Abstract
Black cottonwood (Populus trichocarpa) is a species of economic interest and an outstanding study model. The aspen borer (Saperda calcarata) causes irreversible damage to poplars and other riparian species in North America. The insect can produce multiple effects ranging from the presence of some galleries in the stem to tree death. Despite the ecological and commercial importance of this tree–insect interaction, the genetic mechanisms underlying the response of P. trichocarpa to S. calcarata are scarcely understood. In this study, a common garden trial of P. trichocarpa provenances, established in Davis, California, was assessed at the second year of growth, regarding the infestation of S. calcarata from a natural outbreak. A genome-wide association study (GWAS) was conducted using 629k of exonic SNPs to assess the relationship between genomic variation and insect attack. Tree architecture, in terms of stem number per plant, and the wood metabolome were also included. Insect attack was independent of the number of stems per tree. The performed GWAS identified three significantly associated SNP markers (q-value < 0.2) belonging to the same number of gene models, encoding proteins involved in signal transduction mechanisms and secondary metabolite production, including that of R-mandelonitrile lyase, Chromodomain-helicase-DNA-binding family protein, and Leucine-rich repeat protein. These results are aligned with the current knowledge of defensive pathways in plants and trees, helping to expand the understanding of the defensive response mechanisms of black cottonwood against wood borer insects.
1. Introduction
Populus trichocarpa is a forest tree species of interest as a study model and for breeding purposes, due to its fast growth rate, outbreeding behavior, and worldwide adaptation [1,2,3]. In nature, poplar plantations’ performance and architecture are defined by relayed genetic features, nutrient availability, physiological processes, and biotic and abiotic factors [4,5], including insects or pathogenic fungi, extreme temperatures, or mechanical damage. Populus trichocarpa was the first tree species for which a genome sequence was available [6].
Wood-boring (xylophagous) beetles belong to several genera, including Saperda, Cryptorhynchus, Billaea, Dolichomitus, Paranthrene, and Iphaulax. They cause irreversible damage to forests and timber production due to their impact on the xylem structure and the transport of water and nutrients in stems. The larval ability to tunnel inside wood allows them to successfully evade their natural enemies and access a nutritional source for which there is reduced competition. They are one of the most damaging and costly groups of invasive insects in North American forests [7,8]. For instance, in California, tree borer beetles have caused the injury or mortality of thousands of trees, most of them ornamental species in urban forests and native tree species in adjacent riparian forests located in the wildland–urban interface [9]. In particular, larvae of Saperda calcarata Say (Cerambycidae), a 20–28 mm beetle, are problematic in P. trichocarpa stands, especially those in southern regions of the USA where P. trichocarpa is their principal host. Saperda calcarata is naturally distributed in Canada and the USA [10]. Its larvae tunnel across the xylem, leaving a sawdust-like residue, and causing a loss of height increase, reduced wood quality, and stem breakage under high infestation [11,12].
In woody species, tree borer insects attack the principal stem affecting indirectly the terminal shoot. When this happens, the damaged stem is commonly replaced by the upturning of two or more subtending lateral branches that have been released as a result of apical dominance. Hence, with the destruction of just a single terminal shoot, the growth form of the tree can be permanently and dramatically altered from single-stemmed to multi-stemmed [13]. This increased branching is a key mechanism involved in the increased tolerance to biotic stress, and it is a common response of plants following the removal of apical dominance [14,15]. Pratt et al. (2005) [16] reported clear differences in the herbivore-mediated induction of epicormic bud elongation and reductions in flushing stem length in Melaleuca quinquenervia, leading to weak apical dominance and resulting in altered plant architecture from increased terminal branches. Similar alterations in plant architecture were observed in Pinus edulis (Engelm.) after attack by the stem-boring moth Dioryctria albovitella (Hust.), which resulted in a pattern of increased prostrate growth from increases in lateral rather than terminal branching [17]. Thus, a significant feature of plant–herbivore–xylophage interactions is the influence of feeding on plant growth and architecture [16,18]. On the other hand, García-Cervigón et al. (2021) [19] reported that xylem anatomical and plant architectural traits were the most responsive to environmental conditions, showing the highest mutual coordination. The emergence of wood structures and stem formation is associated with a series of developmental processes as well as the presence and abundance of wood chemical components such as lignin, cellulose and other sugars, and secondary metabolites. Amino acids such as proline or aspartic acid are associated with nitrogen mobilization from the ray parenchyma cells of wood to the growing organs [20]. Variation in metabolite concentrations in the xylem of Populus species has been associated with genetic, seasonal, positional (within the tree), and nutritional variations, with modest genetic control and, correspondingly, a significant effect of the environment or non-genetic factors [2]. The patterns of the relative abundance of metabolites participating in cellular metabolism could be effective indicators of phenotypes related to woody traits [21]. Abreu et al. (2020) [22] analyzed wood formation tissues in Populus tremula, indicating that cambial activity, cell expansion, and second wall thickening are simultaneous processes. Additionally, they explained that the concentration variation of certain metabolites is synchronized to a particular stage of the wood formation process. At the transcriptional level, wounding induces a complete rearrangement of the transcriptional program in the cambial zone close to the injuries. At the first instance, radial growth is stopped, and a complete set of defensive genes, mostly related to biotic stress, are upregulated. Later on, cambial activity is restored in the lateral borders of the wound. During this second stage, certain genes related to early wood formation, including genes controlling cell wall formation, are significantly upregulated, while certain late-wood-related genes are downregulated [23].
Genome-wide association studies using SNP markers have identified several putative loci and/or candidate genes that determine ecophysiological and growth/biomass traits in Populus species [24,25,26,27,28,29,30,31]. Phenological observation and metabolomic profiling, combined with a GWAS, represent an approach for expanding the understanding of metabolites participating in different plant processes [32]. In this context, some studies have combined multi-omics approaches and many data sets at the same time to obtain deeper models that allow a more comprehensive determination of wood dynamics. Wang et al. (2018) [33] improved a quantitative multi-omic integration of the regulation of lignin biosynthesis in wood formation in P. trichocarpa. Furches et al. (2019) [34] revealed nine candidate genes involved in cell wall biosynthesis using intersected omics. Sarkar and Maranas (2020) [35] developed a method that explains the role of SNPs inserted into coding or regulatory regions by associating them with multi-omic networks according to their functional role in P. trichocarpa and Arabidopsis thaliana.
