-mediated selection against inbred Soay sheep in a free-living, island population. Evolution 1999, 53, 1259–1267. [Google Scholar] [CrossRef]
- Amos, W.; Worthington Wilmer, J.; Fullard, K.; Burg, T.M.; Croxall, J.P.; Bloch, D.; Coulson, T. The influence of parental relatedness on reproductive success. Proc. R. Soc. Lond. B Biol. Sci. 2001, 268, 2021–2027. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Li, Y.L.; Liu, J.X. STRUCTURESELECTOR: A web-based software to select and visualize the optimal number of clusters using multiple methods. Mol. Ecol. Resour. 2018, 18, 176–177. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. CLUMPAK: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef]
- Manthos, I.; Sotiropoulos, T.; Karapetsi, L.; Xanthopoulou, A.; Madesis, P. Genetic characterization of Greek chestnut (Castanea sativa Mill.) germplasm collections in Parnon Mountain. Not. Bot. Horti Agrobo. 2025, 53, 14619. [Google Scholar] [CrossRef]
- Petit, R.J.; Hampe, A.; Cheddadi, R. Climate changes and tree phylogeography in the Mediterranean. Taxon 2005, 54, 877–885. [Google Scholar] [CrossRef]
- Hamrick, J.L.; Godt, M.J.W. Effects of life history traits on genetic diversity in plant species. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1996, 351, 1291–1298. [Google Scholar]
- Escudero, A.; Iriondo, J.M.; Torres, M.E. Spatial analysis of genetic diversity as a tool for plant conservation. Biol. Conserv. 2003, 113, 351–365. [Google Scholar] [CrossRef]
- Austerlitz, F.; Garnier-Géré, P. Evolutionary models of seed dispersal and genetic diversity. Heredity 2003, 91, 260–268. [Google Scholar] [CrossRef]
- Ashley, M.V. A quarter century of genetic studies of pollination in oaks. Forests 2021, 12, 575. [Google Scholar] [CrossRef]
- Orman, E.; Çakar, D.; Alkan, M.; Özer, G.; Güler, E.; Gündoğdu, M. Genetic diversity and population structure of Turkish European chestnut (Castanea sativa) genotypes assessed using start codon targeted polymorphism (SCoT) markers. Genet. Resour. Crop Evol. 2025, 72, 6507–6519. [Google Scholar] [CrossRef]
- Cuestas, M.I.; Mattioni, C.; Martín, L.M.; Vargas Osuna, E.; Cherubini, M.; Martin, M.A. Functional genetic diversity of chestnut (Castanea sativa Mill.) in southern Spain: Insights from EST SSRs and geographic structuring. For. Syst. 2017, 26, eSC06. [Google Scholar] [CrossRef]
