Transcriptome Analysis of Biochemistry Responses to Low-Temperature Stress in the Flower Organs of Five Pear Varieties
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cold Treatments
2.3. Determination of Biochemistry Indicators
2.4. Transcriptome Sequencing
2.5. PCA Analysis
2.6. Real-Time PCR Analysis
3. Results
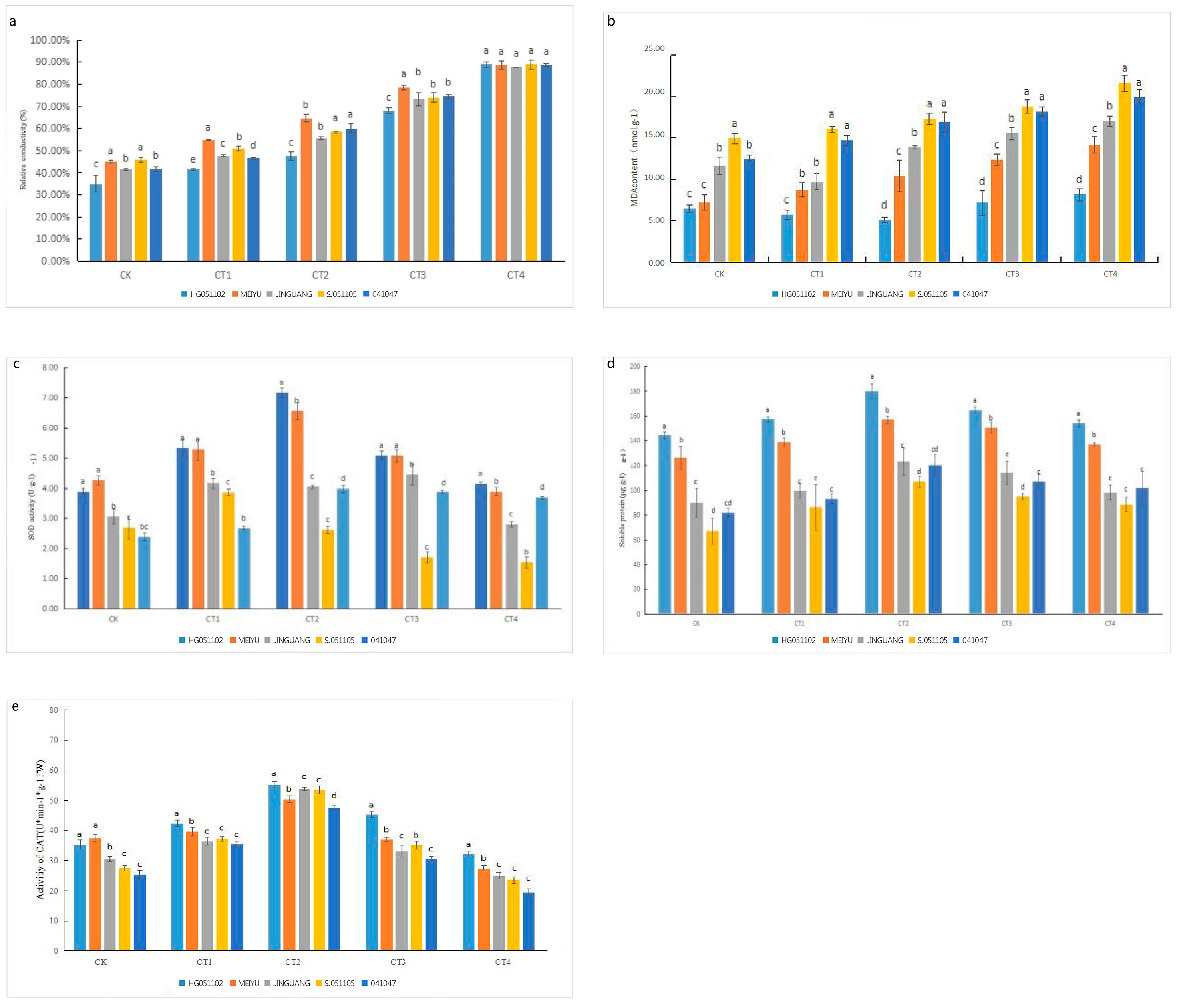
3.1. Biochemistry Characteristics and Cold Resistance of Flower Organs
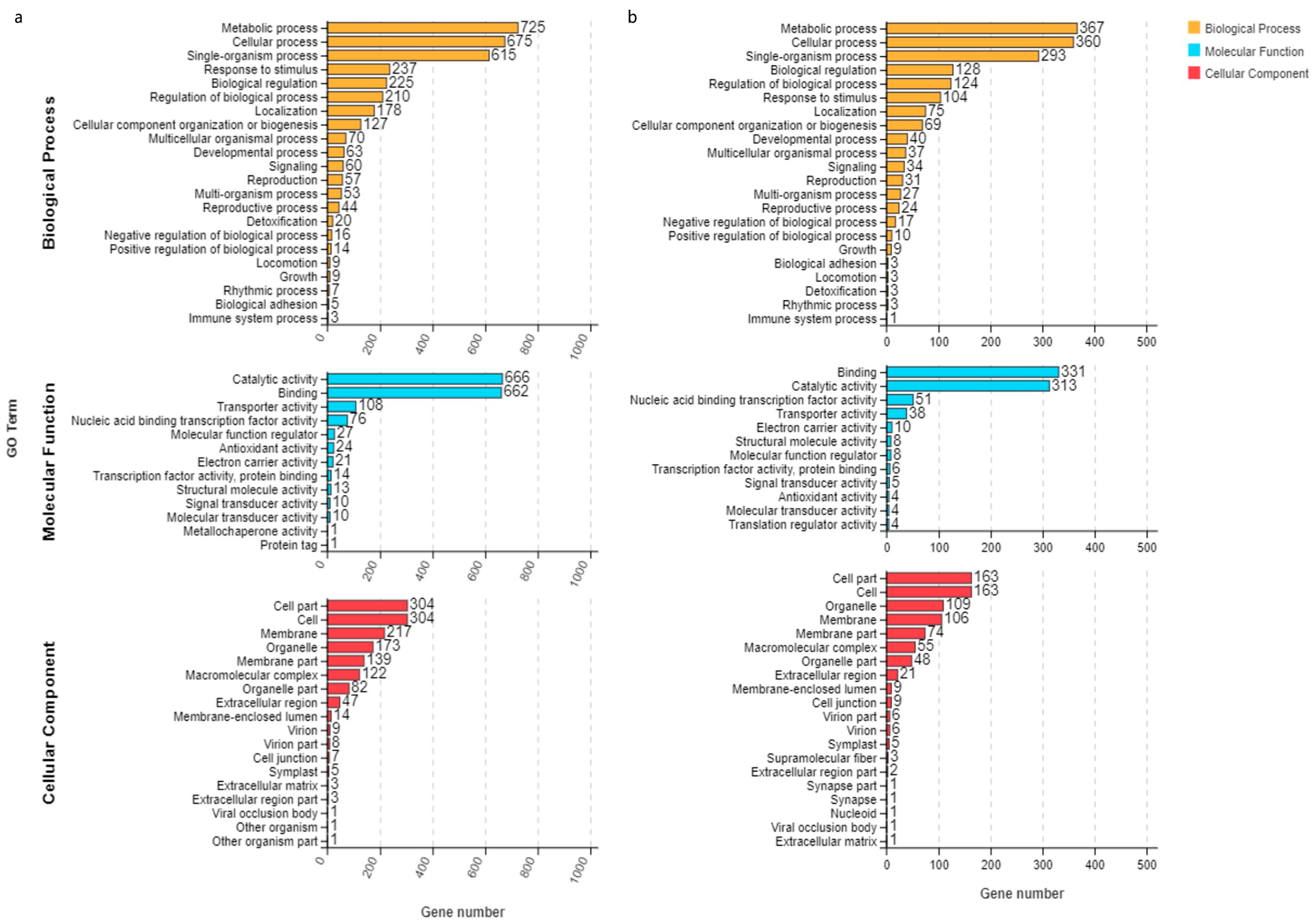
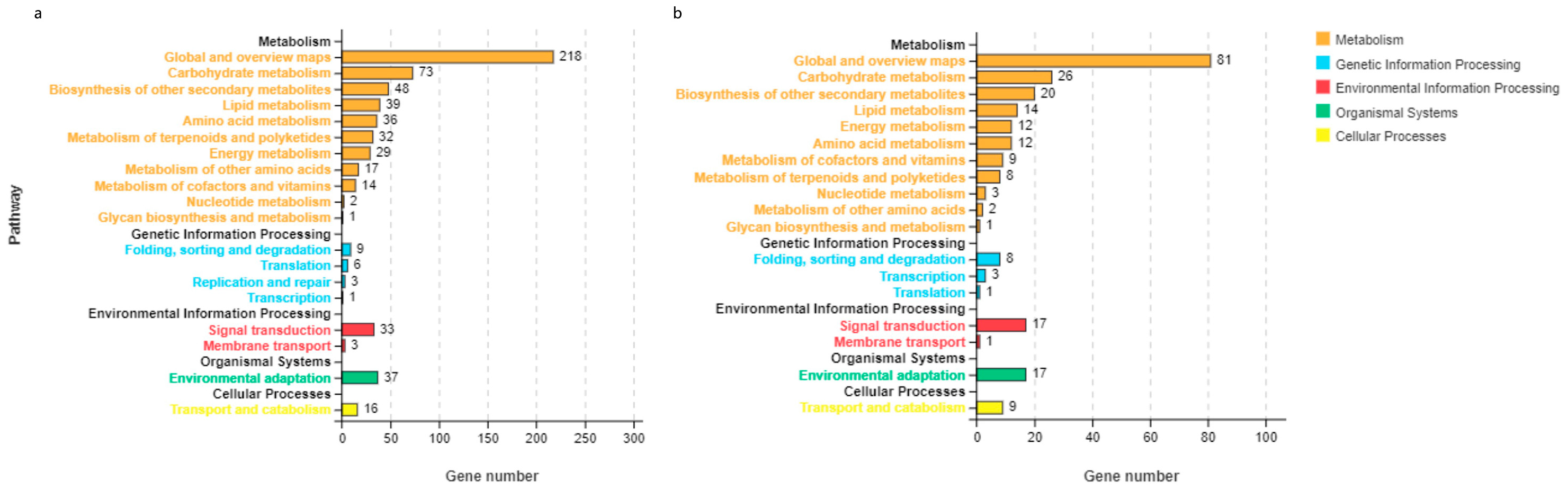
3.2. DEG Analysis for ‘Jinguang’
3.3. Transcription Factors Associated with Chilling Stress in Flowering Organs
3.4. Comparative Analysis of DEGs Treated above and below 0 °C
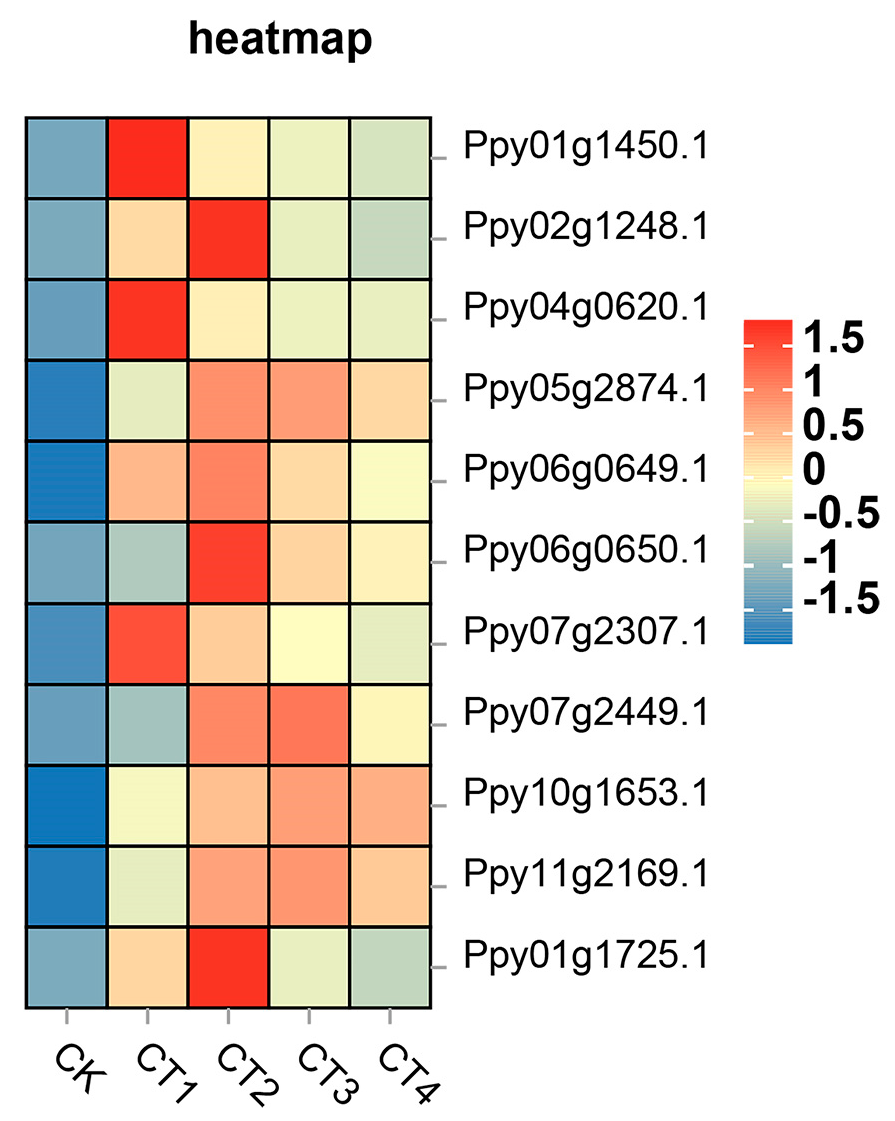
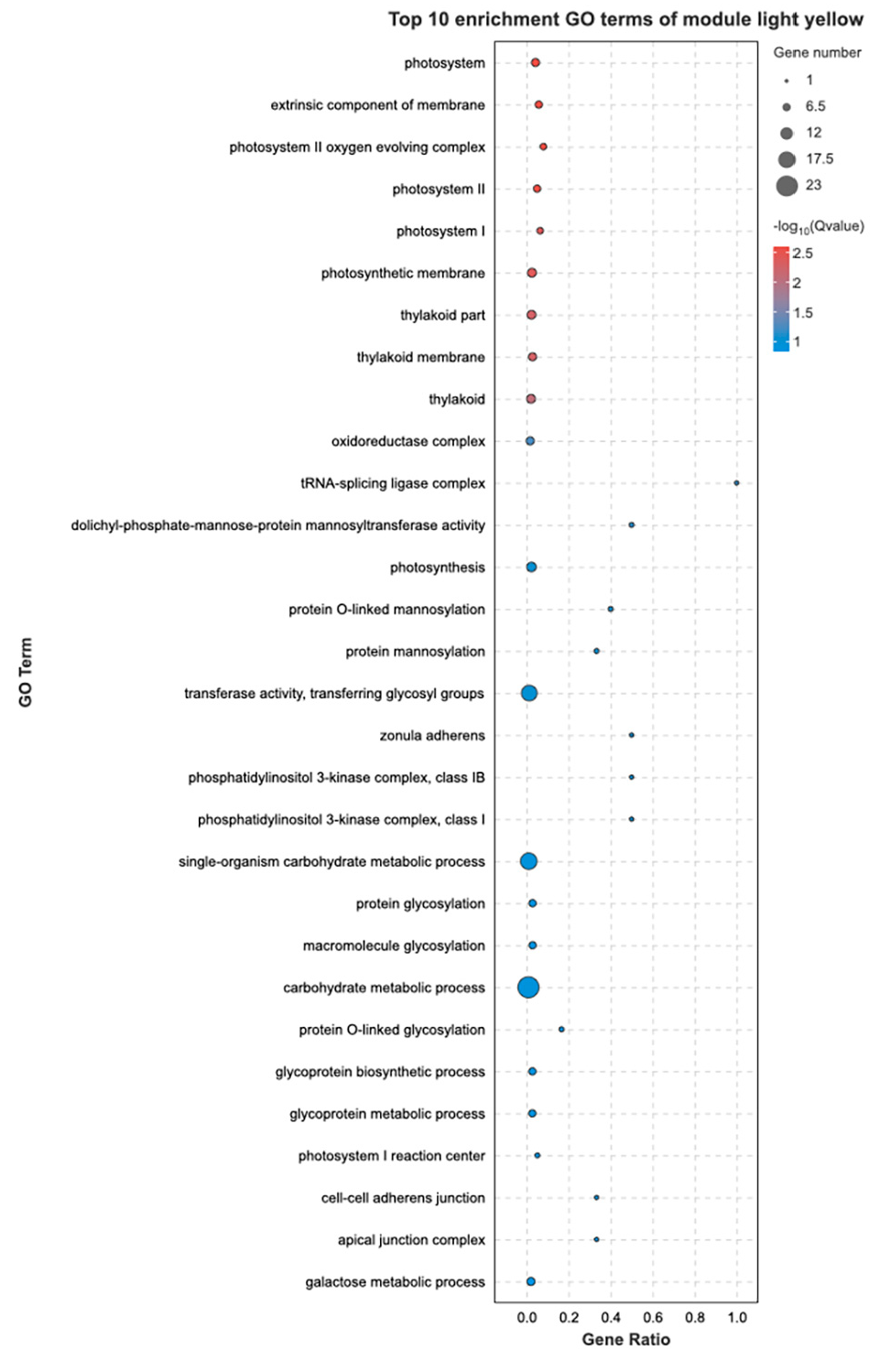
3.5. Analysis of Key DEGs under Low-Temperature Stress
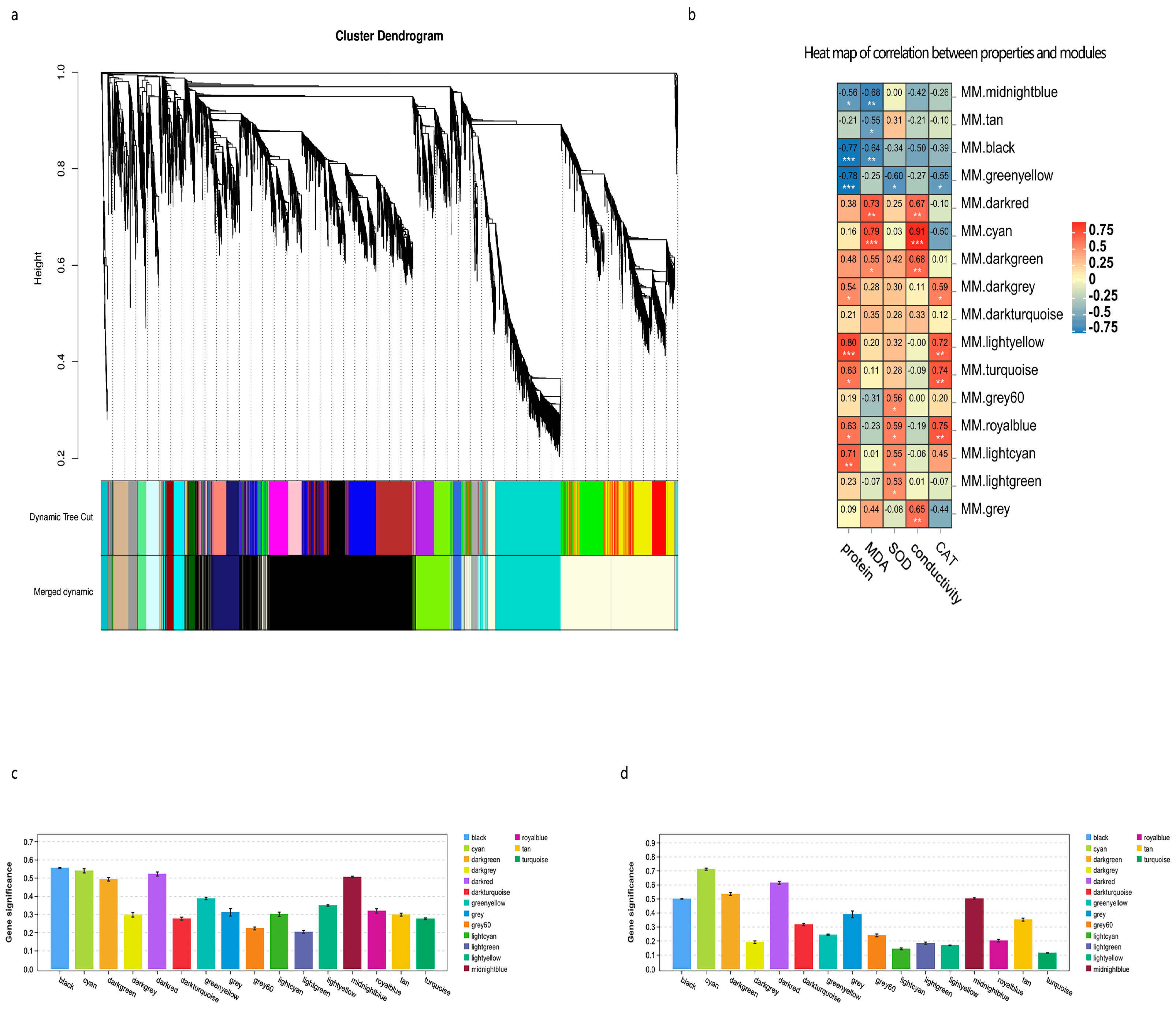
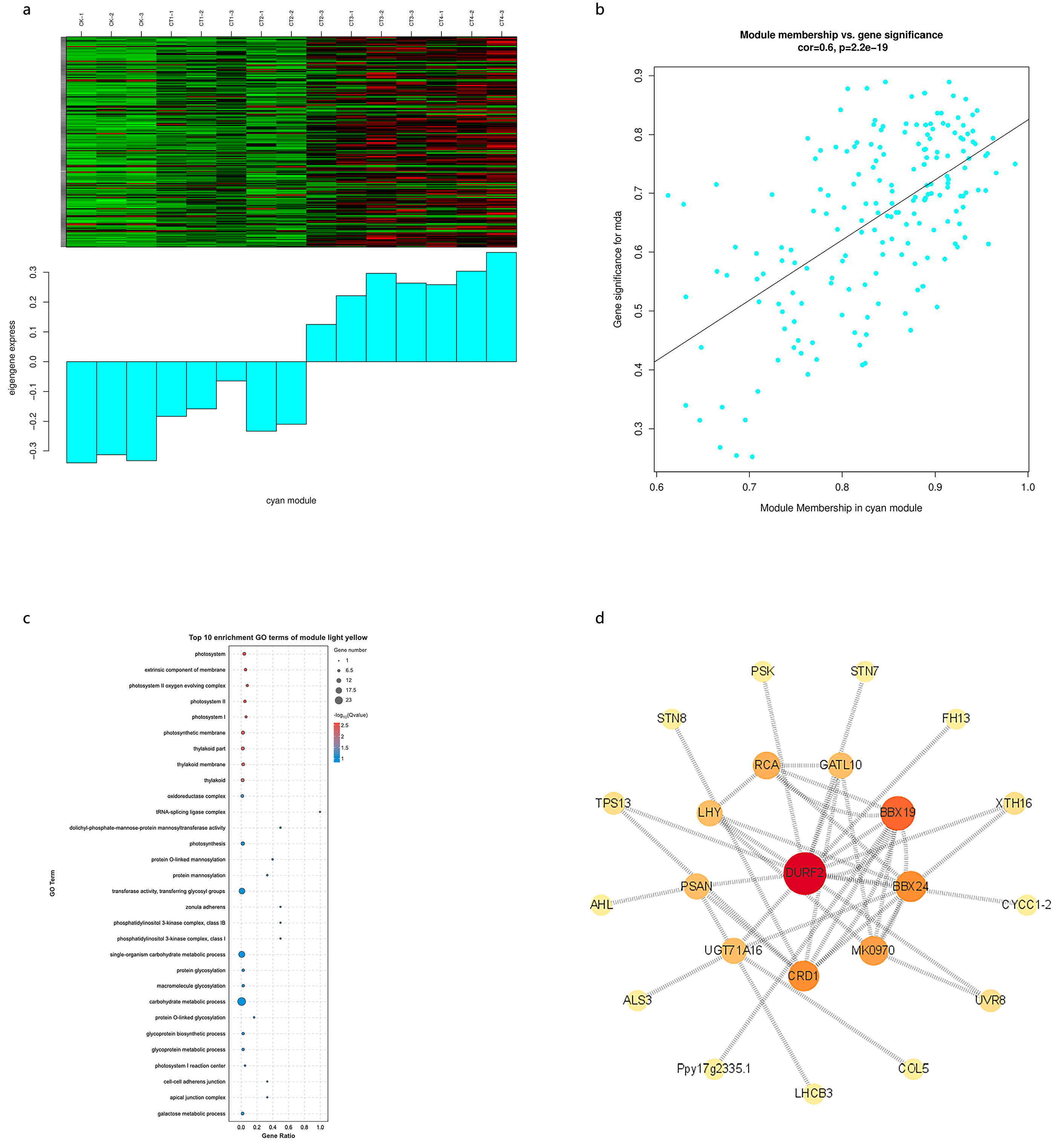
3.6. Co-Expression Network Construction and Identification of Key Genes Induced by Cold Stress
3.7. Analysis of Central Genes Related to MDA Accumulation
3.8. Analysis of Central Genes Related to SP Accumulation
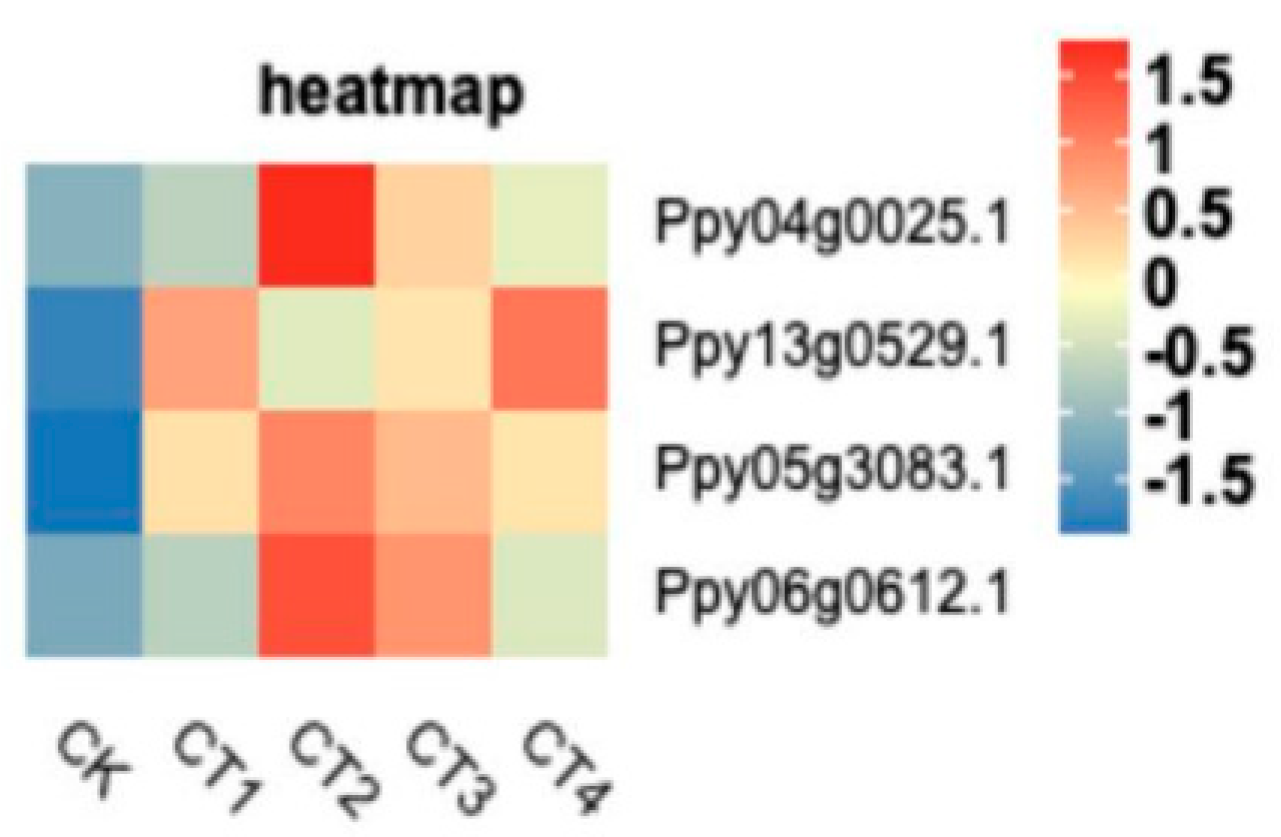
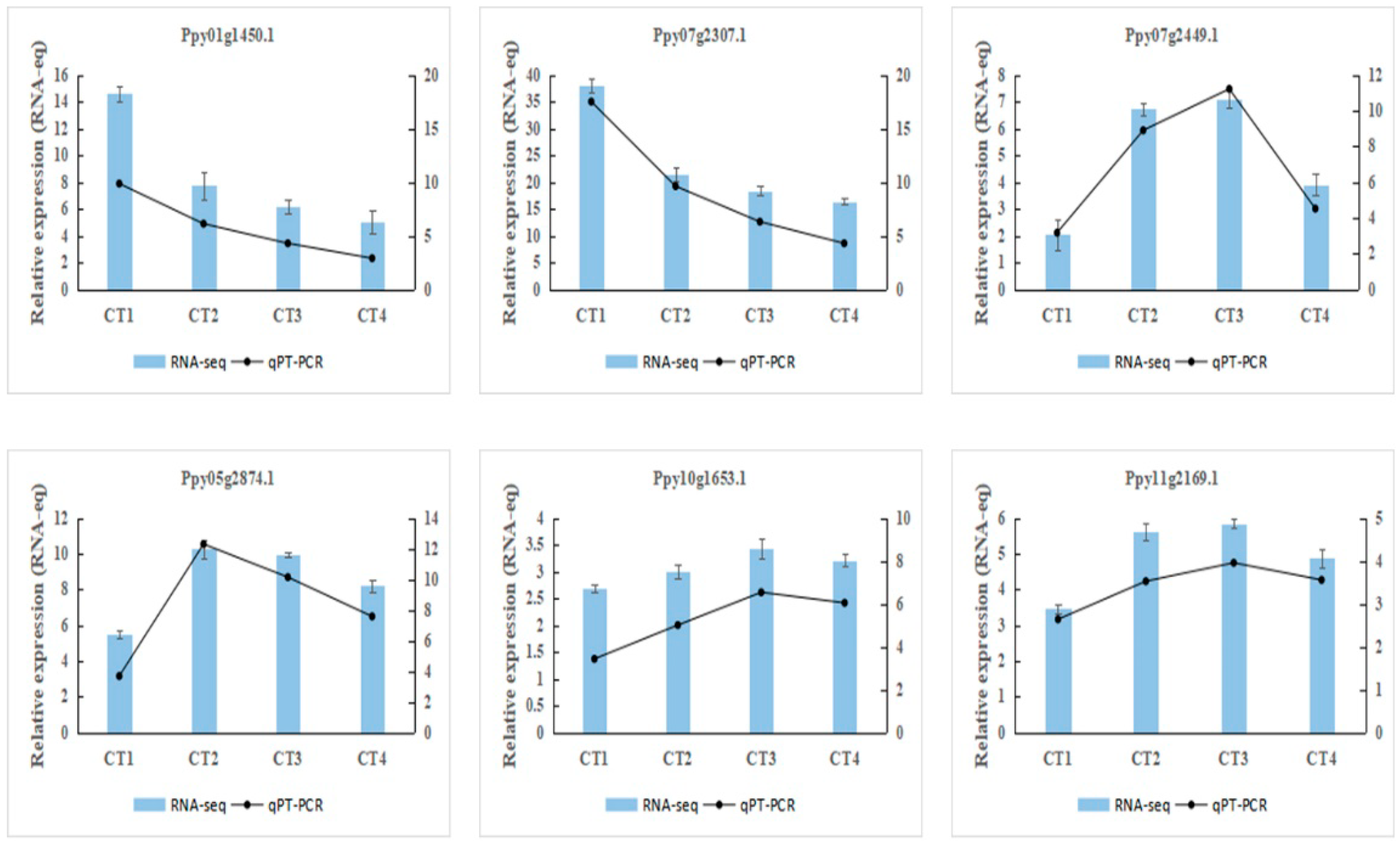
3.9. Verification of Differentially Expressed Genes by qRT-PCR
4. Discussion
4.1. Biochemistry Responses of Pears to Low-Temperature Stress
4.2. Transcriptional Differences in ‘Jinguang’ Pear Organs under Low-Temperature Stress
4.3. Signal Transduction-Related Genes
4.4. Metabolism-Related Genes
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Chen, X.S.; Wang, N.; Zhang, Z.Y.; Feng, S.Q.; Chen, X.L.; Mao, Z.Q. Progress on the Resource and Breeding of Kernel Fruits Ⅰ: Progress on the germplasm resources, quality development and genetics and breeding of pear in China. J. Plant Genet. Resour. 2019, 20, 791–800. [Google Scholar]