In this study, 387 accessions/genotypes of black cottonwood, established in a common garden in Davis, California, were assessed for an aspen borer attack, that emerged from a natural outbreak, using a binomial classification method (attacked/non-attack). A GWAS considering the stem number and relative abundance of metabolites as covariates in a binary-association model was performed to analyze the variation in attack. Thus, this study aimed to analyze the genetic mechanisms underlying the response of this tree species to infection by the aspen borer Saperda calcarata and assess its influence on wood architecture traits and metabolite production.
2. Materials and Methods
2.1. Field Trial Description
Details about experimental design, genotype data acquisition, and chemical and metabolomic analysis are described elsewhere by the authors of [2]. Briefly, a clonal trial was performed at the experimental field of the University of California Davis, in Davis, California. Four hundred and sixty-one P. trichocarpa one-year clones were established in a culture frame of 1.83 × 1.83 m. They corresponded to 101 provenances representing part of the P. trichocarpa natural distribution range, particularly from Oregon and Washington. Three ramets per clone were established in a random block design and they were assessed in the second growing season. At that moment, 1161 trees were assessed for tree borer damage and stem number. The average values of different traits measured in the trial at the time of evaluation are shown in Table 1.

Table 1.
Average values for a set of selected traits measured in the field trial (adapted from [2]).
The chemical composition and metabolome profile of wood samples were characterized as described by the authors of [2]. The contents of 5- and 6-C sugars, lignin, and the syringyl:guaiacyl ratio were determined using high-throughput pyrolysis molecular beam mass spectrometry (pyMBMS) at the National Renewable Energy Laboratory (Golden, CO, USA). Metabolomic analyses were performed at UC Davis West Coast Metabolomics Center (Davis, CA, USA). Wood samples were obtained using a 5 mm Haglöf increment borer (Haglöf, Sweden). Cores were taken 0.3 m above ground level and followed a fixed east-to-west orientation. These were placed in plastic tubes, cooled immediately on dry ice, and stored at −80 °C until analysis. Three wood samples (one per tree) were collected for each clone, representing three biological replicates in the metabolomic analysis. The collected samples were pulverized. In the case of metabolome analyses, they were processed and submitted to an Agilent 6890 N gas chromatograph (Palo Alto, CA, USA) coupled with a time-of-flight Pegasus III mass spectrometer (Leco, MI, USA). Metabolite annotations were confirmed at the MassBank of North America (http://mona.fiehnlab.ucdavis.edu) (accessed on 21 November 2022) databases from mass spectrum profiles.
2.2. Assessment of Wood Borer Attack and Number of Stems
Each tree was inspected for the damage signs associated with S. calcatara (see Figure S1). The presence of tunnels, wet spots, scars on the bark, or swelling around tunnels in the main stems (as well as the presence of larvae or adult individuals in them) were considered indicators of insect attack. The presence or absence of the wood borer was registered using a binomial scale, where “1” indicated attacked and “0” was non-attacked. The presence of the wood borer was part of a natural outbreak observed in the field trial site during its second growing season. No insect control was applied during the duration of the trial. The measurement was carried out at the end of September (beginning of fall), in the last part of the growing season, after the infestation. Each tree was also evaluated for the number of stems. All trees had a main stem at the moment of measurements, which was considered for diameter and height determination. Some trees developed one or two lateral stems, which were also registered. The relationship between borer attack and the number of stems was analyzed using contingency tables and chi2 tests (α = 0.05).
2.3. Wood Metabolite Data Analysis
Six hundred and twenty-five metabolites were used for the analysis. One hundred and sixty-one of them, previously annotated, were analyzed by the authors of [2]. In this study, the whole metabolite set was assessed, independently of their annotation availability. Metabolite profiles were processed using the Metabolanalyst 5.0 platform (https://www.metaboanalyst.ca/ (accessed on 15 January 2023)) to normalize data. Missing values were estimated as 1/5 of the minimum positive value of their corresponding variable, while data were auto-scaled and normalized by the median. The mean relative abundance of individuals’ metabolites was calculated for each genotype and used as a covariate in the association model.
2.4. Genotyping and SNP Data Processing
Sequence capture, genome sequencing, data analysis, and SNP calling are also described in detail by the authors of [2]. Briefly, After DNA isolation, oligonucleotide baits were designed using the Populus trichocarpa genome, v 2.0. Then, the biotinylated RNA bait library and DNA library were synthesized. The prepared libraries were hybridized to the RNA baits and subsequently purified on magnetic beads. The multiplexed libraries were sequenced using Illumina HiSeq 2500 System in a 2 × 100 paired-end format at Virginia Bioinformatics Institute (VBI). Short reads of poplar were filtered, followed by alignment to the P. trichocarpa reference genome. The resulting alignment files were transformed into a binary version for variant calling. Duplicate reads were eliminated, indels were realigned and SNP calling was performed using Genome Analysis Toolkit v3.x (GATK, https://software.broadinstitute.org/gatk/ (accessed on 15 January 2023)). SNP data were filtered considering departures from yjr Hardy–Weinberg equilibrium in GATK, a minor allele frequency (MAF) of < 5%, and individuals with >5% of missing data. Finally, 387 P. trichocarpa accessions and 629.707 SNP markers, distributed within 19 chromosomes, were used for the analyses. SNP data were further transformed into a binary format using the GAPIT3 package (https://zzlab.net/GAPIT/ (accessed on 15 January 2023)) in RStudio [36]. The genetic relationship among P. trichocarpa genotypes was estimated via the kinship matrix from the filtered genotyping data, using the Dominance Centered IBS option in Tassel 5.2 [37].
2.5. Association Analysis
The GWAS involved a generalized linear mixed model association test (GMMAT), including logistic regression, to assess the binomial variable describing the presence of a tree borer attack. The modeling included the number of stems and the relative abundance of individual metabolites as covariates. The analysis was carried out using the GMMAT package [38] in RStudio [36]. The approach included two steps to reduce the computational burden. The first step consisted of performing a score test for the null hypotheses (where the genotype effect was 0) using the following formula:
where (πi0) = P(yi = 1|Xi,bi) is the probability of a binary phenotype for tree i, depending on their covariates (Xi and α) and random effects (bi). After the null hypothesis was tested, a single variant test was performed considering the following model:
where (πi0) = P(yi = 1|Xi, Gi, bi) is the probability of a binary phenotype for tree i, depending on their covariates (Xiα), genotype (Giβ), and random effects (bi). Xi is a row vector of covariates for tree i, and α is a column vector of fixed covariate effects including the intercept. Gi is the genotype of a genetic variant for tree i, and β is the genotypic effect of that genetic variant. bi is a column vector of random effects, accounting for variance component parameters and n x n relatedness matrices (among genetic accessions) [39]. For this study, covariates (Xiα) were represented by some stems and metabolite relative abundance. Each of these variables was added to the formula as a single covariate to observe its effect on the attacked tree’s phenotype. The GMMAT script was set up according to the package’s user manual [38]. p-values obtained for each test were subjected to Bonferroni correction (q-value < 0.1) using the p-adjust function in R. The phenotypic effects of molecular genotypes belonging to the significantly associated markers were compared using Tukey’s honestly significant difference (alpha = 0.05) in RStudio [36].