- El Chami, M.A.; Tourvas, N.; Kazakis, G.; Kalaitzis, P.; Aravanopoulos, F.A. Genetic characterisation of chestnut cultivars in Crete using SSR markers reveals 84.1% variation within populations. Forests 2021, 12, 1659. [Google Scholar] [CrossRef]
- Slatkin, M. Gene flow and the geographic structure of natural populations. Science 1987, 236, 787–792. [Google Scholar] [CrossRef]
- Erichsen, E.O.; Budde, K.B.; Sagheb-Talebi, K.; Vendramin, G.G.; Bagnoli, F.; Fady, B.; Hansen, O.K. Hyrcanian forests—Stable rear-edge populations harbouring high genetic diversity of Fraxinus excelsior, a common European tree species. Divers. Distrib. 2018, 24, 103–114. [Google Scholar] [CrossRef]
- Backs, J.R.; Ashley, M.V. Evolutionary history and gene flow of an endemic island oak: Quercus pacifica. Am. J. Bot. 2019, 106, 339–351. [Google Scholar] [CrossRef]
- Awad, L.; Fady, B.; Khater, C.; Roig, A.; Cheddadi, R. Genetic structure and diversity of the endangered fir tree of Lebanon (Abies cilicica Carr.): Implications for conservation. PLoS ONE 2014, 9, e90086. [Google Scholar] [CrossRef]
- Jarman, R.; Chambers, F.M.; Webb, J. DNA analysis of Castanea sativa (sweet chestnut) in Britain and Ireland: Elucidating European origins and genepool diversity. PLoS ONE 2019, 14, e0222936. [Google Scholar] [CrossRef] [PubMed]
- Jump, A.S.; Peñuelas, J. Running to stand still: Adaptation and the response of plants to rapid climate change. Ecol. Lett. 2005, 8, 1010–1020. [Google Scholar] [CrossRef] [PubMed]
- European Forest Genetic Resources Programme (EUFORGEN). Pan-European Strategy for Genetic Conservation of Forest Trees and Establishment of a Core Network of Dynamic Conservation Units (DCUs); Bioversity International: Rome, Italy, 2015. [Google Scholar]
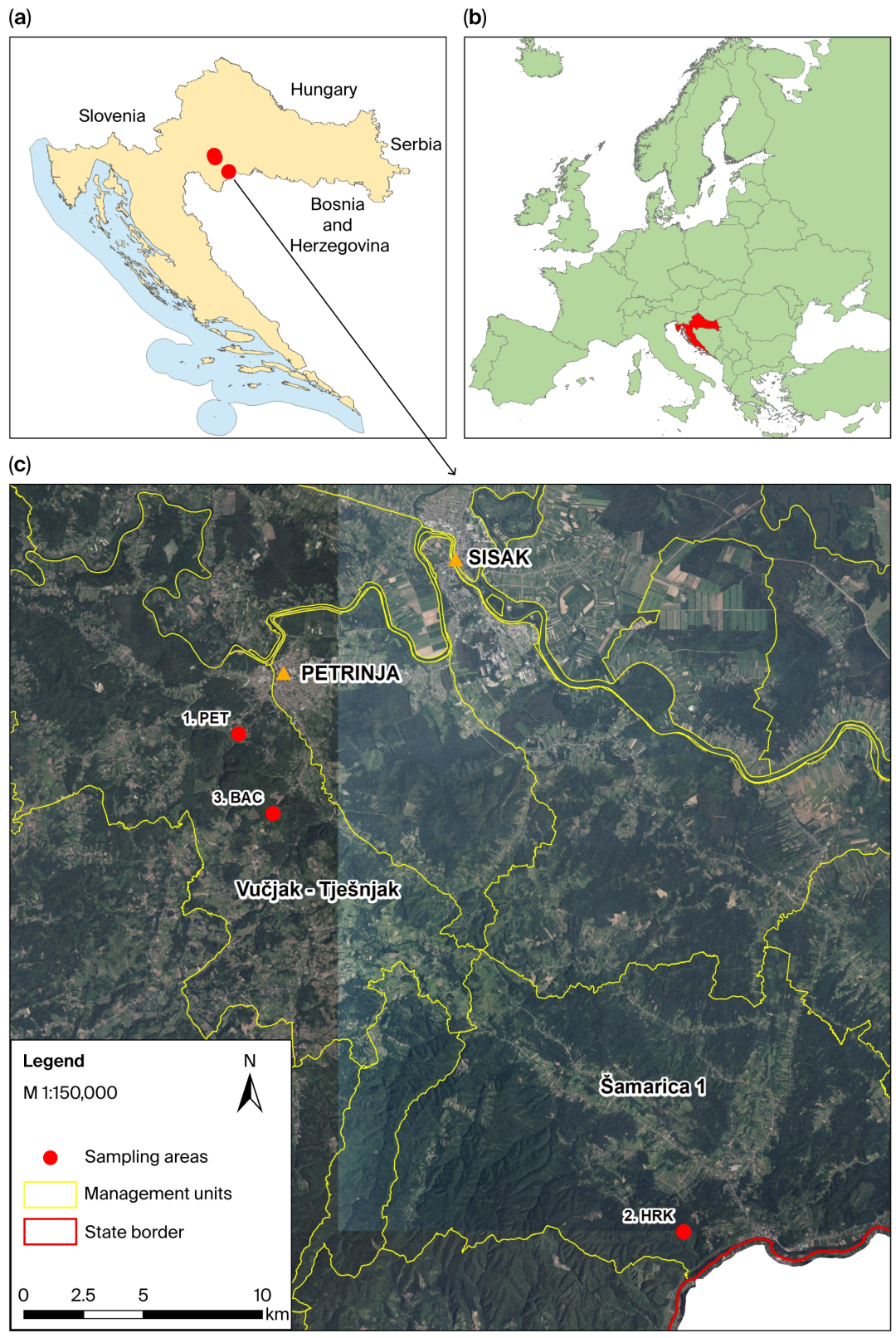

| Code | Location Description | Latitude/ Longitude | Altitude (m) | Ownership | Sampling Year | N |
|---|---|---|---|---|---|---|
| PET | Department 47a, Management Unit Vučjak–Tješnjak, Forest Office Petrinja, Forest Administration Sisak, Croatian Forests Ltd. | 45.416972° N, 16.255232° E | 170–390 | State forest | 2011 | 52 |
| HRK | Department 90a, Management Unit Šamarica I, Forest Office Hrvatska Kostajnica, Forest Administration Sisak, Croatian Forests Ltd. | 45.229502° N, 16.493670° E | 140–240 | State forest | 2013 | 51 |