- Zhang, S.L.; Xie, Z.H. Current status, trends, main problems and the suggestions on the development of pear industry in China. J. Fruit Trees 2019, 36, 1067–1072. [Google Scholar]
- Kunz, A.; Blanke, M. “60 Years on”—Effects of Climatic Change on Tree Phenology—A Case Study Using Pome Fruit. Horticulturae 2022, 8, 110. [Google Scholar] [CrossRef]
- Jiao, H.M.; Zhao, H.; Feng, M.; Xiao, L.J.; Wang, H.Q.; Huang, L.; Chen, G.; Cao, Y.J. Causes and preventive measures of fruit tree freeze injury. Xinjiang Agric. Reclam. Technol. 2022, 45, 36–39. [Google Scholar]
- Xue, Q.Q.; Yuan, J.W.; Zhang, P.F.; Zhang, Z.B.; Duan, G.Q.; Liang, Z.J. Effect of late frost on fruit trees in spring orchards and countermeasures. Shanxi Fruits 2019, 189, 51–53. [Google Scholar]
- Bai, Q.F.; Li, X.M.; Zhu, L. The Changes of the frost-free periods from 1961 to 2010 and its impact on apple industry in Shaanxi province. Resour. Environ. Arid. Areas 2013, 27, 65–70. [Google Scholar]
- Jia, T.K.; Pang, L.X. Investigation on the occurrence of fruit tree late frost damage in Chencang, Shaanxi Province in 2018 and countermeasures. Pract. Technol. Inf. Fruit Trees 2019, 294, 36–38. [Google Scholar]
- Guo, W.Z.; Zhao, J.X. A new mid-early maturing pear variety. China Fruit News 2022, 39, 57–58. [Google Scholar]
- Xu, X.J.; Li, Y.C. Comparison of two methods for measuring relative conductivity of plants. Jiangsu Agric. Sci. 2014, 42, 311–312. [Google Scholar]
- Shukla, N.; Awasthi, R.P.; Rawat, L.; Kumar, J. Biochemical and physiological responses of rice (Oryza sativa L.) as influenced by Trichoderma harzianum under drought stress. Plant Physiol. Biochem. 2012, 54, 78–88. [Google Scholar] [CrossRef]
- Xu, C.; Li, Z.; Wang, J. Linking heat and adaptive responses across temporal proteo-transcriptome and physiological traits of Solidago canadensis. Environ. Exp. Bot. 2020, 175, 102035–104035. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Liu, D.L.; Zhang, B.Y.; Sun, H.M.; Peng, S.B.; Zhu, H.L. Comprehensive evaluation of cold resistance of different varieties of early-bearing walnut. J. Hortic. 2015, 42, 545–553. [Google Scholar]