logit(πi0) = Xiα + bi
logit(πi0) = Xiα + Giβ + bi
2.6. Linkage Disequilibrium Analysis
In Guerra et al. (2016) [2], the extent of linkage disequilibrium (LD) was assessed for each chromosome. On average, the LD on physical distance decreased to below r2 0.2 at 26.9 kbp. Herein, chromosomes containing significantly associated SNPs were submitted to a chromosome-specific LD analysis to gain insight into particular interactions among different SNPs. The LD analysis was performed using the site-by-all method in Tassel 5.2. Only markers in the LD with p-values of less than 0.001 and r2 values greater than 0.2 were included.
2.7. Gene Ontology
Genes containing statistically significant SNP markers were analyzed in terms of their function and coding proteins using different platforms/databases, including Phytozome 13 (using the Populus trichocarpa V. 3.1 genome sequence) [40], quickGO (https://www.ebi.ac.uk/QuickGO/ (accessed on 15 January 2023)), KEGG (https://www.genome.jp/kegg/ (accessed on 15 January 2023)), InterPro (https://www.ebi.ac.uk/interpro/), UniProt (https://www.uniprot.org/ (accessed on 15 January 2023)), Panther (http://pantherdb.org/ (accessed on 15 January 2023)) and Pfam (http://pfam.xfam.org/ (accessed on 15 January 2023)) databases. Upstream or downstream regions around gene models containing significant SNP were analyzed using the Phytozome platform.
3. Results
3.1. Presence of Wood Borer and Number of Stems
S. calcarata attack was found in 15.4% of the assessed trees (Table S2). Concerning the number of stems, the percentage of trees with one, two, and three stems were 82%, 16% and 2%, respectively. The analysis of the phenotypic association between the presence of insect attack and the number of stems indicated that the attack was independent of the number of stems (p-value = 0.244, Chi2 test).
3.2. Association Analysis on S. calcarata Attack
A set of 629.707 SNPs, comprising the whole P. trichocarpa exome (across 19 chromosomes), were assessed regarding their association with the presence of the aspen borer in trees. The top 10 SNP markers more significantly (under the threshold of p-value < 1 × 10−5) associated with attack presence/absence are shown in Figure 1 and Table 2. Two of them remained significant after the Bonferroni correction (q-value < 0.2). Marker Chr03_13378835 in chromosome 3 is part of the gene encoding “Chromodomain-helicase-DNA-binding family protein” (Potri.003G110100) (p-value = 1.61 × 10−8; q-value = 0.0304). This non-synonymous SNP belongs to a codon encoding arginine in its normal sequence and alanine as its variant. Marker Chr10_22235609 in chromosome 10 is located in the “Mandelonitrile lyase/Mandelonitrile benzaldehyde-lyase” (Potri.010G249501) gene (p-value = 3.35 × 10−8; q-value = 0.0632). This SNP is also a non-synonymous one, and the associated codon encodes for isoleucine or aspartic acid, depending on the variant. No significant single-SNP associations were detected when association models included individual wood metabolites as a covariate.
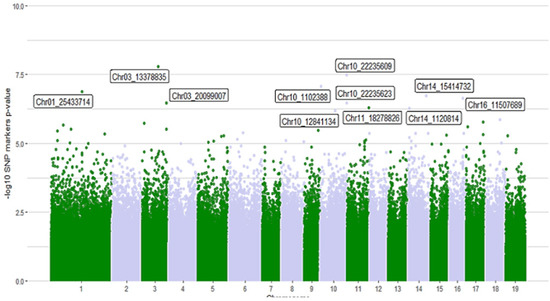
Figure 1.
Manhattan plot for association tests of the presence of S.calcarata in P.trichocarpa stems. p-values < 5.25 × 10−7 are labeled. Labels indicate the SNP marker identification which includes the chromosome number and the position within the chromosome.

Table 2.
Top 10 significant SNP markers identified in association with the presence of S.calcarata in P.trichocarpa stems.
3.3. LD Analysis
Considering the significance level of the SNP markers Chr03_13378835 and Chr10_22235609, a whole chromosome-specific LD analysis was performed for their respective chromosomes. Figure 2A shows the marker Chr03_13378835 and genes in the nearby area. At the 5′ end is located the “similar to Probable elongation factor G; similar to mitochondrial precursor (mEF-G)” (Potri.003G109900) gene and the “Protein kinase family protein” (Potri.003G110000) gene. At the 3′ end is the “PROTEIN FAR1-RELATED SEQUENCE 1” (Potri.003G110300) gene, followed by the uncharacterized protein (Potri.003G110200.1; not depicted in the figure) gene. Marker Chr03_13378835 showed a significant LD with some SNPs located at the farthest 5′ and 3′ regions of chromosome 3, beyond the LD decay region Figure 2 contains.
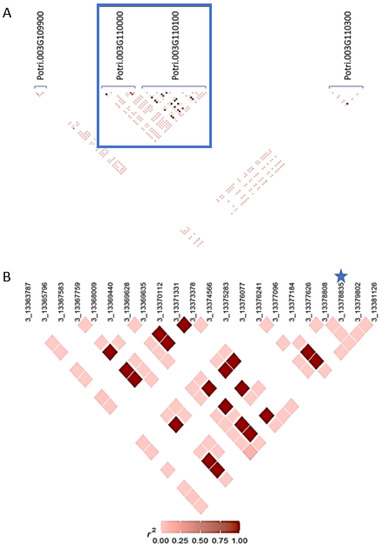
Figure 2.
Linkage disequilibrium heatmaps for SNP marker Chr03_13378835. (A) Overview of genes around gene containing Chr03_13378835. (B) LD of Chr03_13378835 and SNP markers within Potri.003G110100 gene. Gene model names are indicated in (A). The blue square shows the area where the significant marker is located. In (B), significant marker Chr03_13378835 is indicated by a star.