| BAC | Hrastovička Gora | 45.386900° N, 16.273600° E | 374 | Private forest | 2016 | 50 |
| Locus | Fluorescent Dye | 5′-3′ Sequences (F/R) | Expected Length (bp) | Repeat Motif | References |
|---|---|---|---|---|---|
| EMCs2 | NED | GCTGATATGGCAATGCTTTTCCTC/ GCCCTCCAGCCTCACTTCATCAG | 172–178 | (CGG)7 | [30] |
| EMCs10 | PET | GTCTCCCCCAATCATAAGTAGGTC/ TCAAGGGAACATTAGGTCATTTTT | 218–230 | (CA)8 | [30] |
| EMCs13 | VIC | TAGTCGGAGTACGGGCACAG/ TGATATGAGCATTTGACTTTGATT | 158–164 | (GCA)8 | [30] |
| EMCs15 | 6-FAM | CTCTTAGACTCCTTCGCCAATC/ CAGAATCAAAGAAGAGAAAGGTC | 089–095 | (CAC)9 | [30] |
| EMCs17 | 6-FAM | CGCCACGATTAGCTCATTTTCA/ GAGGTAGGGTCTTCTTCGGTCATC | 210–222 | (AGC)4(CCAA)5 | [30] |
| EMCs25 | 6-FAM | ATGGGAAAATGGGTAAAGCAGTAA/ AACCGGAGATAGGATTGAACAGAA | 140–158 | (GA)12 | [30] |
| CsCAT15 | 6-FAM | TTCTGCGACCTCGAAACCGA/ GCTAGGGTTTTCATTTCTAG | 125–160 | (TC)12 | [31] |
| Locus | Number of Alleles (bp) | FIS | FIT | FST | Nm |
|---|---|---|---|---|---|
| EMCs13 | 3 (155, 158, 161) | 0.095 | 0.134 | 0.043 | 5.550 |
| EMCs15 | 3 (79, 82, 88) | 0.130 | 0.213 | 0.096 | 2.360 |
| EMCs2 | 4 (156, 159, 162, 165) | –0.048 | 0.009 | 0.055 | 4.326 |
| EMCs10 | 4 (214, 216, 222, 226) | 0.037 | 0.045 | 0.009 | 27.342 |
| EMCs17 | 4 (205, 209, 213, 217) | 0.130 | 0.221 | 0.104 | 2.146 |
| CsCAT15 | 8 (118, 120, 122, 124, 128, 132, 134, 138) | 0.565 | 0.614 | 0.112 | 1.974 |
| EMCs25 | 10 (138, 140, 144, 146, 148, 150, 154, 156, 158, 160) | −0.033 | −0.004 | 0.028 | 8.689 |
| Total/Mean ± SE | 36 alleles | 0.125 ± 0.078 | 0.176 ± 0.081 | 0.064 ± 0.015 | 7.484 ± 3.432 |
| Population | Locus | Allele | Frequency |
|---|---|---|---|
| HRK | CsCAT15 | 120 | 0.020 |
| HRK | CsCAT15 | 134 | 0.010 |
| HRK | EMCs25 | 154 | 0.029 |
| BAC | CsCAT15 | 118 | 0.010 |
| BAC | EMCs2 | 156 | 0.010 |
| BAC | EMCs25 | 144 | 0.130 |
| BAC | EMCs25 | 150 | 0.010 |
| Pop | Stat | N | Na | Ne | I | Ho | He | F |
|---|---|---|---|---|---|---|---|---|
| PET | Mean | 52 | 4.143 | 2.39 | 1.015 | 0.511 | 0.569 | 0.074 |
| SE | 0.553 | 0.173 | 0.081 | 0.07 | 0.03 | 0.127 | ||
| HRK | Mean | 51 | 4.143 | 2.313 | 0.979 | 0.448 | 0.553 | 0.179 |
| SE | 0.508 | 0.157 | 0.071 | 0.043 | 0.036 | 0.071 | ||
| BAC | Mean | 50 | 4.429 | 2.631 | 1.096 | 0.506 | 0.59 | 0.127 |
| SE | 0.571 | 0.321 | 0.113 | 0.044 | 0.042 | 0.085 | ||
| Total | Mean | 51 | 4.238 | 2.444 | 1.03 | 0.488 | 0.571 | 0.127 |