- Cui, Z.Y.; Kong, F.N.; Mao, Y.X.; Sun, B.; Wang, J.H.; Dong, D.Y. Identification and expression patterns in tubulin and kinesin assays Gene families of Pyropia yezoens respond to dehydration stress. Ocean. Lakes 2021, 52, 673–684. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2011, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Damián, M.T.; Mejía-Muñoz, J.M.; Colinas-León, M.T.; Hernández-Epigmenio, F.; Cruz-Alvarez, O. Nutritio nal value bioactive compounds and capacity ant ioxidant in edible flowers of dahlia. Acta Sci. Pol. Hortorum Cultus 2021, 20, 63–72. [Google Scholar] [CrossRef]
- Ru, J.N.; Yu, T.F.; Chen, J.; Chen, M.; Zhou, Y.B.; Ma, Y.Z.; Xu, Z.S.; Min, D.H. Response of Wheat Zinc-Finger Transcription Factor Ta Di19A to Cold and its Screening of Interacting protein. Sci. Agric. Sin. 2017, 50, 2411–2422. [Google Scholar]
- Zhang, K.; Jiang, L.; Wang, X.; Han, H.; Chen, D.; Qiu, D.; Yang, Y. Transcriptome-wide analysis of AP2/ERF transcription factors involved in regulating taxol biosynthesis in Taxus media. Ind. Crops Prod. 2021, 171, 114373. [Google Scholar] [CrossRef]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA2/ethylene responsive factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; López-Vidriero, I.; Carrasco, J.L.; Godoy, M.; Vera, P.; Solano, R. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. USA 2018, 111, 2367–2372. [Google Scholar] [CrossRef]
- Aliakbari, A. RNA-seq Transcriptome Profiling of the Halophyte Salicornia persica in Response to Salinity. J. Plant Growth Regul. 2021, 40, 707–721. [Google Scholar] [CrossRef]
- Miura, K.; Furumoto, T. Cold signaling and cold response in plants. Int. J. Mol. Sci. 2013, 14, 5312–5337. [Google Scholar] [CrossRef]