Figure 3A shows the marker Chr10_22235609 and the surrounding genes. From the 5′ leftmost region it is a gene encoding an unknown protein (Potri.010G249000), followed by the “Ca2+:H+ antiporter (chaA, CAX)” (Potri.010G249200) and the “mitogen-activated protein kinase 4/5, plant (MKK4_5P)” gene (Potri.010G249300). On the 3′ side is located the gene “Phosphoglycerate dehydrogenase/Phosphoglyceric acid dehydrogenase” (Potri.010G249600), followed by the “RNA binding protein (contains RRM repeats)” (Potri.010G249700) gene.
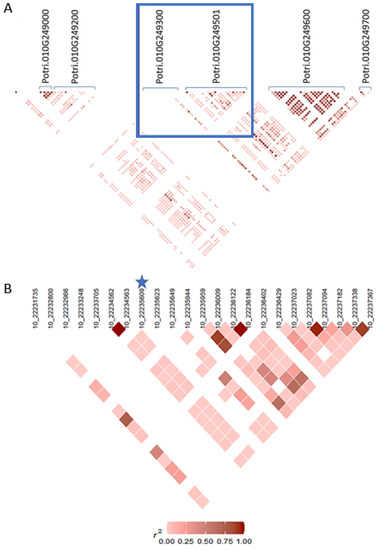
Figure 3.
Linkage disequilibrium heatmaps for SNP marker Chr10_22235609. (A) Overview of genes around gene containing Chr10_22235609. (B) LD of Chr10_22235609 and SNP markers within Potri.010G249501.1 gene. Gene model names are indicated in (A). The blue square shows the area where the significant marker is located. In (B), significant marker Chr03_13378835 is indicated by a star.
A summary of markers of significant LD with marker Chr03_13378835 at chromosome 3 is presented in Table 3.

Table 3.
Summary of SNP markers of significant LD with marker Chr03_13378835.
3.4. Genotype Effects
The effect of molecular genotypes, belonging to the significant SNPs, on the presence of S. calcarata attack is represented in Figure 4. The GG and TT homozygous genotypes are associated with the largest number of attacked trees for the Chr03_13378835 and Chr10_22235609 SNP markers, respectively.
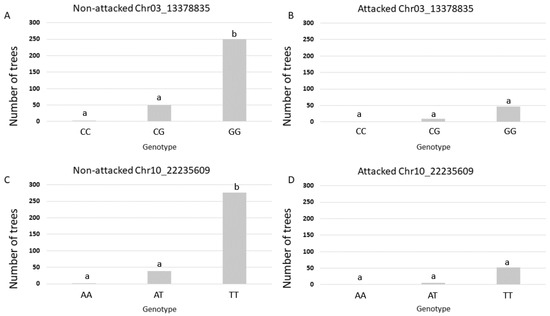
Figure 4.
Relationship between genotypes and the number of attacked and non-attacked trees. (A,B) Marker Chr03_13378835 and its related genotypes. (C,D) Marker Chr10_22235609 and its related genotypes. Different letters above the bars indicate significant differences (Tukey’s honestly significant difference; alpha = 0.05).
4. Discussion
Wood borers such as S. calcarata are one of the most damaging and costly groups of invasive insects in North American forests [7]. Their attack suggests a huge machinery of genetic expression and metabolite production operating in trees to counteract their infestation with a suite of defensive responses, that are similar to what herbivores, fungi, or bacteria can induce. We conducted a GWAS using P. trichocarpa genotypes collected from its natural distribution range and established in a common garden assay to identify the loci underlying the response to S. calcarata attack, to improve our knowledge about this particular insect–tree interaction.
The insect attacks can disrupt the normal development of main stems and may promote the appearance of secondary stems and multi-branched phenotypes. Stem number has been reported to have strong environmental control over plant architecture traits [19,41], which suggests that it has low inheritance. From our results, the relationship between genetic variation and the presence of a wood borer attack seems weak since a reduced quantity of significant SNPs was detected, compared with that in other studies analyzing the interaction between plant–stem borer insects [42,43]. This suggests that this trait would be more responsive to the environment than to genetic factors. However, further analyses such as trait–haplotype association tests may provide complimentary results of the combined effects of SNPs on phenotype. Concerning the stem number and the presence of attack, the analysis of their phenotypic relationship indicated that they have an independent distribution. Similar observations were reported by the authors of [44] in Salix spp., where no underlying relationship was found between growth traits and the proportion of attacked trees by Cryptorhynchus lapathi.
The performed GWAS allowed the identification of significant SNP markers associated with the presence of the aspen borer in the analyzed trees. Marker Chr03_13378835 is in the exonic region of a “Chromodomain-helicase-DNA binding family protein” (CHD) gene (Potri.003G110100), and is in LD with three markers within the same gene, ranging from a 1.74 to 11.08 kbp distance (Chr03_13377096, Chr03_13369635, and Chr03_13367759). Chromatin remodeling enzymes contribute to changes in chromatin during transcription, recombination, repair, and replication [45]. CHD proteins act as important epigenetic regulators to control the expression of a series of genes participating in plant development and stress response [46,47,48]. Linkage disequilibrium analysis also showed significant r2 values with SNP markers present in other genes such as “similar to probable elongation factor G; similar to mitochondrial precursor (mEF-G)” (Potri.003G109900). Mitochondrial elongation factor G is an essential protein with central roles in both the elongation and ribosome recycling phases of protein synthesis [49]. Another gene with markers of LD was “Protein kinase family protein” (Potri.003G110000). Protein kinases are regulatory components in plant responses to environmental stresses such as drought, high salinity, cold, and pathogen attack [50]. Another gene containing SNPs in LD was the “Protein FAR1-Related sequence 1” (Potri.003G110300). Recent studies have demonstrated that FAR1 plays multiple roles in development, oxidative stress responses, and plant immunity [51]. In the farthest regions at the 3′ end from marker Chr03_13378835, markers Chr03_17107069 and Chr03_17107097 are part of the “Heparan-alpha-glucosaminide N-acetyltransferase” (HGSNAT) gene (Potri.003G159200). The encoded enzyme catalyzes the acetylation of the terminal glucosamine residues of heparan sulfate, a process related to the virus’s recognition in humans [52,53]. In plants, a non-synonymous mutation of a histidine residue in the HGSNAT gene was suggested as a molecular marker for breeding sudden death syndrome resistance in soybean [54]. Another marker in LD was Chr03_284474, which is part of the “MLO-LIKE PROTEIN 3” gene (Potri.003G002500), involved in immune responses and cell death mediated by cytosolic-free calcium [55,56].