| SE | 0.3 | 0.129 | 0.051 | 0.03 | 0.02 | 0.054 |
| Source | df | SS | MS | Est. Var. | % |
|---|---|---|---|---|---|
| Among Populations | 2 | 1542.097 | 771.048 | 5.480 | 3% |
| Among Individuals | 150 | 31,826.374 | 212.176 | 50.300 | 30% |
| Within Pops Within pops-Within individuals | 153 | 17,071.000 | 111.575 | 111.575 | 67% |
| Total | 305 | 50,439.471 | 167.355 | 100% |
| Statistic | Value | p (Rand ≥ Data) | Interpretation |
|---|---|---|---|
| RST | 0.033 | 0.001 | Among-population differentiation (stepwise model) |
| RIS | 0.311 | 0.001 | Within-individual diversity relative to subpopulations |
| RIT | 0.333 | 0.001 | Within-individual diversity relative to total population |
| Nm | 7.385 | — | Estimated gene flow (number of migrants per generation) |
| Source | df | SS | MS | Est. Var. | % |
|---|---|---|---|---|---|
| Among Pops | 2 | 90.659 | 45.330 | 0.798 | 15% |
| Within Pops | 150 | 698.903 | 4.659 | 4.659 | 85% |
| Total | 152 | 789.562 | 5.457 | 100% |
| Stat | Value | p (Rand ≥ Data) |
|---|---|---|
| PhiPT | 0.146 | 0.001 |
| Nm | 1.461 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ćelepirović, N.; Novak Agbaba, S.; Bogunović, S.; Ivanković, M.; Kandemir, G.; Karija Vlahović, M.; Gradečki-Poštenjak, M. Population Structure and Genetic Diversity of Castanea sativa Mill. Genotypes in the Republic of Croatia. Forests 2025, 16, 1534. https://doi.org/10.3390/f16101534
Ćelepirović N, Novak Agbaba S, Bogunović S, Ivanković M, Kandemir G, Karija Vlahović M, Gradečki-Poštenjak M. Population Structure and Genetic Diversity of Castanea sativa Mill. Genotypes in the Republic of Croatia. Forests. 2025; 16(10):1534. https://doi.org/10.3390/f16101534
Chicago/Turabian StyleĆelepirović, Nevenka, Sanja Novak Agbaba, Sanja Bogunović, Mladen Ivanković, Gaye Kandemir, Monika Karija Vlahović, and Marija Gradečki-Poštenjak. 2025. "Population Structure and Genetic Diversity of Castanea sativa Mill. Genotypes in the Republic of Croatia" Forests 16, no. 10: 1534. https://doi.org/10.3390/f16101534
APA StyleĆelepirović, N., Novak Agbaba, S., Bogunović, S., Ivanković, M., Kandemir, G., Karija Vlahović, M., & Gradečki-Poštenjak, M. (2025). Population Structure and Genetic Diversity of Castanea sativa Mill. Genotypes in the Republic of Croatia. Forests, 16(10), 1534. https://doi.org/10.3390/f16101534