- Bai, X.X.; Run, X.; Yao, Y.C.; Ning, L.; Wang, S.G.; Sun, D.Z. The role and research progress of DREB/CBF transcription factors in plant abiotic stress. J. Biol. 2017, 34, 88–93. [Google Scholar]
- Zhang, C.; Ding, Z.; Wu, K.; Yang, L.; Li, Y.; Yang, Z.; Shi, S.; Liu, X.J.; Zhao, S.S.; Yang, Z.R.; et al. Suppression ofjasmonic acid-mediated defenseby vira-inducible microRNA319 faciitates virus infection in rice. Mol Plant. 2016, 9, 1302–1314. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.T.; Sun, X.L.; Hoshino, Y.; Yu, Y.; Jia, B.; Sun, Z.W.; Duan, X.B.; Zhu, Y.M. MicroRNA319 positively regulates cold tolerance bytargetingOsPCF6 and OsTCP21 in rice (Oryza sativa L.). PLoS ONE 2014, 9, e91357. [Google Scholar]
- Lv, K.W.; Li, J.; Zhao, K.; Chen, S.; Nie, J.; Zhanq, W.L.; Liu, G.F.; Wei, H. Overexpression of an AP2/ERF family gene, BpERF13.in birch enhances cold tolerance through upregulating CBF genes and mitigating reactive oxygen species. Plant Sci. 2019, 292, 110375. [Google Scholar] [CrossRef]
- Zhang, Y.; Ming, R.; Khan, M.; Wang, Y.; Dahro, B.; Xiao, W.; Li, C.; Liu, J.H. ERF9 of Poncirus trifoliata (L.) Raf. undergoes feedback regulation by ethylene and modulates cold tolerance via regulating a glutathione S-transferase U17 gene. Plant Biotechnol. 2022, 20, 183–200. [Google Scholar] [CrossRef]
- Li, M.Y. Chilling injury and freezing injury of melons hazard symptim recognition. Vegetables 2021, 361, 81–83. [Google Scholar]
- Song, J.W.; Yin, D.S.; Zhao, D.G.; Qi, B.B.; Chen, Y.Y.; Zhou, L.; Peng, L. The response of flavonoid and polyphenols in juglans regia leaves to low temperature stress. Study Hebei For. Fruit 2017, 32, 34–41. [Google Scholar]
- Jiang, Y.X.; Zhang, H.N.; Guo, P.; Hu, H.C.; Li, T.; Lai, B.; Du, L.N. Genome-wide identification and expression analysis of chalcone synthase (CHS) gene family in longan. Mol. Plant Breed. 2021. [Google Scholar]