In chromosome 10, the significant SNP marker Chr10_22235609 is located in an exonic region of an “R-mandelonitrile lyase” gene (Potri.010G249501.1), a flavoprotein that plays a key role in cyanogenesis. This process consists of the release of hydrogen cyanide from damaged plant tissues [57], which is consistent with the defense against insect attack. Mandelonitrile has been suggested as a starter molecule of an auxiliary pathway for salicylic acid production in Prunus [58], and protection against insects in wheat seeds [59]. The position of a marker, near an exon–intron limit, may be indicative of its participation in splicing during mRNA maturation. Marker Chr10_22235609 shows LD with markers in other genes in proximity to it and considering their function and relationship with defensive responses, they are worth explaining briefly (Figure 3A). The “Ca2+:H+ antiporter (chaA, CAX)” gene (Potri.010G249200) is important to cell regulation against developmental cues and environmental challenges. Ca2+ has a versatile role in cellular signaling. H+/Ca2+ antiporter CAX proteins facilitate cellular Ca2+ homeostasis [60]. Moving toward the 3′ end, the gene “mitogen-activated protein kinase 4/5, plant (MKK4_5P)” (Potri.010G249300) is closely related to the mitogen-activated protein kinase (MAPK) cascades, that transduce environmental and developmental cues into intracellular responses. MAPK cascades are activated in the presence of PAMPs (pathogen-associated molecular patterns) to induce defensive responses against pathogenic agents such as plant-triggered immunity and effector-triggered immunity, and ethylene production [61]. Another gene is “Phosphoglycerate dehydrogenase” (PGDH) (Potri.010G249600), which encodes an enzyme related to plant development and stress tolerance [62]. Thus, PGDH is highly induced under biotic or abiotic stress conditions [63]. Additionally, the gene “RNA binding protein (contains RRM repeats)” (Potri.010G249700) encodes RNA binding proteins, key regulators of gene expression under stress conditions [64]. Finally, the GWAS also detected two SNPs associated with the presence of the insect, which were located within intergenic regions of chromosomes 1 and 14 (Table 2). These polymorphisms could be important for gene expression, in case they are part of transcriptional regulatory elements located upstream of the coding region of some genes.
Regarding the models utilized for the GWAS, when the wood metabolite abundance was used as a covariate, none of the individually assessed metabolites influenced the single-trait SNP marker association (at q-value < 0.1). This would have been indicative of the negligible effect of the variation in specific wood metabolites on the presence of S. calcarata. Nevertheless, some markers attained significant p-values (near 1 × 10−7) when some metabolites were tested (Table S1). The lack of significant metabolites could be explained by the fact that insect attack can be an independent phenomenon regarding the production of some specific wood metabolites by the trees. In that sense, Broberg et al. (2010) [65] reported atypical results in Populus maximowiczii hybrids that exhibited antixenotic resistance to Cryptorhynchus lapathi independently of a high-metabolite concentration such as that of condensed tannins and catechin, suggesting the possibility of an alternative defense mechanism.
Considering that the aspen borer attack analyzed in this study was from a natural outbreak, the infestation dynamics of the insect could be different from that which would follow in an experiment in which the presence of the insect is experimentally directed. In this sense, this study represents an initial approach to understanding the molecular mechanisms operating in the response of P. trichocarpa to the aspen borer, which are currently poorly understood. Further studies that consider, for example, a larger number of infected trees, would contribute to the corroboration of our observed results. Similarly, an assessment of the level of attack on a quantitative measure, or a broader categorical scale of damage, would allow a more precise characterization of the damage produced by the insect. This could also extend to the detection of molecular processes, genes, and SNP markers involved in the defensive response of poplars, using a new GWAS or complimentary omics approaches.
Altogether, the significant SNP markers and genes detected in this study, as well as those in LD with them, seems geared toward a general defensive response to stress caused by S. calcarata attack. Association analyses of P. trichocarpa individuals affected by the borer larvae revealed that the responses were independent of wood metabolite production and new stem formation. The results are consistent with the elicited-defensive mechanism against biotic stress, suggesting the participation of signaling mechanisms regulating secondary metabolite production. Further studies with P. trichocarpa under directed infestation environments and the application of complementary methodological approaches can contribute to the confirmation or broadening of the understanding of the participation of genes, SNPs, and secondary metabolites in its defensive response, as well as to identifying resistant genotypes, and supporting the design of strategies against tree borers beetles.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/f14061129/s1, Figure S1: Damage produced by S. calcarata in the assessed P.trichocarpa trees.; Table S1: Metabolites with significant effect as a covariate on association models, and SNP-markers significantly associated with the presence of S. calcarata in stems; Table S2: General statistics for the presence of S. calcarata in the trial.
Author Contributions
Conceptualization, F.P.G., D.B.N., J.A.H., B.J.S. and S.L.S.; methodology, S.L.S., D.B.N., J.A.H., B.J.S., O.F. and R.F.; software, S.L.S.; formal analysis, S.L.S., J.A.H., O.F. and R.F.; investigation, D.B.N., J.A.H., B.J.S., O.F. and F.P.G.; writing—original draft preparation, S.L.S. and F.P.G.; writing—review and editing, S.L.S., F.P.G., D.B.N., J.A.H., B.J.S., O.F. and R.F.; supervision, D.B.N. and F.P.G.; funding acquisition, D.B.N., J.A.H. and F.P.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Advanced Hardwood Biofuels Northwest Project, supported by Agriculture and Food Research Initiative Competitive (grant no. 2011–68005-30407, USDA National Institute of Food and Agri-culture), the National Science Foundation Plant Genome Research Program (IOS grant no. 1054444 to JAH), and ANID FONDECYT Program, Project FONDECYT Regular 1201740.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Sequences of the prepared libraries for exome capture are available at Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/ (accessed on 15 January 2023)) under the accession number SRA058855.
Acknowledgments
We would like to thank the California Agricultural Experiment Station for the provided support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Geraldes, A.; Pang, J.; Thiessen, N.; Cezard, T.; Moore, R.; Zhao, Y.; Tam, A.; Wang, S.; Friedmann, M.; Birol, I.; et al. SNP discovery in black cottonwood (Populus trichocarpa) by population transcriptome resequencing. Mol. Ecol. Resour. 2011, 11, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Guerra, F.P.; Richards, J.H.; Fiehn, O.; Famula, R.; Stanton, B.J.; Shuren, R.; Sykes, R.; Davis, M.F.; Neale, D.B. Analysis of the genetic variation in growth, ecophysiology, and chemical and metabolomic composition of wood of Populus trichocarpa provenances. Tree Genet. Genomes 2016, 12, 1–16. [Google Scholar] [CrossRef]
- AHB. Advances Hardwood Biofuels Northwest. Available online: https://hardwoodbiofuels.org/topics/fuel-and-chemicals/ (accessed on 20 December 2020).