- Zhang, N.; Bao, H.; Cui, B.L.; Chen, K.L. Genomic analysis of chalcone synthase gene family of Phyllostachys edulis. Mol. Plant Breed. 2022, 20, 817–825. [Google Scholar]
- Qin, X.Y.; Qiao, J.J.; Li, Y.N. Structure, function and application of hydroxycinnamoyl transferase. Chin. J. Biochem. Mol. Biol. 2019, 35, 1058–1066. [Google Scholar]
- Long, X.Q.; Nie, C.; He, J.Z.; Tian, Y.; Li, X.R.; Pi, X.Y.; Wang, W.J.; Zhu, Q.K. Bioinformatics and expression analysis of hydroxycinnamoyl transferase gene family in Dendrobium candidum. J. Fujian Agric. 2022, 37, 492–502. [Google Scholar]
- Li, J.; An, Y.Y.; Wang, L.J. Transcriptomic analysis of Ficus carica Peels with a focus on the key genes for anthocyanin bi osynthesis. Int. J. Mol. Sci. 2020, 21, 1245. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.P.; Ding, Z.J.; Wang, F.P.; Liu, Y.; Liu, A.M.; Liu, K.Q.; Cai, H. Molecular characteristion, prokaryotic expression and expression pattern analysis of an anthocyanin synthase gene from Chrysanthemum morifolium ‘Chuju’. J. Plant Physiol. 2022, 58, 677–686. [Google Scholar]
- Li, J.; Liang, Y.M.; Zhao, A.C.; Liu, C.Y.; Lv, R.H.; Liu, X.Q.; Yu, M.D. Cloning and expression analysis of anthocyanin reductase gene from Mulberry. Seric. Sci. 2016, 42, 570–575. [Google Scholar]
- Xu, Y.; Chen, J.H.; Zhu, A.G.; Luan, M.B.; Wang, X.F.; Sun, Z.M. Research progress on response mechanism of plant under low temperature stress. Chin. Hemp Ind. Sci. 2015, 37, 40–49. [Google Scholar]
- Luo, P.; He, J.J.; Yao, Y.L.; Mo, Y.W. Effect of chilling stress on leaf antioxidative abilities of rubble trees with different chilling tolerance. J. Northwest Bot. 2014, 34, 311–317. [Google Scholar]
- Wu, Y.X.; Tian, J.B.; Qiao, Y.S.; Shi, M.J.; Tian, X.; Liu, Z.H.; Ru, H.L.; Liang, Q.; Cheng, P.H. Analysis of physiological and biochemical index for cold resistance of walnut leaf under low temperature stress. China Agron. Bull. 2016, 32, 101–106. [Google Scholar]
- Ren, J.J.; Zhao, S.; Su, Y.P.; Qi, G.H.; Li, B.G. Effect of low temperature stress in spring on antioxidase indexes of walnuts. J. Northwest Agric. For. Univ. (Nat. Sci. Ed.) 2016, 44, 75–81. [Google Scholar]
- Ai, P.F.; Jin, X.J.; Jin, Z.Z.; Yan, F.Q.; Li, K.W. Evaluation of cold-Resistant physiological of Kernel Apricot. J. Hebei Univ. Sci. Technol. 2013, 34, 48–53. [Google Scholar]
- Shen, H.; Meng, J.L.; Wu, S.J.; Yu, X. Screen of low temperature tolerant germplasm and the effect of low temperature on the physiological indexes in watermelon. J. North. Agric. 2021, 49, 115–123. [Google Scholar]
- Shalmani, A.; Jing, X.Q.; Shi, Y.; Muhammad, I.; Chen, K.M. Characterization of B-BOX gene family and their expression profiles under hormonal, abiotic and metal stresses in Poaceae plants. BMC Genom. 2019, 20, 27. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, J.; Fischer, C.; Dietrich, P.; Sauter, M. Kinase activity and calmodulin binding are essential for growth signaling by the phytosulfokine receptor PSKR1. PIant J. 2014, 78, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Wu, Y. The research progress on the functions of plant phospholipase C. Life Sci. 2017, 29, 575–581. [Google Scholar]
- Hong, Y.; Zhao, J.; Guo, L.; Kim, S.C.; Deng, X.; Wang, G.; Zhang, G.; Li, M.; Wang, X. Plant phospholipases D and C and their diverse functions in stress responses. Prog. Lipid Res. 2016, 62, 55–74. [Google Scholar] [CrossRef]
- Wang, Y.T.; Guo, B.S.; Zhang, M.X.; Zhu, H.H.; Xu, J.; Liu, H.W.; Wang, F.Y.; Wang, H. Methods for determining hydrolysis activity of phospholipase D. Food Res. Dev. 2022, 43, 189–194. [Google Scholar]