- Moreno-Cortés, A.; Ramos-Sánchez, J.M.; Hernández-Verdeja, T.; González-Melendi, P.; Alves, A.; Simões, R.; Rodrigues, J.C.; Guijarro, M.; Canellas, I.; Allona, I. Impact of RAV1-engineering on poplar biomass production: A short-rotation coppice field trial. Biotechnol. Biofuels 2017, 10, 110. [Google Scholar] [CrossRef]
- Xu, D.; Qi, X.; Li, J.; Han, X.; Wang, J.; Jiang, Y.; Tian, Y.; Wang, Y. PzTAC and PzLAZY from a narrow-crown poplar contribute to regulation of branch angles. Plant Physiol. Biochem. 2017, 118, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Tuskan, G.A.; DiFazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The Genome of Black Cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [CrossRef]
- Aukema, J.E.; Leung, B.; Kovacs, K.; Chivers, C.; Britton, K.O.; Englin, J.; Frankel, S.J.; Haight, R.G.; Holmes, T.P.; Liebhold, A.M.; et al. Economic Impacts of Non-Native Forest Insects in the Continental United States. PLoS ONE 2011, 6, e24587. [Google Scholar] [CrossRef] [PubMed]
- Kundanati, L.; Chahare, N.R.; Jaddivada, S.; Karkisaval, A.G.; Sridhar, R.; Pugno, N.M.; Gundiah, N. Cutting mechanics of wood by beetle larval mandibles. J. Mech. Behav. Biomed. Mater. 2020, 112, 104027. [Google Scholar] [CrossRef]
- Coleman, T.W.; Poloni, A.L.; Chen, Y.; Thu, P.Q.; Li, Q.; Sun, J.; Rabaglia, R.J.; Man, G.; Seybold, S.J. Hardwood injury and mortality associated with two shot hole borers, Euwallacea spp., in the invaded region of southern California, USA, and the native region of Southeast Asia. Ann. For. Sci. 2019, 76, 61. [Google Scholar] [CrossRef]
- U.S. Department of Agriculture; Forest Service. Insect of Eastern Forests. N° 1426; U.S. Department of Agriculture: Washington, DC, USA, 1985; pp. 295–297. Available online: https://archive.org/details/insectsofeastern1426unit/page/n5/mode/2up (accessed on 15 January 2023).
- Tsankov, G.; Georgiev, G. Records on parasitoids of smaller poplar borer, Saperda populnea [Coleoptera, Cerambycidae] along the Danube in Bulgaria. Biocontrol 1991, 36, 493–498. [Google Scholar] [CrossRef]
- Ke, J.; Laskar, D.D.; Chen, S. Biodegradation of hardwood lignocellulosic by the western poplar clearwing borer, Paranthrene robiniae (Hy. Edwards). Biomacromolecules 2011, 12, 1610–1620. [Google Scholar] [CrossRef]
- Chamberlin, E.A.; Aarssen, L.W. The Cost of Apical Dominance in White Pine (Pinus strobus L.): Growth in Multi-Stemmed Versus Single-Stemmed Trees. Bull. Torrey Bot. Club 1996, 123, 268. [Google Scholar] [CrossRef]
- Stephens, A.E.A.; Westoby, M. Effects of insect attack to stems on plant survival, growth, reproduction and photosynthesis. Oikos 2014, 124, 266–273. [Google Scholar] [CrossRef]
- Puettmann, K.J.; Saunders, M.R. Patterns of growth compensation in eastern white pine (Pinus strobus L.): The influence of herbivory intensity and competitive environments. Oecologia 2001, 129, 376–384. [Google Scholar] [CrossRef]
- Pratt, P.D.; Rayamajhi, M.B.; Van, T.K.; Center, T.D.; Tipping, P.W. Herbivory alters resource allocation and compensation in the invasive tree Melaleuca quinquenervia. Ecol. Èntomol. 2005, 30, 316–326. [Google Scholar] [CrossRef]
- Whitham, T.G.; Mopper, S. Chronic Herbivory: Impacts on Architecture and Sex Expression of Pinyon Pine. Science 1985, 228, 1089–1091. [Google Scholar] [CrossRef]
- Sacchi, C.F.; Connor, E.F. Changes in Reproduction and Architecture in Flowering Dogwood, Cornus florida, after Attack by the Dogwood Club Gall, Resseliella clavula. Oikos 1999, 86, 138. [Google Scholar] [CrossRef]
- García-Cervigón, A.I.; García-López, M.A.; Pistón, N.; Pugnaire, F.I.; Olano, J.M. Co-ordination between xylem anatomy, plant architecture and leaf functional traits in response to abiotic and biotic drivers in a nurse cushion plant. Ann. Bot. 2021, 127, 919–929. [Google Scholar] [CrossRef]
- Sauter, J.J.; van Cleve, B. Seasonal variation of amino acids in the xylem sap of “Populus × canadensis” and its relation to protein body mobilization. Trees 1992, 7, 26–32. [Google Scholar] [CrossRef]
- Robinson, A.R.; Gheneim, R.; Kozak, R.A.; Ellis, D.D.; Mansfield, S.D. The potential of metabolite profiling as a selection tool for genotype discrimination in Populus. J. Exp. Bot. 2005, 56, 2807–2819. [Google Scholar] [CrossRef]