- Li, H.; Yang, N.; Liu, R.R.; Zhou, Y.P.; Gao, R.; Du, Y.Q. The role of phosphoinositide- specific PLC in plant growth and development. Chin. J. Cell Biol. 2020, 42, 2215–2226. [Google Scholar]
- Peters, C.; Kim, S.C.; Devaiah, S.; Li, M.; Wang, X. Non-specific phospholipase C5 and diacylglycerol promote lateral root development under mild salt stress in Arabidopsis. Plant Cell Environ. 2014, 37, 2002–2013. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Pandey, A.; Baranwal, V.; Kapoor, S.; Pandey, G.K. Comprehensive expression analysis of rice phospholipase D gene family during abiotic stresses and development. Plant Signal. Behav. 2013, 7, 847–854. [Google Scholar] [CrossRef]
- Krčková, Z.; Brouzdová, J.; Daněk, M.; Kocourková, D.; Rainteau, D.; Ruelland, E.; Valentová, O.; Pejchar, P.; Martinec, J. Arabidopsis non-specific phospholipase C1: Characterization and its involvement in response to heat stress. Front. Plant Sci. 2015, 6, 928. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, S.K.; Wang, C.W.; Li, J.W.; Li, H.F. Genome-wide identification, phylogeny and Expression analysis of MYB-MYC transcription factors in rice. J. Northwest Agric. 2019, 28, 1790–1800. [Google Scholar]
- Cheng, L.Q.; Li, X.X.; Huang, X.; Ma, T.; Liang, Y.; Ma, X.Y.; Peng, X.J.; Jia, J.T.; Chen, S.Y.; Chen, Y.; et al. Overexpression of sheepgrass R1-MYB transcription factor LcMYB1 confers salt tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2013, 70, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, S.; Gao, Y.; Huang, H.J. Functional study of BpMYB4 in birch response to low temperature stress. J. Nanjing For. Univ. (Nat. Sci. Ed.) 2019, 43, 25–31. [Google Scholar]
- Wang, F.B.; Kong, W.L.; Wong, G.; Fu, L.F.; Peng, R.H.; Li, Z.J.; Yao, Q.H. AtMYB12 regulates flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis thaliana. Mol. Genet. Genom. 2016, 291, 1545–1559. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.J.; She, L.L.; Lan, X.Z.; Lu, Y.N.; Yang, J.H.; Liu, H.; Lu, C.F.; Chen, Y.Z. Bioinformatics and expression analysis of MYB gene familybased on transcriptome of cold-acclimated Mirabilis himalaica callus. China Agron. Bull. 2020, 36, 54–61. [Google Scholar]