- Abreu, I.N.; Johansson, A.I.; Sokołowska, K.; Niittylä, T.; Sundberg, B.; Hvidsten, T.R.; Street, N.R.; Moritz, T. A metabolite roadmap of the wood-forming tissue in Populus tremula. New Phytol. 2020, 228, 1559–1572. [Google Scholar] [CrossRef]
- Chano, V.; Collada, C.; Soto, A. Transcriptomic analysis of wound xylem formation in Pinus canariensis. BMC Plant Biol. 2017, 17, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Wegrzyn, J.L.; Eckert, A.J.; Choi, M.; Lee, J.M.; Stanton, B.J.; Sykes, R.; Davis, M.F.; Tsai, C.-J.; Neale, D.B. Association genetics of traits controlling lignin and cellulose biosynthesis in black cottonwood (Populus trichocarpa, Salicaceae) secondary xylem. New Phytol. 2010, 188, 515–532. [Google Scholar] [CrossRef] [PubMed]
- McKown, A.D.; Guy, R.D.; Quamme, L.; Klápště, J.; La Mantia, J.; Constabel, C.P.; El-Kassaby, Y.A.; Hamelin, R.C.; Zifkin, M.; Azam, M.S. Association genetics, geography and ecophysiology link stomatal patterning in Populus trichocarpa with carbon gain and disease resistance trade-offs. Mol. Ecol. 2014, 23, 5771–5790. [Google Scholar] [CrossRef] [PubMed]
- McKown, A.D.; Klápště, J.; Guy, R.D.; Geraldes, A.; Porth, I.; Hannemann, J.; Friedmann, M.; Muchero, W.; Tuskan, G.A.; Ehlting, J.; et al. Genome-wide association implicates numerous genes underlying ecological trait variation in natural populations of Populus trichocarpa. New Phytol. 2014, 203, 535–553. [Google Scholar] [CrossRef] [PubMed]
- Porth, I.; El-Kassaby, Y.A. Using Populus as a lignocellulosic feedstock for bioethanol. Biotechnol. J. 2015, 10, 510–524. [Google Scholar] [CrossRef]
- Guerra, F.P.; Suren, H.; Holliday, J.; Richards, J.H.; Fiehn, O.; Famula, R.; Stanton, B.J.; Shuren, R.; Sykes, R.; Davis, M.F.; et al. Exome resequencing and GWAS for growth, ecophysiology, and chemical and metabolomic composition of wood of Populus trichocarpa. BMC Genom. 2019, 20, 1–14. [Google Scholar] [CrossRef]
- Chhetri, H.B.; Macaya-Sanz, D.; Kainer, D.; Biswal, A.K.; Evans, L.M.; Chen, J.G.; Collins, C.; Hunt, K.; Mohanty, S.S.; DiFazio, S.P.; et al. Multitrait genome-wide association analysis of Populus trichocarpa identifies key polymorphisms controlling morphological and physiological traits. New Phytol. 2019, 223, 293–309. [Google Scholar] [CrossRef]
- Chhetri, H.; Furches, A.; Macaya-Sanz, D.; Walker, A.R.; Kainer, D.; Jones, P.; Harman-Ware, A.E.; Tschaplinski, T.J.; Jacobson, D.; Tuskan, G.A.; et al. Genome-Wide Association Study of Wood Anatomical and Morphological Traits in Populus trichocarpa. Front. Plant Sci. 2020, 11, 545748. [Google Scholar] [CrossRef] [PubMed]
- Mähler, N.; Schiffthaler, B.; Robinson, K.M.; Terebieniec, B.K.; Vučak, M.; Mannapperuma, C.; Street, N.R. Leaf shape in Populus tremula is a complex, omnigenic trait. Ecol. Evol. 2020, 10, 11922–11940. [Google Scholar] [CrossRef]
- Zhou, S.; Kremling, K.A.; Bandillo, N.; Richter, A.; Zhang, Y.K.; Ahern, K.R.; Artyukhin, A.B.; Hui, J.X.; Younkin, G.C.; Schroeder, F.C.; et al. Metabolome-Scale Genome-Wide Association Studies Reveal Chemical Diversity and Genetic Control of Maize Specialized Metabolites. Plant Cell 2019, 31, 937–955. [Google Scholar] [CrossRef]
- Wang, J.P.; Matthews, M.L.; Williams, C.M.; Shi, R.; Yang, C.; Tunlaya-Anukit, S.; Chen, H.-C.; Li, Q.; Liu, J.; Lin, C.-Y.; et al. Improving wood properties for wood utilization through multi-omics integration in lignin biosynthesis. Nat. Commun. 2018, 9, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Furches, A.; Kainer, D.; Weighill, D.; Large, A.; Jones, P.; Walker, A.; Romero, J.; Gazolla, J.G.F.M.; Joubert, W.; Shah, M.; et al. Finding New Cell Wall Regulatory Genes in Populus trichocarpa Using Multiple Lines of Evidence. Front. Plant Sci. 2019, 10, 1249. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, D.; Maranas, C.D. SNPeffect: Identifying functional roles of SNPs using metabolic networks. Plant J. 2020, 103, 512–531. [Google Scholar] [CrossRef] [PubMed]
- RStudio Team. RStudio: Integrated Development for R. RStudio; Posit Software, PBC formerly RStudio: Boston, MA, USA, 2020; Available online: https://posit.co/download/rstudio-desktop/ (accessed on 25 May 2023).
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Chen, H. GMMAT: Generalized linear Mixed Model Association Tests Version 1.3.2; The University of Texas Health Science Center at Houston: Huston, TX, USA, 2021; Available online: https://cran.r-project.org/web/packages/GMMAT/index.html/ (accessed on 10 December 2021).