| Variety | PCA Results | Rank | ||||
|---|---|---|---|---|---|---|
| PC1 | PC2 | PC3 | Composite Score | Average Affiliation | ||
| ‘HG051102′ | −0.107723 | 0.513327 | 0.114686 | 0.0005 | 0.76127 | 1 |
| ‘Meiyu’ | 0.025991 | −0.070941 | 0.720783 | 0.0029 | 0.64523 | 2 |
| ‘Jinguang’ | 0.254036 | 0.072755 | −0.018173 | 0.0023 | 0.35742 | 3 |
| ‘SJ051105′ | 0.260128 | 0.081443 | 0.030126 | 0.0129 | 0.17582 | 5 |
| ‘041047′ | 0.072619 | 0.083559 | 0.001793 | 0.0154 | 0.29127 | 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, S.; Li, Y.; Zhao, J.; Guo, W.; Jiang, M.; Li, X.; Liu, W.; Zhang, J.; Yang, M. Transcriptome Analysis of Biochemistry Responses to Low-Temperature Stress in the Flower Organs of Five Pear Varieties. Forests 2023, 14, 490. https://doi.org/10.3390/f14030490
Lin S, Li Y, Zhao J, Guo W, Jiang M, Li X, Liu W, Zhang J, Yang M. Transcriptome Analysis of Biochemistry Responses to Low-Temperature Stress in the Flower Organs of Five Pear Varieties. Forests. 2023; 14(3):490. https://doi.org/10.3390/f14030490
Chicago/Turabian StyleLin, Shun, Yongtan Li, Jingxian Zhao, Weizhen Guo, Min Jiang, Xinman Li, Weiping Liu, Jun Zhang, and Minsheng Yang. 2023. "Transcriptome Analysis of Biochemistry Responses to Low-Temperature Stress in the Flower Organs of Five Pear Varieties" Forests 14, no. 3: 490. https://doi.org/10.3390/f14030490
APA StyleLin, S., Li, Y., Zhao, J., Guo, W., Jiang, M., Li, X., Liu, W., Zhang, J., & Yang, M. (2023). Transcriptome Analysis of Biochemistry Responses to Low-Temperature Stress in the Flower Organs of Five Pear Varieties. Forests, 14(3), 490. https://doi.org/10.3390/f14030490