- Chen, H.; Wang, C.; Conomos, M.P.; Stilp, A.M.; Li, Z.; Sofer, T.; Szpiro, A.A.; Chen, W.; Brehm, J.M.; Celedón, J.C.; et al. Control for Population Structure and Relatedness for Binary Traits in Genetic Association Studies via Logistic Mixed Models. Am. J. Hum. Genet. 2016, 98, 653–666. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Marron, N.; Ricciotti, L.; Bastien, C.; Beritognolo, I.; Gaudet, M.; Paolucci, I.; Fabbrini, F.; Salani, F.; Dillen, S.Y.; Ceulemans, R.; et al. Plasticity of growth and biomass production of an intraspecific Populus alba family grown at three sites across Europe during three growing seasons. Can. J. For. Res. 2010, 40, 1887–1903. [Google Scholar] [CrossRef]
- Samayoa, L.F.; Malvar, R.A.; Olukolu, B.A.; Holland, J.B.; Butrón, A. Genome-wide association study reveals a set of genes associated with resistance to the Mediterranean corn borer (Sesamia nonagrioides L.) in a maize diversity panel. BMC Plant Biol. 2015, 15, 35. [Google Scholar] [CrossRef]
- Tamiru, A.; Paliwal, R.; Manthi, S.J.; Odeny, D.A.; Midega, C.A.O.; Khan, Z.R.; Pickett, J.A.; Bruce, T.J.A. Genome wide association analysis of a stemborer egg induced “call-for-help” defence trait in maize. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Broberg, C.L.; Borden, J.H.; Humble, L.M. Distribution and abundance of Cryptorhynchus lapathi on Salix spp. in British Columbia. Can. J. For. Res. 2002, 32, 561–568. [Google Scholar] [CrossRef]
- Marfella, C.G.; Imbalzano, A.N. The Chd family of chromatin remodelers. Mutat. Res./Fundam. Mol. Mech. Mutagen. 2007, 618, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Liu, D.; Zhong, X.; Zhang, C.; Zhang, Q.; Zhou, D.-X. CHD3 protein recognizes and regulates methylated histone H3 lysines 4 and 27 over a subset of targets in the rice genome. Proc. Natl. Acad. Sci. USA 2012, 109, 5773–5778. [Google Scholar] [CrossRef]
- Hu, Y.; Lai, Y.; Zhu, D. Transcription regulation by CHD proteins to control plant development. Front. Plant Sci. 2014, 5, 223. [Google Scholar] [CrossRef]
- Perruc, E.; Kinoshita, N.; Lopez-Molina, L. The role of chromatin-remodeling factor PKL in balancing osmotic stress responses during Arabidopsis seed germination. Plant J. 2007, 52, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, G.C.; Baldauf, S. Evolution of Elongation Factor G and the Origins of Mitochondrial and Chloroplast Forms. Mol. Biol. Evol. 2010, 28, 1281–1292. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Hsu, C.-C.; Du, Y.; Zhu, P.; Zhao, C.; Fu, X.; Zhang, C.; Paez, J.S.; Macho, A.P.; Tao, W.A.; et al. Mapping proteome-wide targets of protein kinases in plant stress responses. Proc. Natl. Acad. Sci. USA 2020, 117, 3270–3280. [Google Scholar] [CrossRef]
- Ma, L.; Li, G. FAR1-RELATED SEQUENCE (FRS) and FRS-RELATED FACTOR (FRF) Family Proteins in Arabidopsis Growth and Development. Front. Plant Sci. 2018, 9, 692. [Google Scholar] [CrossRef]
- Klein, U.; Kresse, H.; Von Figura, K. Sanfilippo syndrome type C: Deficiency of acetyl-CoA: Alpha-glucosaminide N-acetyltransferase in skin fibroblasts. Proc. Natl. Acad. Sci. USA 1978, 75, 5185–5189. [Google Scholar] [CrossRef]
- Kim, K.; Choi, D.; Kim, S.-M.; Kwak, D.-Y.; Choi, J.; Lee, S.; Lee, B.-C.; Hwang, D.; Hwang, I. A Systems Approach for Identifying Resistance Factors to Rice stripe virus. Mol. Plant-Microbe Interact. 2012, 25, 534–545. [Google Scholar] [CrossRef]
- Swaminathan, S.; Das, A.; Assefa, T.; Knight, J.M.; Da Silva, A.F.; Carvalho, J.P.S.; Hartman, G.L.; Huang, X.; Leandro, L.F.; Cianzio, S.R.; et al. Genome wide association study identifies novel single nucleotide polymorphic loci and candidate genes involved in soybean sudden death syndrome resistance. PLoS ONE 2019, 14, e0212071. [Google Scholar] [CrossRef]
- Devoto, A.; Piffanelli, P.; Nilsson, I.; Wallin, E.; Panstruga, R.; von Heijne, G.; Schulze-Lefert, P. Topology, Subcellular Localization, and Sequence Diversity of the Mlo Family in Plants. J. Biol. Chem. 1999, 274, 34993–35004. [Google Scholar] [CrossRef]
- Kim, M.C.; Lee, S.H.; Kim, J.K.; Chun, H.J.; Choi, M.S.; Chung, W.S.; Cho, M.J. Mlo, a modulator of plant defense and cell death, is a novel calmodulin-binding protein: Isolation and characterization of a rice Mlo homologue. J. Biol. Chem. 2002, 277, 19304–19314. [Google Scholar] [CrossRef]
- Hughes, M.A. 1.31—Biosynthesis and Degradation of Cyanogenic Glycosides Eds: Sir Derek Barton, Koji Nakanishi, Otto Meth-Cohn; Comprehensive Natural Products Chemistry; Elsevier Science Ltd.: Oxford, UK, 1999. [Google Scholar]
- Bernal-Vicente, A.; Petri, C.; Hernández, J.A.; Diaz-Vivancos, P. Biochemical study of the effect of stress conditions on the mandelonitrile-associated salicylic acid biosynthesis in peach. Plant Biol. 2020, 22, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Halter, J.G.; Chen, W.D.; Hild, N.; Mora, C.A.; Stoessel, P.R.; Koehler, F.M.; Stark, W.J. Induced cyanogenesis from hydroxy nitrile lyase and mandelonitrile on wheat with polylactic acid multilayer-coating produces self-defending seeds. J. Mater. Chem. A 2014, 2, 853–858. [Google Scholar] [CrossRef]
- Lu, S.; Li, Z.; Gorfe, A.A.; Zheng, L. Intracellular Ca2+ regulation of H+/Ca2+ antiporter YfkE mediated by a Ca2+ mini-sensor. Proc. Natl. Acad. Sci. USA 2020, 117, 10313–10321. [Google Scholar] [CrossRef] [PubMed]
- Cristina, M.S.; Petersen, M.; Mundy, J. Mitogen-Activated Protein Kinase Signaling in Plants. Annu. Rev. Plant Biol. 2010, 61, 621–649. [Google Scholar] [CrossRef]
- Kishor, P.B.K.; Suravajhala, R.; Rajasheker, G.; Marka, N.; Shridhar, K.K.; Dhulala, D.; Scinthia, K.P.; Divya, K.; Doma, M.; Edupuganti, S.; et al. Lysine, Lysine-Rich, Serine, and Serine-Rich Proteins: Link Between Metabolism, Development, and Abiotic Stress Tolerance and the Role of ncRNAs in Their Regulation. Front. Plant Sci. 2020, 11, 546213. [Google Scholar] [CrossRef] [PubMed]
- Casatejada-Anchel, R.; Muñoz-Bertomeu, J.; Rosa-Téllez, S.; Anoman, A.D.; Nebauer, S.G.; Torres-Moncho, A.; Fernie, A.R.; Ros, R. Phosphoglycerate dehydrogenase genes differentially affect Arabidopsis metabolism and development. Plant Sci. 2021, 306, 110863. [Google Scholar] [CrossRef]
- Alves, L.R.; Goldenberg, S. RNA-binding proteins related to stress response and differentiation in protozoa. World J. Biol. Chem. 2016, 7, 78. [Google Scholar] [CrossRef]
- Broberg, C.L.; Inkster, J.A.H.; Borden, J.H. Phenological and chemical differences among hybrid poplar clones (Salicaceae) varying in resistance to Cryptorhynchus lapathi (L.) (Coleoptera: Curculionidae). Biochem. Syst. Ecol. 2010, 38, 29–48. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).